Abstract
Seladin-1 is a neuroprotective protein selectively down-regulated in brain regions affected in Alzheimer disease (AD). Seladin-1 protects cells against β-amyloid (Aβ) peptide 42- and oxidative stress-induced apoptosis activated by caspase-3, a key mediator of apoptosis. Here, we have employed RNA interference to assess the molecular effects of seladin-1 down-regulation on the β-secretase (BACE1) function and β-amyloid precursor protein (APP) processing in SH-SY5Y human neuroblastoma cells in both normal and apoptotic conditions. Our results show that ∼60% reduction in seladin-1 protein levels, resembling the decrease observed in AD brain, did not significantly affect APP processing or Aβ secretion in normal growth conditions. However, under apoptosis, seladin-1 small interfering RNA (siRNA)-transfected cells showed increased caspase-3 activity on average by 2-fold when compared with control siRNA-transfected cells. Increased caspase-3 activity coincided with a significant depletion of the BACE1-sorting protein, GGA3 (Golgi-localized γ-ear-containing ADP-ribosylation factor-binding protein), and subsequently augmented BACE1 protein levels and activity. Augmented BACE1 activity in turn correlated with the enhanced β-amyloidogenic processing of APP and ultimately increased Aβ production. These adverse changes associated with decreased cell viability in seladin-1 siRNA-transfected cells under apoptosis. No changes in GGA3 or BACE1 levels were found after seladin-1 knockdown in normal growth conditions. Collectively, our results suggest that under stress conditions, reduced seladin-1 expression results in enhanced GGA3 depletion, which further leads to augmented post-translational stabilization of BACE1 and increased β-amyloidogenic processing of APP. These mechanistic findings related to seladin-1 down-regulation are important in the context of AD as the oxidative stress-induced apoptosis plays a key role in the disease pathogenesis.
Introduction
Alzheimer disease (AD)2 is the most common neurodegenerative disorder leading to dementia. It is neuropathologically characterized by extracellular amyloid plaques as well as intraneuronal neurofibrillary tangles, composed of β-amyloid peptide (Aβ) and hyper-phosphorylated Tau, respectively. Aβ is generated from the β-amyloid precursor protein (APP) after sequential cleavages by β- and γ-secretases (1). Importantly, mutations in APP and PSEN1 and -2 genes have been shown to increase the production of the 42-amino acid-long Aβ42 and to cause the familial autosomal dominant form of AD. Based on this notion, it has been proposed that aberrant metabolism of APP is the initiating event in AD pathogenesis, which is followed by other adverse events, such as inflammation, oxidative stress, and neurofibrillary tangle formation (2). Recently, this model was challenged by a dual pathway hypothesis, which proposed that Aβ and Tau can actually be linked by separate mechanisms driven by common upstream initiators, such as apolipoprotein E (apoE) or glycogen synthase kinase-3 (3). Therefore, considering the complexity of AD pathogenesis, it is evident that also other AD-related risk genes exist in addition to the already established ones, such as APOE. Functional characterization of these novel risk genes is an important task as it may reveal new avenues to understand and design therapeutic approaches to AD.
An interesting candidate from a genetic and functional point of view in AD pathogenesis is seladin-1 (selective Alzheimer disease indicator-1), also known as 3-β-hydroxysteroid-Δ-24 reductase (DHCR24) (4, 5). Seladin-1 was initially found to be down-regulated in vulnerable brain regions in AD (4), and subsequently decreased seladin-1 mRNA levels were shown to inversely correlate with hyper-phosphorylated Tau in AD brain (6). Functional characterization of seladin-1 has revealed that this ER resident protein protects cells against oxidative stress- and Aβ42-induced toxicity in vitro (4, 7). More recently, it was shown that seladin-1 down-regulation leads to increased cleavage of APP and elevated Aβ levels in vivo by contributing to the formation of detergent-resistant membrane domains (8), which are known to modulate APP cleavage by β- and γ-secretases (9). This is an important finding considering that seladin-1 catalyzes the reduction of Δ24 double bond of desmosterol to form cholesterol (5). Furthermore, it has been shown previously that specific seladin-1 gene variants genetically associate with increased AD risk, suggesting that genetic alterations could also affect expression of this gene (10). Collectively, these data strongly support the idea that seladin-1 plays a pivotal role in AD pathogenesis, although the exact molecular mechanisms are still elusive.
Given the established down-regulation of seladin-1 in AD brain (4) and its association with neuronal degeneration (6), we set out to assess the molecular mechanisms related to reduced expression of seladin-1 on β-secretase (BACE1; β-site APP-cleaving enzyme) expression and stability as well as APP processing using RNA interference. To mimic the prevailing stress conditions observed in AD brain, we treated SH-SY5Y human neuroblastoma cells overexpressing the APP751 isoform (SH-SY5Y-APP751) or C-terminally Myc-tagged BACE1 (SH-SY5Y-BACE1-Myc) with staurosporine (STS) to induce apoptosis. Although seladin-1 knockdown under normal growth conditions did not significantly affect APP processing, BACE1 levels were increased on average 1.5-fold after seladin-1 down-regulation under apoptotic conditions. The increase in BACE1 levels after seladin-1 down-regulation coincided with increased caspase-3 activation, decreased cell viability, and enhanced depletion of the BACE1-sorting protein, GGA3 (Golgi-localized γ-ear-containing ARF-binding protein), which is responsible for sorting BACE1 to the lysosomes for degradation (11). This in turn correlated with increased BACE1 activity, APP β-CTF, and Aβ production. Finally, confocal microscopy analysis suggested that down-regulation of seladin-1 reinforced BACE1 accumulation to early endosomes and the plasma membrane under apoptosis. Taken together, our results suggest that under apoptotic conditions seladin-1 down-regulation increases post-translational stabilization of BACE1, which may accumulate in early endosomes and/or plasma membrane. This takes place through increased caspase-3 activation and subsequent depletion of GGA3, ultimately leading to increased β-amyloidogenic processing of APP.
EXPERIMENTAL PROCEDURES
siRNA and Plasmid Constructs
SMARTselectionTM predesigned ON-TARGETplus SMARTpool small interfering RNAs (siRNAs) (Thermo Scientific Dharmacon) targeted to human DHCR24/seladin-1 gene (GGAGUACAUUCCCUUGAGA, AGAACUAUCUGAAGACAAA, GCACAGGCAUCGAGUCAUC, and GAAAUGAGGCAGAGCUCUA) were used to down-regulate seladin-1. Negative control siRNA duplex with 3′-Alexa Fluor 488 (Qiagen) was used as a control in RNA interference experiments. cDNAs for human APP751 isoform and BACE1 (3′-tagged with Myc; BACE1-Myc) cloned into pcDNA 3.1 vector were used to create stable cell lines.
