Abstract
The mitochondrial pyruvate dehydrogenase complex (PDC) is down-regulated by phosphorylation catalyzed by pyruvate dehydrogenase kinase (PDK) isoforms 1–4. Overexpression of PDK isoforms and therefore reduced PDC activity prevails in cancer and diabetes. In the present study, we investigated the role of the invariant C-terminal DW-motif in inhibition of human PDK2 by dichloroacetate (DCA). Substitutions were made in the DW-motif (Asp-382 and Trp-383) and its interacting residues (Tyr-145 and Arg-149) in the other subunit of PDK2 homodimer. Single and double mutants show 20–60% residual activities that are not stimulated by the PDC core. The R149A and Y145F/R149A mutants show drastic increases in apparent IC50 values for DCA, whereas binding affinities for DCA are comparable with wild-type PDK2. Both R149A and Y145F variants exhibit increased similar affinities for ADP and ATP, mimicking the effects of DCA. The R149A and the DW-motif mutations (D382A/W383A) forestall binding of the lipoyl domain of PDC to these mutants, analogous to wild-type PDK2 in the presence of DCA and ADP. In contrast, the binding of a dihydrolipoamide mimetic AZD7545 is largely unaffected in these PDK2 variants. Our results illuminate the pivotal role of the DW-motif in mediating communications between the DCA-, the nucleotide-, and the lipoyl domain-binding sites. This signaling network locks PDK2 in the inactive closed conformation, which is in equilibrium with the active open conformation without DCA and ADP. These results implicate the DW-motif anchoring site as a drug target for the inhibition of aberrant PDK activity in cancer and diabetes.
Introduction
The pyruvate dehydrogenase complex (PDC)2 catalyzes the oxidative decarboxylation of pyruvate to produce acetyl-CoA, linking glycolysis to the Krebs cycle (1–3). The PDC is a 9.5-megadalton catalytic machine comprising multiple copies of the three catalytic components pyruvate dehydrogenase (E1p), dihydrolipoyl transacetylase (E2p), and dihydrolipoamide dehydrogenase (E3); as well as the two regulatory enzymes pyruvate dehydrogenase kinase and pyruvate dehydrogenase phosphatase. The PDC is organized around a structural core comprising multiple subunits of E2p and a non-catalytic component that specifically binds E3 (i.e. the E3-binding protein (E3BP)). Each E2p subunit contains two consecutive N-terminal lipoic acid-bearing domains, termed L1 and L2 (beginning from the N terminus), followed by the E1p-binding domain and the C-terminal inner core/catalytic domain, with these independent domains connected by unstructured linker regions. By analogy, each E3BP subunit consists of a single N-terminal lipoic acid-bearing domain (referred to as L3), the E3-binding domain, and the non-catalytic inner core domain. Together, the inner core domains of E2p and E3BP assemble to form the pentagonal dodecahedral 60-meric E2p/E3BP core.
The mammalian PDC is tightly regulated by reversible phosphorylation. The phosphorylation of E1p by PDK isoforms inactivates the PDC, whereas dephosphorylation by pyruvate dehydrogenase phosphatase isoforms restores PDC activity (1, 4, 5). Phosphorylation of E1p occurs at three serine residues (Ser-264 (site 1), Ser-271 (site 2), and Ser-203 (site 3)) in the α subunit (6–8), although phosphorylation of site 1 alone inactivates PDC (9). To date, four mammalian PDK isoforms (isoforms 1–4) in the mitochondrion have been identified (10). Each PDK isoform exhibits different site specificity; all four isoforms phosphorylate sites 1 and 2 at different rates, but only PDK1 modifies site 3 (10, 11). Phosphorylation at site 1 prevents cofactor thiamine diphosphate-induced ordering of the loop conformations in the E1p active site, which interrupts lipoic acid-bearing domain binding, resulting in the inactivation of PDC (12).
PDKs are recruited to the PDC by preferentially binding to the inner lipoyl (L2) domain of the E2p subunit in the E2p/E3BP core (2, 13). Binding of PDKs to the L2 domain requires a lipoyl group covalently attached to the Lys-173 of L2 (i.e. lipoylated L2 (lipL2)) (14). Individual isoforms exhibit different binding affinities for lipL2 with PDK3 > PDK1 ≈ PDK2 > PDK4 (15). PDK3 is robustly activated by the E2p/E3BP core; the majority of this activation is achieved by binding to isolated lipL2 (16, 17). PDK2 activity is augmented only slightly by lipL2 but by as much as 2-fold by the E2p/E3BP core (18). In contrast, PDK4, with the highest basal activity among PDK isoforms, and PDK1 are not stimulated by either lipL2 or the E2p/E3BP core (18) .