Cell Culture and Transfection
SH-SY5Y human neuroblastoma cells were transfected with APP751 and BACE1-Myc plasmid constructs using Effectene transfection reagent (Qiagen) to create stable cells lines overexpressing APP751 (SH-SY5Y-APP751) and BACE1-Myc (SH-SY5Y-BACE1-Myc). After the transfections, cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin, and 2 mm l-glutamine (Dulbecco's modified Eagle's medium-C). Two days after the transfection, 800 μg/ml G418 (Invitrogen) was added to induce selection of APP751- and BACE1-Myc-overexpressing cell clones. Three weeks after selection, clones overexpressing either APP751 or BACE1-Myc were identified using APP-, BACE1-, and Myc-specific antibodies in Western blotting. Selected clones were subsequently cultured in Dulbecco's modified Eagle's medium-C containing 200 μg/ml G418. Seladin-1 and control siRNAs were reversely transfected into SH-SY5Y cells overexpressing APP751 or BACE1-Myc using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions in the RNA interference experiments.
Western Blot Analysis
For Western blot analysis, total proteins were extracted from SH-SY5Y cells using TPER extraction buffer (Pierce) containing EDTA-free protease inhibitor mixture (Thermo Scientific). After protein quantification using the BCA protein assay kit (Pierce), 10–30 μg of total protein lysates were subjected to 4–12% BisTris-PAGE (Invitrogen) and subsequently blotted onto Immun-Blot polyvinylidene fluoride membranes (Bio-Rad). Primary antibodies against seladin-1/DHCR24 (C59D8, Cell Signaling), GGA3 (BD Biosciences), Myc (clone 4A6, Millipore), BACE1 (PA1–757, Affinity Bioreagents), caspase-3 (detecting the activated caspase-3 fragments; Cell Signaling), APP C terminus (A8717, Sigma), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Ab8245, Abcam) were used for immunoblotting. After incubation with appropriate horseradish peroxidase-conjugated secondary antibodies (GE Healthcare), enhanced chemiluminescence substrate (Amersham Biosciences) was applied to membranes, and detection of protein bands was performed with ImageQuant RT ECL Imager (GE Healthcare). Western blot images were quantified using Quantity One software (Bio-Rad).
RNA Extraction and Real Time Quantitative PCR Analysis
Total RNA was extracted from transfected SH-SY5Y cells using TRIzol® reagent (Invitrogen). Equal amounts (100 ng) of total RNA samples were subjected to cDNA synthesis using Superscript III reverse transcriptase (Invitrogen). Subsequently, SYBR Green Master PCR Mix (Applied Biosystems) and target-specific PCR primers for seladin-1 (5′-CAAGCCGTGGTTCTTTAAGC-3′ and 5′-CATCCAGCCAAAGAGGTAGC-3′) and GAPDH (5′-GATCATTCAGCTCAGCAAACA-3′ and 5′-GTATTCAAACCCAAGCTACTCAGA-3′) were used for amplification of cDNA samples by using real time quantitative PCR machine (7500 Fast Real Time PCR System, Applied Biosystems). PCR primers were designed to amplify a region flanking at least two different exons. A standard curve method was used to obtain seladin-1 and GAPDH mRNA levels. Seladin-1 mRNA levels were normalized to those of GAPDH from the same samples.
Secreted APP and Aβ Measurements in Conditioned Media
sAPPα (secreted N-terminal APP fragments) and sAPP total (= sAPPα + sAPPβ) levels were detected from cell culture media using Western blot analysis with 6E10 (Signet Laboratories) and 22C11 (Mab348, Millipore), respectively. The data were expressed as sAPP levels normalized to total protein. Aβ x-40 (Aβ40) levels were determined by using hAmyloid β40 enzyme-linked immunosorbent assay kit (The Genetics Co.).
Caspase-3 Activity Assay
Caspase-3 activity assay was performed in total protein lysates (0.6 μg/μl) in 280-μl reaction mixtures containing 20 mm HEPES (pH 7.5), 10% glycerol, 2 mm dithiothreitol, and Ac-DEVD-aminomethylcoumarin fluorogenic substrate (Pharmingen). After 60 min of incubation at 37 °C, production of aminomethylcoumarin as an indication of caspase-3 activity was measured at 30-min intervals for 60 min at 37 °C in a Victor 1420 multilabel counter 96-well plate enzyme-linked immunosorbent assay reader (Wallac) using excitation wavelength of 380 nm and emission wavelength of 460 nm. The data are shown as relative fluorescence units.
Staurosporine Treatment and Cell Viability Assay
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide assay was used to assess cell viability after treatment of the cells with 1 μm staurosporine (STS; Sigma) for 6 h. After the addition of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide, cells were incubated for 60 min at 37 °C, after which the formazan granules were solubilized in 99.7% DMSO (Hybri-MaxTM, Sigma). Finally, cell viability was determined as the absorbance (A) at 595 nm with Victor 1420 enzyme-linked immunosorbent assay reader (Wallac) and expressed as percentage of untreated control cells.
Confocal Microscopy
For confocal microscopy analysis, SH-SY5Y cells stably overexpressing BACE1-Myc were plated on sterile coverslips coated with 100 μg/ml poly-d-lysine (Sigma), transfected, and treated with or without STS. The cells were fixed in 4% paraformaldehyde for 15 min at room temperature. Next, the cells were incubated in blocking and permeabilization buffer containing 5% bovine serum albumin (fraction V, A9647, Sigma) and 0.1% Triton X-100 (BHD Laboratory Supplies) in phosphate-buffered saline (Invitrogen) for 30 min and stained with primary antibodies against Myc (clone 4A6, Millipore; 1:100), lysosomal marker LAMP2a (Ab18528, Abcam; 1:200), early endosomal marker EEA1 (clone 14, BD Biosciences; 1:200), or transferrin receptor (TfR, 13–6800, Zymed Laboratories Inc.; 1:200) as a marker for early endosomes and plasma membrane. Alexa Fluor® 488 goat anti-mouse, Alexa Fluor® 568 goat anti-rabbit (Invitrogen), or Cy3 sheep anti-mouse (C-2181, Sigma) were used as secondary antibodies. Single optical z-sections were obtained with Nikon Eclipse-TE300 microscope and Ultra VIEW laser scanning confocal unit (PerkinElmer Life Sciences) at ×60 magnification, and the images were processed with Adobe Photoshop software.
Statistical Analyses
Statistical analyses were performed using SPSS program version 14.0. Independent samples t test (equal variances assumed) or Mann-Whitney U test (equal variances not assumed) was used to test statistical significance between sample groups. Values are indicated as means ± S.D. Level of statistical significance was set to p < 0.05.
RESULTS
Down-regulation of Seladin-1 Does Not Significantly Affect APP Processing in Human SH-SY5Y Cells Overexpressing APP under Normal Growth Conditions
RNA interference was used to assess the effects of seladin-1 knockdown on APP processing under normal growth conditions in SH-SY5Y-APP751 cells. Using different siRNA concentrations, seladin-1 mRNA and protein levels normalized to GAPDH were specifically down-regulated by approximately 40–60% in SH- SY5Y-APP751 cells transfected with seladin-1 siRNA (siSel) when compared with control siRNA (siCtrl)-transfected cells (Fig. 1A). Down-regulation of seladin-1 increased APP C-terminal fragment (CTF; C83 and C99) levels on average 1.3-fold, albeit these increases did not reach statistical significance (Fig. 1B). APP immature (APPim), APP mature (APPm), and total APP (APP total = APPim + APPm) levels as well as the APPm/APPim ratio remained unchanged after down-regulation of seladin-1 (Fig. 1B). Analysis of the levels of secreted APP forms (sAPPα and sAPP total) and Aβ40 normalized to total protein from the conditioned SH-SY5Y-APP751 cell media did not reveal changes after seladin-1 down-regulation (supplemental Fig. 1). Even though our focus in the subsequent seladin-1 knockdown experiments was to elucidate the role of GGA3, a BACE1-sorting protein, in post-translational stabilization of BACE1 under apoptotic conditions, we first determined whether seladin-1 down-regulation affects GGA3 levels under normal growth conditions (Fig. 1B). Western blot analysis did not show changes in the GAPDH-normalized GGA3 levels after seladin-1 knockdown in SH-SY5Y-APP751 cells (Fig. 1B). Collectively, these data suggest that down-regulation of seladin-1 in the normal growth conditions does not significantly affect APP processing in SH-SY5Y-APP751 cells.
FIGURE 1.
Down-regulation of seladin-1 does not significantly affect APP processing in SH-SY5Y human neuroblastoma cells stably overexpressing APP751 (SH-SY5Y-APP751). A, quantitative PCR was used to assess seladin-1 knockdown efficiency at the mRNA level using two different seladin-1 siRNA concentrations (siSel, 0.65- and 1.3-μg total amounts of siRNA used in transfections) in SH-SY5Y-APP751 cells. On average, a 60% decrease in GAPDH-normalized seladin-1 mRNA levels was observed with both siRNA concentrations. B, APPim, APPm, and total APP and APP CTF (C83 and C99) levels were determined using Western blotting with APP C-terminal antibody (A8717). Blots were reprobed with antibodies against seladin-1, GGA3, and GAPDH. On average, a 40% decrease in GAPDH-normalized seladin-1 protein levels was detected. The GAPDH-normalized levels of different forms and fragments of APP or GGA3 were not significantly altered. ***, p < 0.001; **, p < 0.01, n ≥ 4, S.D. Ctrl, control.
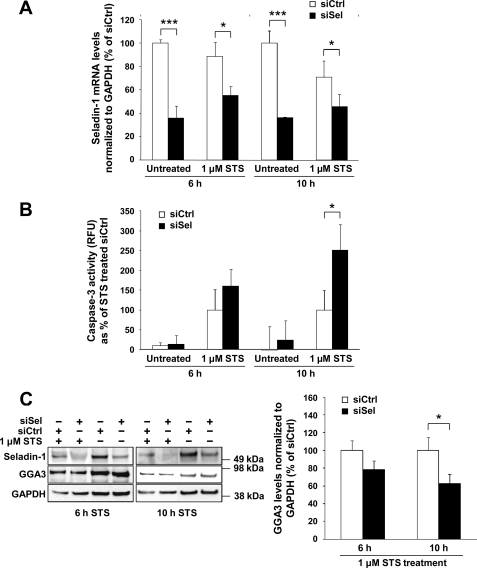
Down-regulation of Seladin-1 Increases Caspase-3 Activity and Leads to Enhanced Depletion of GGA3 during Apoptosis
To study the effects of seladin-1 knockdown on caspase-3 activity and GGA3 levels during apoptosis, siSel- and siCtrl-transfected SH-SY5Y-APP751 cells were treated with 1 μm STS for 6 or 10 h to induce apoptosis. As described previously, seladin-1 mRNA levels normalized to GAPDH were significantly down-regulated in both the untreated and STS-treated cells after siSel transfection (Fig. 2A). Coinciding with the down-regulated seladin-1 levels, we observed on average 1.5- and 2.0-fold augmentation of caspase-3 activity in siSel-transfected cells treated with STS for 6 and 10 h, respectively, when compared with STS-treated cells transfected with siCtrl (Fig. 2B). Subsequently, we assessed whether increased caspase-3 activity due to seladin-1 knockdown correlated with decreased GGA3 full-length protein levels in siSel-transfected cells. After GAPDH normalization, an ∼30–40% decrease in GGA3 full-length protein levels was observed in seladin-1 knockdown cells, in which seladin-1 protein levels were decreased by 60%, when compared with the STS-treated siCtrl-transfected cells (Fig. 2C). Consistent with the more pronounced caspase-3 activation after 10-h STS treatment as compared with the 6-h time point (Fig. 2B), a more robust decrease in GGA3 full-length protein levels was detected at 10 h (Fig. 2C). However, it should be noted that the 10-h STS treatment resulted in a noticeable cell loss when compared with the 6-h STS-treated cells (data not shown). Accordingly, this prolonged apoptotic induction may also have affected general protein degradation status. The fact that we observed an inverse correlation between caspase-3 activity and GGA3 full-length protein levels is consistent with a previous study, which demonstrated that caspase-3 cleaves GGA3 during apoptosis (11). Taken together, these data suggest that down-regulation of seladin-1 increases caspase-3 activity during apoptosis.
FIGURE 2.