The PDK3-L2 structure revealed that PDK3 exists in equilibrium between the inactive closed and the active open conformations (19). The closed conformation with clamped active-site clefts and disordered C-terminal cross-tails (supplemental Fig. S1, left model) is present in the rat PDK2-ADP structure (20). Nucleotide-free apo-PDK2 (21) and apo-PDK1 (22) show open active site clefts with partially ordered C-terminal cross-tails, which is dubbed the “intermediate open” conformation (supplemental Fig. S1, middle model) (18). The partially ordered C-terminal cross-tails are achieved through the binding of the conserved “DW-motif” from the C-terminal tail of one subunit to the N-terminal domain of the other. It is suggested that an apo-PDK isoform shuttles between the inactive closed conformation and the active “intermediate open” conformation; the equilibrium between these two conformational species determines the basal activity level of each isoform (18). Negative ligands, such as ADP, shift the equilibrium to the inactive closed conformation, whereas positive ligands, including small molecule activators and lipL2, lock the PDK in the intermediate-open (supplemental Fig. S1, middle) and the open (supplemental Fig. S1, right) conformations, respectively. The PDK4-ADP structure is present in a DW-motif-mediated metastable intermediate-open conformation (supplemental Fig. S1, middle) (18). The metastable intermediate-open conformation allows PDK4 to remove ADP efficiently without reverting to the inactive closed conformation, accounting for the robust basal activity in the absence of the E2p/E3BP core.
The expression of PDK2 and PDK4 is induced in starvation and diabetes, which is reversed by insulin treatment (4, 23). Impaired insulin-induced down-regulation of PDK4 (due to the lack of insulin or insensitivity to insulin) leads to the overexpression of PDK4, impeding glucose oxidation in diabetic animals (5, 17). Therefore, PDK4 is a potential drug target for the treatment of type 2 diabetes. Among known synthetic inhibitors for PDK, pyruvate analog dichloroacetate (DCA) is a common classic inhibitor for PDK isoforms (24, 25) and has shown beneficial effects in diabetes (24), lactic acidosis and myocardial ischemia (26, 27). Moreover, recent reports showed that PDK1 (28–30) and PDK3 (31) are up-regulated by HIF-1 in hypoxic cancer cells, which inhibits PDC activity, effectively shutting off the Krebs cycle in tumor cells (i.e. the Warburg effect) (32). Selective inactivation of PDK isoforms by DCA derepresses the expression of a mitochondrial potassium ion channel axis, which triggers apoptosis in cancer cells and leads to the inhibition of tumor growth (33, 34). Thus, DCA is proposed as a potential metabolism-targeting therapy for cancer (35).
The recent structural and mutational analyses showed that DCA binds to a hydrophobic pocket in the N-terminal domain of PDK2 (21, 36). However, the interplays between different ligand-binding sites that lead to PDK inhibition by DCA are not known. In the present study, we chose human PDK2 as a model to dissect the DCA-mediated abrogation of kinase activity. The results show that the conserved DW-motif (Asp-382/Trp-383) plays a pivotal role in mediating communications between DCA-binding, lipoyl domain-binding, and nucleotide-binding sites, resulting in the inactivation of PDK2 by DCA. These findings offer a paradigm for structure-based designs of new inhibitors for this highly conserved family of the mitochondrial PDK isoforms.
EXPERIMENTAL PROCEDURES
Materials
Compound AZD7545 (37) was kindly provided by Dr. Rachel Mayers (AstraZeneca, UK). Dichloroacetate was purchased from Sigma. The expression plasmids for human E1p protein (15) and the E2p/E3BP core (38) were generous gifts from Dr. Krill Popov (University of Alabama at Birmingham). The small ubiquitin modifier (SUMO) plasmid was purchased from Lifesensors Inc. (Malvern, PA).