Down-regulation of seladin-1 increases caspase-3 activity and decreases GGA3 full-length protein levels under apoptosis in SH-SY5Y-APP751 cells. A, quantitative PCR analysis of GAPDH-normalized seladin-1 mRNA levels in untreated and STS (1 μm)-treated cells transfected with seladin-1 (siSel) or control (siCtrl) siRNA after 6 and 10 h, indicating significantly decreased seladin-1 mRNA levels after siSel transfection in all conditions. B, caspase-3 activity was measured using fluorogenic caspase-3 substrate in STS-treated and -untreated SH-SY5Y-APP751 cells transfected with seladin-1 or control siRNA. Caspase-3 activity increased by ∼1.5–2.0-fold in siSel-transfected cells after 6 and 10 h of STS treatment, respectively, when compared with siCtrl-transfected cells. Background-corrected relative fluorescence units are indicated. RFU, relative fluorescence units; Ctrl, control. C, assessment of seladin-1, GGA3, and GAPDH levels in STS-treated SH-SY5Y-APP751 cells transfected with seladin-1 or control siRNA using Western blotting. Approximately 30–40% decrease in GAPDH-normalized GGA3 full-length levels was observed after seladin-1 down-regulation. ***, p < 0.001; *, p < 0.05, n ≥ 3, S.D.
Down-regulation of Seladin-1 Increases BACE1 Levels through Post-translational Stabilization during Apoptosis
Because seladin-1 down-regulation resulted in a decrease in GGA3 levels during apoptosis, we next assessed whether the decrease in GGA3 levels affected BACE1 protein levels and activity. As the endogenous BACE1 levels in SH-SY5Y cells were low (Fig. 3A), we created an SH-SY5Y cell line stably overexpressing BACE1, which encompassed a C-terminal Myc-tag (SH-SY5Y-BACE1-Myc) (Fig. 3A). This cDNA construct did not include endogenous promoter or 5′-untranslated regions of BACE1, thus allowing us to elucidate the underlying molecular mechanisms of seladin-1 knockdown on the post-translational stability of BACE1. Western blot analysis revealed a significant increase in BACE1 protein levels in SH-SY5Y-BACE1-Myc cells as compared with the endogenous BACE1 levels in SH-SY5Y-APP751 cells (Fig. 3A). The increased protein levels of BACE1 correlated with an increase in BACE1 enzymatic activity, as indicated by augmented β-secretase-mediated cleavage of endogenous APP leading to an extensive APP C89 and C99 production at the expense of generation of APP C83 (Fig. 3A and supplemental Fig. 2). The fact that we observed a robust increase in C89 and C99 levels and a significant decrease in C83 production is in line with the previous studies related to BACE1 overexpression both in vitro (12) and in vivo (13). We also studied the subcellular localization of overexpressed BACE1 and endogenous seladin-1 in SH-SY5Y-BACE1-Myc cells using confocal microscopy. Microscopy revealed that BACE1 and seladin-1 did not significantly co-localize under the normal growth conditions, even though a minor co-localization of the two proteins was detectable in subcellular structures resembling ER (Fig. 3B). This is consistent with the previous studies showing seladin-1 as an ER resident protein (4). STS treatment of these BACE1-overexpressing cells did not change subcellular localization of seladin-1 (data not shown). Accordingly, overexpressed BACE1 in SH-SY5Y-BACE1-Myc cells localized intracellularly mostly in the trans-Golgi network and endosomal compartments as shown previously (14).
FIGURE 3.
Characterization of human SH-SY5Y cells overexpressing BACE1-Myc (SH-SY5Y-BACE1-Myc). A, Western blot analysis related to the functional characterization of SH-SY5Y cells overexpressing BACE1-Myc (SH-SY5Y-BACE1-Myc; left column) compared with the SH-SY5Y cells overexpressing APP751 isoform (SH-SY5Y-APP751; right column). Endogenous BACE1 expression was low in SH-SY5Y-APP751 cells (detected by Ab757; right column). In SH-SY5Y-BACE1-Myc cells, BACE1-Myc was abundantly overexpressed, and this increase was correlated with an enhanced β-secretase cleavage of endogenous APP augmenting the APP C89 and C99 levels. The blot was reprobed with GAPDH antibody to verify equal loading. B, confocal microscopy of SH-SY5Y-BACE1-Myc cells revealed that overexpressed BACE1 (anti-Myc; green) was mostly localized in the trans-Golgi network with some expression at or near the plasma membrane (endosomes) under normal conditions. Seladin-1 (anti-seladin-1; red) showed a granular intracellular localization resembling the ER. Endogenous seladin-1 showed only little intracellular co-localization with BACE1-Myc under normal growth conditions consistent with the previous data showing seladin-1 predominantly as an ER resident protein (4). Images show single optical z-sections.
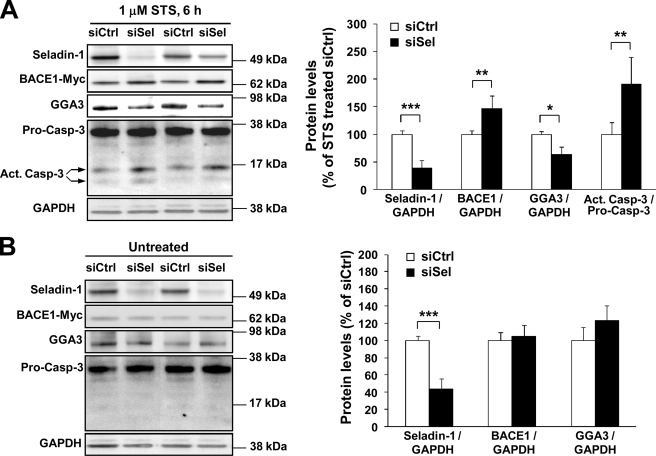
Next, we treated siSel- and siCtrl-transfected SH-SY5Y-BACE1-Myc cells with STS and studied caspase-3 activation using Western blotting (Fig. 4). To avoid adverse effects that we observed in the previous experiments in SH-SY5Y-APP751 cells (Fig. 2C) related to prolonged apoptotic induction, we treated SH-SY5Y-BACE1-Myc cells for 6 h instead of 10 h with 1 μm STS. Similar to SH-SY5Y-APP751 cells, we detected an augmentation of caspase-3 activity, as indicated by the ∼2.0-fold increase in active caspase-3 cleavage fragments (∼14 and 17 kDa) as well as an ∼40% decrease in GGA3 full-length levels in seladin-1 down-regulated cells when compared with siCtrl-transfected cells (Fig. 4A). A more prominent decrease in GGA3 full-length levels in SH-SY5Y-BACE1-Myc cells when compared with SH-SY5Y-APP751 cells (Fig. 2C) was likely associated with more pronounced caspase-3 activation and subsequently enhanced cleavage of full-length GGA3 in SH-SY5Y-BACE1-Myc cells after seladin-1 down-regulation. Importantly, these alterations coincided with an average 1.5-fold increase in BACE1 levels in SH-SY5Y-BACE1-Myc cells. Conversely, no differences in BACE1 or GGA3 full-length protein levels were found between siSel- and siCtrl-transfected cells under normal growth conditions. Moreover, cleaved caspase-3 fragments were not observed under normal conditions (Fig. 4B). Together, these results suggest that seladin-1 enhances the caspase-3-mediated cleavage of GGA3 leading to post-translational stabilization of BACE1 during apoptosis.
FIGURE 4.
Down-regulation of seladin-1 increases BACE1 levels under apoptotic conditions through enhanced depletion of GGA3 in SH-SY5Y-BACE1-Myc cells. A, Western blot analysis of BACE1, GGA3, and caspase-3 levels normalized to GAPDH in staurosporine (1 μm STS, 6 h)-treated SH-SY5Y-BACE1-Myc cells transfected with seladin-1 (siSel) or control (siCtrl) siRNA. Caspase-3 activation was determined as a ratio of the levels of active caspase-3 fragments (14 and 17 kDa) versus those of pro-caspase-3 (Act. Casp-3/Pro-Casp-3). Seladin-1 knockdown during apoptosis significantly depleted full-length GGA3 protein levels, which coincided with enhanced caspase-3 activation. Depletion of GGA3 in turn inversely correlated with significantly increased BACE1-Myc levels. Ctrl, control. B, Western blot analysis of GAPDH-normalized BACE1-Myc and GGA3 levels did not show alterations between siSel- and siCtrl-transfected SH-SY5Y-BACE1-Myc cells. ***, p < 0.001; **, p < 0.01; *, p < 0.05, n = 3–6, S.D.
Down-regulation of Seladin-1 Increases Production of APP β-CTF and Aβ40 through Increased BACE1 Activity and Decreases Cell Viability under Apoptosis
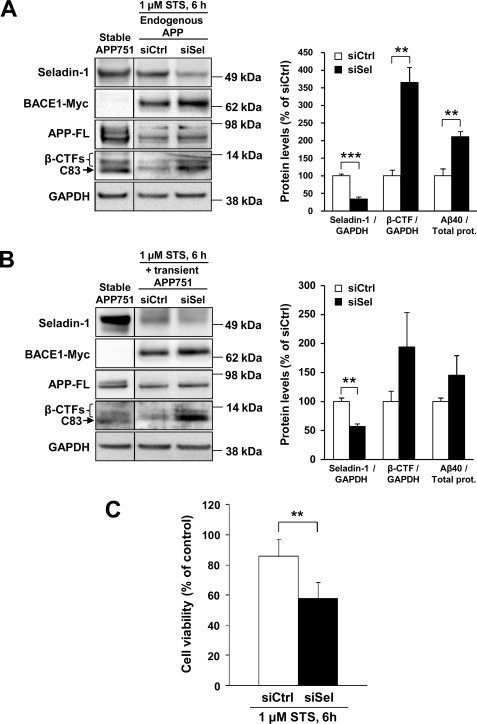
As we detected a prominent increase in BACE1 levels after seladin-1 knockdown, we next wanted to verify the anticipated β-secretase effect on APP processing under apoptotic conditions. After a 48-h siSel and siCtrl transfection of SH-SY5Y-BACE1-Myc cells followed by a 6-h treatment with 1 μm STS, we discovered a significant increase in both endogenous APP β-CTF and Aβ40 levels in cells transfected with siSel, whereas the C83 levels remained undetectable (Fig. 5A, 2nd and 3rd lanes). We also co-transfected SH-SY5Y-BACE1-Myc cells with APP751 cDNA construct together with siSel or siCtrl and subsequently detected an increase in both the APP β-CTF and Aβ40 levels in siSel-transfected cells, although less pronounced than with endogenous APP (Fig. 5B). Furthermore, the effect of seladin-1 knockdown on the viability of SH-SY5Y-BACE1-Myc cells after STS treatment (1 μm, 6 h) was studied using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide assay. A significant decrease in cell viability (∼30%) was observed in cells transfected with siSel when compared with siCtrl-transfected cells (Fig. 5C). These data indicate that down-regulation of seladin-1 increases BACE1 levels and activity during apoptosis leading to enhanced production of APP β-CTFs and Aβ, which also parallel with decreased cell viability in SH-SY5Y-BACE1-Myc cells.
FIGURE 5.
Down-regulation of seladin-1 increases the production of APP β-CTFs and Aβ40 due to increased BACE1 activity and decreases cell viability under apoptosis in SH-SY5Y-BACE1-Myc cells. Western blot and Aβ40 enzyme-linked immunosorbent assay analyses showing that post-translationally stabilized BACE1 increased the production of APP β-CTFs and Aβ40 from endogenously expressed APP (A) and transiently overexpressed APP751 (B) in staurosporine-treated (1 μm STS, 6 h) SH-SY5Y-BACE1-Myc cells after seladin-1 knockdown. Seladin-1 (siSel) or control (siCtrl) siRNAs was transfected alone (A) or in combination with APP751 cDNA construct (B). Increases in transiently overexpressed APP751 cells showed borderline significance in APP β-CTFs (p = 0.06) and Aβ40 (p = 0.08) levels after seladin-1 down-regulation. Total protein (prot.) lysate from SH-SY5Y cells overexpressing APP751 (APP751) was used as a control to assess APP CTF profiles (see also supplemental Fig. 2). C, seladin-1 knockdown significantly decreased cell viability after a 6-h staurosporine (1 μm STS) treatment in SH-SY5Y-BACE1-Myc cells as assessed by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay. Cell viability is expressed as a percentage from the untreated cells. ***, p < 0.001, and **, p < 0.01, n = 3 (A and B), n = 5 (C), S.D.