Protein Expression and Purification
Wild-type and variant human PDK2 proteins were expressed as N-terminal His6-tagged SUMO fusions. A tobacco etch virus protease sequence (Glu-Asn-Leu-Tyr-Phe-Gln↓Ala, with the arrow indicating the cleavage site) was engineered into the linker region between SUMO and PDK2. Single and double variants of SUMO-PDK2 were produced using the QuikChangeTM site-directed mutagenesis kit from Stratagene (La Jolla, CA), with the mutations confirmed by DNA sequencing. All SUMO-PDK2 proteins were co-expressed with chaperonins GroEL and GroES and purified with nickel-nitrilotriacetic acid resin and subsequently on a Superdex S200 column by fast protein liquid chromatography. Untagged PDK2 proteins were prepared by incubating the His6-tagged tobacco etch virus protease with SUMO-PDK2 (mass ratio of 1:25) for 48 h at 4 °C. The reaction mixture was applied to a nickel-nitrilotriacetic acid column; the flow-through portion containing untagged PDK2 was further purified on a Superdex S200 column equilibrated with 50 mm potassium phosphate (pH 7.5), 200 mm KCl, 5% (v/v) glycerol, and 20 mm β-mercaptoethanol. Concentrations of SUMO-PDK2 and untagged PDK2 were determined using calculated extinction coefficients (in mg−1 ml·cm−1) of 0.85 and 1.03, respectively. His6-tagged human lipL2 (residues 128–266) and the E2p/E3BP core were generated and purified as described previously (19).
Microcentrifuge-based Kinase Activity Assay
The microcentrifuge-based assay for PDK2 activity was modified from that described previously (18). The assay mixture (total volume of 25 μl) in a 0.5-ml centrifuge tube contained 20 mm Tris-HCl (pH 7.5), 10 mm KCl, 2 mm dithiothreitol, 2 mm MgCl2, 30 μm E1p (based on the tetramer), and 0.1 μm wild-type SUMO-PDK2 or the Y145F variant (based on the dimer) or a 0.2 μm concentration of the R149A or Y145F/R149A mutant, without or with 1.2 μm E2p/E3BP (based on the 60-meric core). The phosphorylation reaction was initiated by adding 0.5 mm [γ-32P]ATP (specific activity 200–300 cpm/pmol). The reaction was allowed to proceed for 1 min at 23 °C and stopped by adding 20% trichloroacetic acid and 50 mm sodium pyrophosphate. Following centrifugation and washing with 10% trichloroacetic acid, 32P incorporation into the E1p α-subunit was determined by scintillation counting. DCA inhibition of core-free basal (without E2/E3BP) or core-dependent (with E2/E3BP) PDK2 activity was conducted by including DCA (in a concentration range between 0.1 μm and 500 mm) in the assay mixture. Relative activity levels expressed as percentage of controls (activities without DCA) were plotted against increasing DCA concentrations. Curve fitting with the GraphPad Prism program (GraphPad Software Inc., La Jolla, CA) was done to obtain apparent half-maximal inhibition (IC50) values.
Tryptophan Fluorescence Quenching
Tryptophan fluorescence quenching of Trp-383 in the DW-motif was monitored by emission spectra (averaged from three consecutive scans) at 23 °C between 320 and 420 nm in a Fluorolog 3 spectrofluorometer (HORIBA Jobin Yvon, Edison, NJ), with excitation wavelength set at 290 nm. The slit width for both emission and excitation was set at 5 nm. Untagged PDK2 concentrations were 0.5 μm (based on the dimer) in 50 mm potassium phosphate buffer (pH 7.5) containing 0.2 mm EDTA and 2 mm MgCl2. After initial scans, spectra were obtained by adding DCA in aliquots of 1–10 μl from a 40 mm stock. The fluorescence intensities at 350 nm were used to calculate fluorescence quenching. The %Qmax, the maximal quenching at saturating DCA concentration, was an extrapolated value from a double-reciprocal plot. The L0.5 values, representing DCA concentrations causing 50% of maximal fluorescence quenching, were calculated as previously described (18, 39).