Down-regulation of Seladin-1 May Affect BACE1 Subcellular Localization during Apoptosis
Finally, because GGA3 is an adaptor protein responsible for sorting of BACE1 to lysosomal degradation (11) and because BACE di-leucine sorting signal mutant has been shown to reduce the accumulation of BACE in LAMP2-positive compartments after inhibition of lysosomal hydrolases (15), we explored whether depletion of GGA3 due to seladin-1 knockdown affects BACE1 subcellular localization. We hypothesized that BACE1 localization might be altered more toward endosomal compartments and less to lysosomes during apoptosis. After siSel and siCtrl transfection and STS treatment, SH-SY5Y-BACE1-Myc cells were stained with anti-Myc to analyze subcellular localization of overexpressed BACE1-Myc using confocal microscopy (Figs. 6 and 7). The cells were also co-stained with antibodies against TfR as a marker for early endosomes and the plasma membrane (Fig. 6) and LAMP2a as a marker for lysosomes (Fig. 7). Confocal microscope analysis showed that during apoptosis, more BACE1-Myc appeared to co-localize with TfR in the subcellular compartments near or at the plasma membrane in SH-SY5Y-BACE1-Myc cells transfected with siSel when compared with siCtrl-transfected cells (Fig. 6). A similar result was also obtained using co-staining with an antibody against EEA1, another marker for early endosomes (data not shown). Furthermore, STS treatment induced pronounced blebbing of the plasma membrane in siSel-transfected cells, and TfR and BACE1-Myc co-localized in these membrane blebs, further implying that more BACE1-Myc may be present at or near the plasma membrane in apoptotic cells. Some blebbing was also detectable in siCtrl-transfected cells treated with STS, but to a lesser extent than in siSel-transfected cells. This agrees with our observation that there was an ∼2-fold increase in caspase-3 activity in cells transfected with siSel under apoptosis as compared with siCtrl-transfected cells (Fig. 4A). In cells transfected with siCtrl with or without STS treatment, some co-localization of BACE1-Myc with LAMP2a was detectable in intracellular structures, most likely lysosomes, which is in line with previous reports on BACE1 subcellular localization (14) (Fig. 7). However, there was no apparent co-localization between BACE1-Myc and LAMP2a during apoptosis after seladin-1 knockdown. Rather, the co-localization appeared to decrease in siSel-transfected cells treated with STS as compared with the siCtrl-transfected cells with or without STS treatment, and more BACE1-Myc seemed to reside near or at the plasma membrane. Collectively, the data presented here suggest that down-regulation of seladin-1 reinforces BACE1 stabilization during apoptosis and may alter BACE1 subcellular localization more toward TfR-positive compartments, such as early endosomes and the plasma membrane.
FIGURE 6.
Down-regulation of seladin-1 alters BACE1 subcellular localization to TfR-positive early endosomes and plasma membrane in SH-SY5Y-BACE1-Myc cells under apoptosis. Confocal microscopy of BACE1-Myc subcellular localization in untreated (UNT) and staurosporine (1 μm STS, 6 h)-treated cells transfected with seladin-1 (siSel) or control (siCtrl) siRNA. During apoptosis, more BACE1-Myc (anti-Myc; green) is localized in the TfR-positive (anti-TfR; red) compartments near or at the plasma membrane (arrow) in SH-SY5Y-BACE1-Myc cells transfected with siSel when compared with siCtrl-transfected cells. Images show single optical z-sections.
FIGURE 7.
Down-regulation of seladin-1 decreases co-localization of BACE1-Myc with the lysosomal marker LAMP2a. Confocal microscopy of subcellular localization of BACE1-Myc was assessed using anti-Myc (green) and LAMP2a (red) antibody staining in untreated (UNT) and STS-treated SH-SY5Y-BACE1-Myc cells transfected with seladin-1 (siSel) or control (siCtrl) siRNA. In siCtrl-transfected cells (with or without STS), there is some intracellular co-localization (yellow) of the two proteins, most likely in lysosomes. After seladin-1 knockdown, no apparent co-localization of BACE1-Myc with LAMP2a was detected, and more BACE1-Myc was localized near or at the plasma membrane (arrow) during apoptosis. Images show single optical z-sections.
DISCUSSION
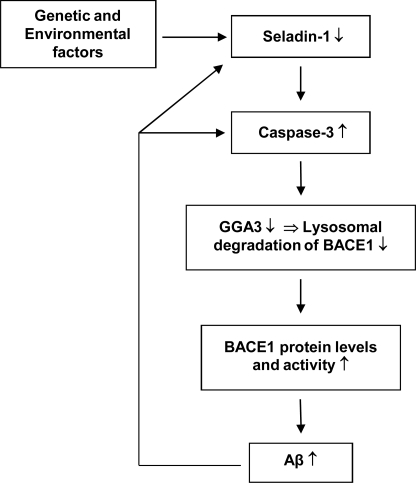
Here we have studied the underlying molecular mechanisms related to seladin-1 down-regulation in the context of β-secretase-mediated processing of APP in SH-SY5Y human neuroblastoma cells. Our results show that down-regulation of seladin-1 expression under apoptotic conditions resulted in augmented BACE1 levels and activity through increased caspase-3 activation and enhanced depletion of the BACE1-sorting protein, GGA3, which is known to be responsible for sorting of BACE1 to the lysosomes for degradation (11). Consistent with this, depletion of GGA3 after seladin-1 knockdown in turn may associate with an altered subcellular localization of BACE1, as some BACE1 appeared to accumulate in early endosomes and the plasma membrane in apoptotic conditions. Based on these observations, it is plausible that lysosomal sorting and thereby the degradation of BACE1 is reduced under apoptosis leading to increased levels and activity of this protease by means of post-translational stabilization. These adverse effects increased BACE1-mediated cleavage of APP leading to elevated production of APP β-CTFs and Aβ as well as decreased cell viability of SH-SY5Y cells overexpressing BACE1 under apoptosis. The above-mentioned cascade is likely linked to AD pathogenesis due to the fact that seladin-1 is down-regulated in the temporal cortex of AD patients by ∼40% (4, 6) and that the decreased seladin-1 mRNA levels correlate with increased phosphorylated Tau levels in AD brain (6). Furthermore, a recent genetic study revealed that genetic alterations in the DHCR24/seladin-1 gene increased the risk of AD, suggesting that genetic predisposition may also contribute to the observed down-regulation of seladin-1 expression in AD brain (10). Collectively, these results indicate that seladin-1 plays an important role in AD pathogenesis and thereby even a moderate decrease in seladin-1 expression due to genetic and/or environmental factors (oxidative stress-induced apoptosis, ER stress, etc.) may initiate a vicious cascade resulting in increased BACE1 activity and ultimately augmented Aβ production (Fig. 8). Increased production of APP β-CTFs and Aβ again may further reinforce seladin-1 down-regulation and/or caspase-3 activation, possibly augmenting the perpetual cycle of pathogenic events.
FIGURE 8.
Suggested mechanistic model related to the effects of reduced seladin-1 expression on BACE1 stabilization and BACE1-mediated processing of APP under apoptosis.