Isothermal Titration Calorimetry
SUMO-PDK2 and lipL2 were dialyzed overnight against 50 mm potassium phosphate buffer (pH 7.5) containing 50 mm KCl, 2 mm MgCl2 and 20 mm β-mercaptoethanol. To measure binding affinities of PDK2 for lipL2, 500 μm lipL2 in the syringe was injected into the reaction cell containing 30 μm SUMO-PDK2 (based on the monomer) at 15 °C in a VP-ITC microcalorimeter (MicroCal, Northampton, MA), in the absence or presence of ligands (DCA, ADP, and ATP individually or in combination) at specified concentrations. AZD7545 titrations of SUMO-PDK2 were conducted in a similar fashion except that the syringe contained 100 μm AZD7545, and the reaction cell contained 10 μm SUMO-PDK2. For nucleotide titrations of SUMO-PDK2, 150 μm ADP or ATP in the syringe was injected into the reaction cell containing 20 μm SUMO-PDK2 (based on the monomer) in the absence or presence of 0.3 mm DCA. Dissociation constants (Kd) were calculated with Origin version 7.0 software (OriginLab Corp., Northampton, MA).
RESULTS
Interactions of the DW-motif from Subunit II with Tyr145 and Arg149 in Subunit I
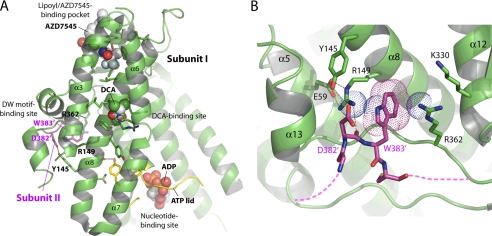
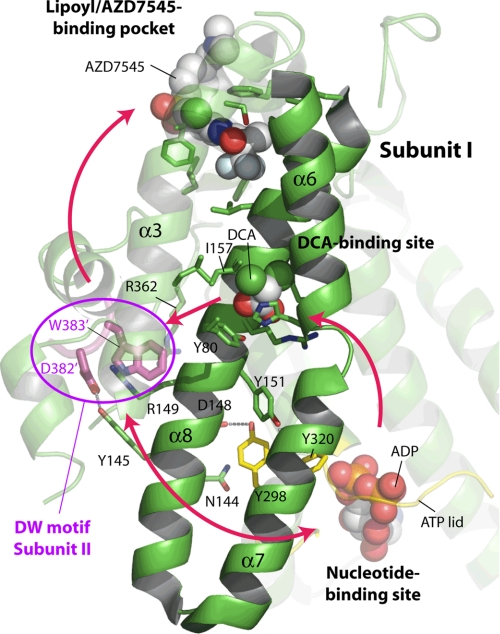
Fig. 1A shows the conserved DW-motif (Asp-382′ and Trp-383′) from subunit II of the PDK2 homodimer anchoring to the binding site in the N-terminal domain of subunit I. The DW-motif-binding site consists of Tyr-145 and Arg-149, with the side chain of Arg-149 forming a hydrogen bond and a salt bridge with the side chain of Glu-59 (Fig. 1B) (39). Moreover, the side chain of a stabilized Arg-149 forms a cation-π interaction with Trp-383′ to secure this residue of the DW-motif. The side chain of Arg-362 further provides van der Waals contacts with the indole ring of Trp-383′. The main-chain amino group of Asp-382′ also forms a hydrogen bond with the side chain of Arg-149. The other residue of the DW-motif, Asp-382′, is hydrogen-bonded to the side chain of Tyr-145. The result shows a more extensive role of Arg-149 than Tyr-145 of subunit I in stabilizing the DW-motif residues from subunit II.
FIGURE 1.
Crystal structure of human PDK2 with different bound ligands and the DW-motif anchoring site. A, ligand-binding sites in the N-terminal domain of human PDK2. The structure of human PDK2 bound to ADP and DCA (Protein Data Bank code 2BU8) (21) was superimposed on that of human PDK1 in complex with AZD7545 (Protein Data Bank code 2Q8G) (22), and only AZD7545 was shown in the PDK2 structure. The N-terminal domain (green) of PDK2 is shown in front of the C-terminal domain. The bound ligands, ADP, DCA, and AZD7545, are shown as sphere models. The residues involved in ligand binding and putative communicating residues between the ligand-binding sites are shown as stick models. The ATP lid is shown in yellow, and the DW-motif in the C-terminal tail from subunit II in the PDK2 dimer is shown in magenta. For clarity, α-helix 5 of PDK2 is removed from the figure. B, close-up view of the DW-motif anchoring site (adapted from 39). The side chains of Asp-382′ and Trp-383′ (DW, in magenta) and the interacting residues in the N-terminal domain (green) are shown as stick models. The dashed lines in magenta indicate disordered regions upstream and downstream of the DW-motif. The gray dashed lines indicate hydrogen bonds. The cation-π interaction between the side chain of Try-383 and two arginine residues (Arg-149 and Arg-362) are represented by dotted spheres. Molecular graphics and superimpositions of the structures were carried out using PyMOL (DeLano Scientific, LLC, Palo Alto, CA).