Functional characterization of seladin-1 has revealed that it confers resistance against Aβ-mediated and oxidative stress-induced apoptosis by inhibiting caspase-3 activity, which is the key mediator of the apoptosis (4). The anti-apoptotic property of seladin-1 was observed in rat PC12 cell clones selected for their resistance for Aβ-induced toxicity, in which case both seladin-1 mRNA and protein levels were significantly elevated. Subsequently, other studies (16, 17) demonstrated similar findings, suggesting that seladin-1 protein by definition could encompass specific protective properties. Related to this issue, it was recently reported that the protective function of seladin-1 could be partially associated with its scavenging activity toward reactive oxygen species in the ER (18). This study demonstrated that seladin-1 diminished H2O2-induced generation of reactive oxygen species in mouse embryonic fibroblasts expressing seladin-1 when compared with mouse embryonic fibroblasts lacking seladin-1 expression, indicating that seladin-1 possesses the capacity to protect cells from H2O2-induced apoptosis. The reactive oxygen species scavenging activity domain was subsequently narrowed down to the C-terminal portion of seladin-1, as the N-terminal deletion mutant of seladin-1 failed to exert reactive oxygen species-scavenging activity (18). Alternatively, the protective effect of seladin-1 against H2O2-induced toxicity has been suggested to be related to its function in a p53-dependent manner (7). Seladin-1 is also suggested to play a key role in preventing Ras-induced senescence through association with p53, and thus regulating cellular response against oncogenic and oxidative stress in fibroblasts (19). These data strongly support the anti-apoptotic role of seladin-1 in stress-induced conditions, such as oxidative stress-induced apoptosis.
Although it is possible that seladin-1 protein itself may encompass anti-apoptotic functions during different stress conditions as discussed above, it should be emphasized that seladin-1 has specific enzymatic activity, which catalyzes the reduction of the Δ24 double bond of sterol intermediates in the cholesterol biosynthesis pathway (5). The fact that seladin-1 catalyzes reduction of the Δ24 double bond of desmosterol to form cholesterol indicates that seladin-1 has a role in the formation of cholesterol-rich detergent-resistant membrane domains, also known as lipid rafts (9). On the other hand, lipid rafts have been found to affect β- and γ-secretase-mediated APP cleavage and Aβ generation in vitro and in vivo (8, 9). Related to this, seladin-1 heterozygous knock-out mice (50% reduction in seladin-1 levels) showed decreased cholesterol levels, which were accompanied by the disorganization of lipid raft domains in brain (8). This defect led to the displacement of β-secretase from the detergent-resistant membrane domains to the APP-containing membrane fractions increasing APP C99 and Aβ production in the brain of seladin-1-deficient mice. Similar to this study, we also observed a moderate increase (∼1.3-fold) in the levels of APP C99 after seladin-1 knockdown in neuroblastoma cells overexpressing APP under normal conditions. In addition to the increase in APP C99 levels, however, we also observed augmented APP C83 levels, suggesting that APP processing in general was enhanced rather than the β-secretase-mediated cleavage of APP exclusively. In this context, it should be noted that the seladin-1 heterozygous knock-out mice also showed a moderate increase in APP C83 levels (8). The fact that we could not detect an increase in Aβ levels due to increased BACE1 activity under normal growth conditions, however, could be related to underlying differences between the in vitro (transient) and in vivo (stable) knockdown models of seladin-1 in general. Interestingly, recent findings related to lipid rafts and S-palmitoylation-deficient BACE1 mutants have revealed surprising results because the non-raft-localized BACE1 could also process APP and enhance secretion of Aβ in human neuroblastoma cells, suggesting that BACE1 is able to cleave APP efficiently in both lipid raft and non-raft microdomains (12). Although there are several lines of evidence supporting the important role of cholesterol in the β-amyloidogenic processing of APP, it is possible that cholesterol also modulates Aβ-induced neurotoxicity in AD. As an example of this, mitochondrial cholesterol loading was shown to exacerbate Aβ-induced neurotoxicity by depleting the mitochondrial glutathione levels and thereby increasing the susceptibility to Aβ-(1–42)-induced oxidative stress and the release of apoptotic proteins in vivo (20). These data together indicate that there are different plausible mechanisms underlying the down-regulation of seladin-1 during apoptosis. In this study, however, it is more likely that the pro-apoptotic effects of seladin-1 down-regulation are linked to the scavenging activity of seladin-1 protein rather than depletion of de novo-synthesized cholesterol. This suggestion is based on the fact that the serum-containing cell culture medium contained extracellular cholesterol and thereby supplemented the reduced de novo synthesis of cholesterol during the acute STS treatment (6 h).
It is well known that BACE1 is a stress-induced protease, whose activity has been shown to increase after e.g. traumatic brain injury (21), oxidative stress (22), and cerebral ischemia (11, 23, 24). Recently, it was demonstrated that GGA3 is an adaptor protein responsible for sorting of BACE to the lysosome for degradation (11). Importantly, both in vitro and in vivo experiments revealed that GGA3 is cleaved by activated caspase-3 and that the depletion of GGA3 resulted in an increase in BACE1 levels and activity through post-translational stabilization of this protease. This finding has important significance in the context of AD, because oxidative stress-induced apoptosis plays an essential role in the disease pathogenesis (2), emphasizing the importance of post-translational events in the regulation of BACE1 levels and activity. In addition to this, it has been recently shown that BACE1 expression is also regulated in a transcriptional (25) and translational manner (26) during different stress conditions. Energy deprivation induced phosphorylation of the translation initiation factor eIF2α leading to increased BACE1 levels both in vitro and in vivo (26). This study showed that BACE1 is translationally controlled by its complex 5′-untranslated region encompassing three upstream open reading frames, which are able to regulate the translation of BACE1 mRNA depending on the eIF2α phosphorylation status. In this study, however, transcriptional or translational regulation of BACE1 was not expected to take place because we used a BACE1-Myc cDNA construct, which did not include the above-mentioned upstream open reading frames, enabling us to study the post-translational stability of BACE1 after seladin-1 knockdown. Nevertheless, our present data together with previous studies suggest that the expressional regulation of BACE1 in AD is a complex process, which likely involves several overlapping regulatory mechanisms simultaneously affecting BACE1 levels and activity.
Taken together, we have elucidated here the possible molecular mechanisms related to seladin-1 down-regulation known to take place in the AD brain (4, 6). Our findings suggest that seladin-1 down-regulation increases caspase-3 activity sensitizing neuroblastoma cells to apoptosis and consequently augmenting BACE1 levels and activity through depletion of the BACE1-sorting protein, GGA3. Although the initial cause(s) for seladin-1 down-regulation in AD is still elusive, our experimental model supports the idea that seladin-1 is a plausible target for disease-modifying therapies for AD as well as other conditions involving oxidative stress-induced apoptosis, such as stroke.
Supplementary Material
Acknowledgments
We thank Drs. Doo Yeon Kim, Dora M. Kovacs, and Rudolph E. Tanzi (Massachusetts General Hospital and Harvard Medical School) for their generous gifts of the BACE1-Myc and APP751 cDNA constructs and Dr. Antero Salminen (University of Kuopio) for kindly providing the naive SH-SY5Y cells.