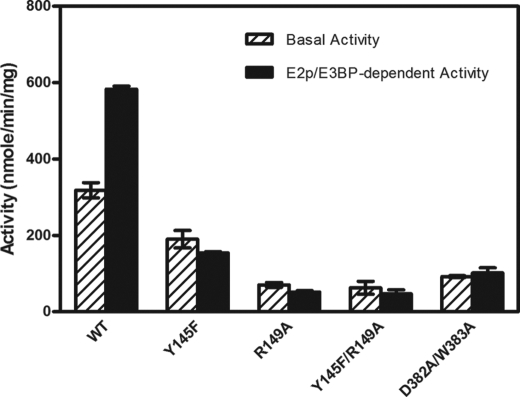
Mutations in the DW-motif and Its Interacting Residues Reduce Basal Activities and Revoke Stimulation by the E2p/E3BP Core
To probe the role of the DW-motif in ligand-mediated regulation of PDK activity, Arg-149 and Tyr-145 that interact with the DW-motif as well as residues of the DW-motif (Asp-382′ and Trp-383′) were altered. As shown in Fig. 2, the basal activity of wild-type PDK2 is stimulated 1.8-fold by the E2p/E3BP core. Y145F, R149A, and Y145F/R149A mutants exhibit 59.7, 22.2, and 19.7% of wild-type basal activity, respectively. Significantly, none of basal activities of these three PDK2 variants are stimulated by E2p/E3BP core. Likewise, the basal activity of the DW-motif double mutant D382A/W383A is reduced to 29% of the wild-type and is also not activated by the E2p/E3BP core. These results lend credence to the notion that the DW-motif is critical for PDK2 basal activity as well as communications between the active site and the lipoyl domain-binding site.
FIGURE 2.
Kinase activities in wild-type and variant PDK2. The phosphorylation reaction was carried out in the absence and presence of the 60-meric E2p/E3BP core as described under “Experimental Procedures.” The specific activities were expressed as nmol of 32P incorporated/min/mg of kinase. Bars, averages ± S.D. (n = 3–5).
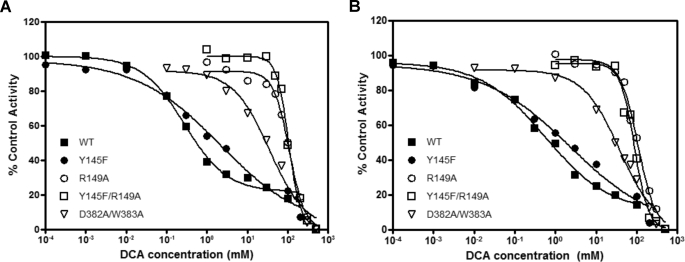
Alterations in the DW-motif and Its Interacting Residues Thwart Inhibition of PDK2 by DCA
Apparent IC50 values for the inhibition of wild-type and mutant PDK2 were determined both in the absence (Fig. 3A) and presence (Fig. 3B) of the E2p/E3BP core. The Y145F variant shows 10- and 6.8-fold increases in apparent IC50 for DCA for basal and E2p/E3BP-dependent activities, respectively (Table 1). More drastic increases (ranging from 74- to 352-fold) in apparent IC50 over the wild-type were obtained with R149A, Y145F/R149A, and D382A/W383A mutants, when measured both in the absence and presence of the E2p/E3BP core. The results indicate that the integrity of the DW-motif anchoring site is critical for the robust inhibition of PDK2 by DCA.
FIGURE 3.
Inhibition of PDK2 basal and E2p/E3BP-dependent kinase activities by DCA. A, inhibition of PDK2 core-free basal activities (without E2/E3BP) by DCA (in a concentration range from 0.1 μm to 500 mm), which is expressed as percentage of control activities (measured without DCA). The phosphorylation reaction was conducted essentially as described in the legend to Fig. 2 except for the inclusion of DCA at different concentrations. Each point represents the average of two independent reactions. B, inhibition of PDK2 core-dependent activities (with 60-meric E2/E3BP) by DCA (in a concentration range from 0.1 μm to 500 mm), expressed as percentage of control activities (measured without DCA). All data were plotted with the GraphPad Prism program.