This work was supported by the Health Research Council of the Academy of Finland, EVO Grant 5772708 of Kuopio University Hospital, and the Nordic Centre of Excellence in Neurodegeneration.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- AD
- Alzheimer disease
- APP
- β-amyloid precursor protein, β-secretase
- BACE
- β-site APP-cleaving enzyme
- STS
- staurosporine
- siRNA
- small interfering RNA
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- Aβ
- β-amyloid peptide
- TfR
- transferrin receptor
- CTF
- C-terminal fragment
- siSEL
- seladin-1 siRNA
- ER
- endoplasmic reticulum
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- sAPP
- secreted APP
- APPm
- APP mature
- APPim
- APP immature.
REFERENCES
- 1.Bertram L., Tanzi R. E. (2004) Hum. Mol. Genet. 13, Spec. No. 1:R135–R141 [DOI] [PubMed] [Google Scholar]
- 2.Hardy J., Selkoe D. J. (2002) Science 297, 353–356 [DOI] [PubMed] [Google Scholar]
- 3.Small S. A., Duff K. (2008) Neuron 60, 534–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greeve I., Hermans-Borgmeyer I., Brellinger C., Kasper D., Gomez-Isla T., Behl C., Levkau B., Nitsch R. M. (2000) J. Neurosci. 20, 7345–7352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waterham H. R., Koster J., Romeijn G. J., Hennekam R. C., Vreken P., Andersson H. C., FitzPatrick D. R., Kelley R. I., Wanders R. J. (2001) Am. J. Hum. Genet. 69, 685–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iivonen S., Hiltunen M., Alafuzoff I., Mannermaa A., Kerokoski P., Puoliväli J., Salminen A., Helisalmi S., Soininen H. (2002) Neuroscience 113, 301–310 [DOI] [PubMed] [Google Scholar]
- 7.Kuehnle K., Crameri A., Kälin R. E., Luciani P., Benvenuti S., Peri A., Ratti F., Rodolfo M., Kulic L., Heppner F. L., Nitsch R. M., Mohajeri M. H. (2008) Mol. Cell. Biol. 28, 539–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crameri A., Biondi E., Kuehnle K., Lütjohann D., Thelen K. M., Perga S., Dotti C. G., Nitsch R. M., Ledesma M. D., Mohajeri M. H. (2006) EMBO J. 25, 432–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abad-Rodriguez J., Ledesma M. D., Craessaerts K., Perga S., Medina M., Delacourte A., Dingwall C., De Strooper B., Dotti C. G. (2004) J. Cell Biol. 167, 953–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lämsä R., Helisalmi S., Hiltunen M., Herukka S. K., Tapiola T., Pirttilä T., Vepsäläinen S., Soininen H. (2007) Am. J. Med. Genet. B. Neuropsychiatr. Genet. 144B, 906–910 [DOI] [PubMed] [Google Scholar]
- 11.Tesco G., Koh Y. H., Kang E. L., Cameron A. N., Das S., Sena-Esteves M., Hiltunen M., Yang S. H., Zhong Z., Shen Y., Simpkins J. W., Tanzi R. E. (2007) Neuron 54, 721–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vetrivel K. S., Meckler X., Chen Y., Nguyen P. D., Seidah N. G., Vassar R., Wong P. C., Fukata M., Kounnas M. Z., Thinakaran G. (2009) J. Biol. Chem. 284, 3793–3803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rockenstein E., Mante M., Alford M., Adame A., Crews L., Hashimoto M., Esposito L., Mucke L., Masliah E. (2005) J. Biol. Chem. 280, 32957–32967 [DOI] [PubMed] [Google Scholar]
- 14.Huse J. T., Pijak D. S., Leslie G. J., Lee V. M., Doms R. W. (2000) J. Biol. Chem. 275, 33729–33737 [DOI] [PubMed] [Google Scholar]
- 15.Koh Y. H., von Arnim C. A., Hyman B. T., Tanzi R. E., Tesco G. (2005) J. Biol. Chem. 280, 32499–32504 [DOI] [PubMed] [Google Scholar]
- 16.Luciani P., Gelmini S., Ferrante E., Lania A., Benvenuti S., Baglioni S., Mantovani G., Cellai I., Ammannati F., Spada A., Serio M., Peri A. (2005) J. Clin. Endocrinol. Metab. 90, 6156–6161 [DOI] [PubMed] [Google Scholar]
- 17.Di Stasi D., Vallacchi V., Campi V., Ranzani T., Daniotti M., Chiodini E., Fiorentini S., Greeve I., Prinetti A., Rivoltini L., Pierotti M. A., Rodolfo M. (2005) Int. J. Cancer 115, 224–230 [DOI] [PubMed] [Google Scholar]
- 18.Lu X., Kambe F., Cao X., Kozaki Y., Kaji T., Ishii T., Seo H. (2008) Endocrinology 149, 3267–3273 [DOI] [PubMed] [Google Scholar]
- 19.Wu C., Miloslavskaya I., Demontis S., Maestro R., Galaktionov K. (2004) Nature 432, 640–645 [DOI] [PubMed] [Google Scholar]
- 20.Fernández A., Llacuna L., Fernández-Checa J. C., Colell A. (2009) J. Neurosci. 29, 6394–6405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blasko I., Beer R., Bigl M., Apelt J., Franz G., Rudzki D., Ransmayr G., Kampfl A., Schliebs R. (2004) J. Neural Transm. 111, 523–536 [DOI] [PubMed] [Google Scholar]
- 22.Tong Y., Zhou W., Fung V., Christensen M. A., Qing H., Sun X., Song W. (2005) J. Neural Transm. 112, 455–469 [DOI] [PubMed] [Google Scholar]
- 23.Wen Y., Onyewuchi O., Yang S., Liu R., Simpkins J. W. (2004) Brain Res. 1009, 1–8 [DOI] [PubMed] [Google Scholar]
- 24.Hiltunen M., Mäkinen P., Peräniemi S., Sivenius J., van Groen T., Soininen H., Jolkkonen J. (2009) Neurobiol. Dis. 35, 103–113 [DOI] [PubMed] [Google Scholar]
- 25.Guglielmotto M., Aragno M., Autelli R., Giliberto L., Novo E., Colombatto S., Danni O., Parola M., Smith M. A., Perry G., Tamagno E., Tabaton M. (2009) J. Neurochem. 108, 1045–1056 [DOI] [PubMed] [Google Scholar]
- 26.O'Connor T., Sadleir K. R., Maus E., Velliquette R. A., Zhao J., Cole S. L., Eimer W. A., Hitt B., Bembinster L. A., Lammich S., Lichtenthaler S. F., Hébert S. S., De Strooper B., Haass C., Bennett D. A., Vassar R. (2008) Neuron 60, 988–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.