TABLE 1.
Apparent IC50 values for inhibition of wild-type and mutant PDK2 by DCA
For apparent IC50 determinations, basal and E2p/E3BP-dependent PDK2 activities were assayed in the presence of DCA with concentrations ranging from 0.1 μm to 500 mm. Apparent IC50 values were expressed as average ± S.D. (n = 3–5).
| PDK2 | Apparent IC50 for basal activity | Apparent IC50 for E2p/E3BP-dependent activity |
|---|---|---|
| mm | mm | |
| WT | 0.29 ± 0.02 | 0.48 ± 0.08 |
| Y145F | 3.04 ± 0.16 | 3.24 ± 0.83 |
| R149A | 98.7 ± 8.3 | 108 ± 3 |
| Y145F/R149A | 102 ± 3 | 86.3 ± 7.5 |
| D382A/W383A | 37.9 ± 4.9 | 35.3 ± 4.2 |
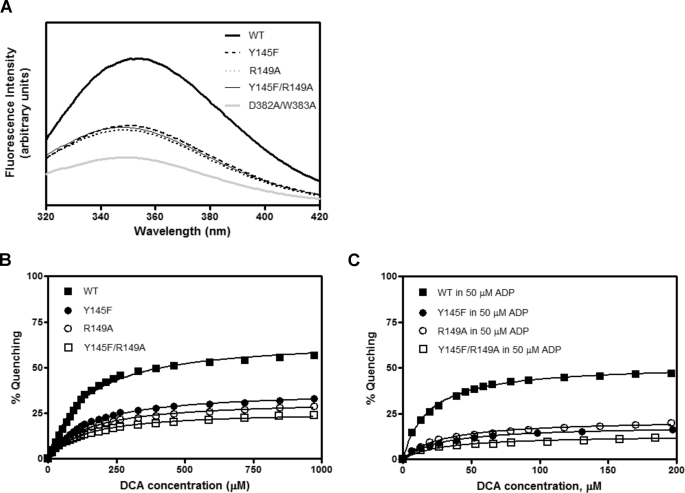
The Binding of DCA Is Not Affected in the PDK2 Mutants
The binding affinities of wild type and the PDK2 mutants for DCA were measured by tryptophan fluorescence quenching as described previously (18, 39). Tryptophan fluorescence quenching results from the solvent exposure of the Trp residue in the DW-motif; the latter dissociates from the DW-motif anchoring site in response to ligand binding. Binding affinities of these proteins for DCA are too low (in the submillimolar range) to allow for direct binding measurement by isothermal titration calorimetry (ITC). Alterations in Tyr-145 and Arg-149, which bind the DW-motif, reduce tryptophan fluorescence significantly (Fig. 4A), indicating a solvent exposure of Trp-383 caused by modifications at the interface between the DW-motif and the N-terminal domain (Fig. 4A). The residual tryptophan fluorescence in D382A/W382A mutant is contributed by other tryptophan residues Trp-79 and Trp-371 in PDK2 (39). Titrations of wild-type and mutant PDK2 with DCA lead to fluorescence quenching of Trp-383 (39) (Fig. 4B). The DCA concentration for the half-maximal quenching (L0.5) provides an approximation of affinity for the inhibitor. Wild-type PDK2 shows L0.5 of 132 μm when titrated with DCA (Table 2). In the presence of 50 μm ADP (Fig. 4C), the affinity of wild-type PDK2 for DCA is increased by 7.9-fold to L0.5 = 16.8 μm (Table 2). Y145F, R149A, and Y145F/R149A mutants uniformly show L0.5 values that are similar to that of wild-type PDK2. In the presence of ADP, the affinities of Y145F, R149A, and Y145F/R149A mutants increase by 4.8-, 4.5-, and 3.3-fold, respectively. The maximal tryptophan fluorescence quenching (Qmax) in the D382A/W383A mutant is too low to allow for L0.5 determination. Taken together, these results indicate that the abated DCA inhibition in the DW-motif and its interacting residue mutants is not caused by impeded binding of DCA to these PDK2 mutants.
FIGURE 4.
Tryptophan fluorescence quenching in wild-type and mutant PDK2. Tryptophan fluorescence quenching from the DW-motif was monitored by recording emission spectra (averaged from three consecutive scans) between 320 and 420 nm in a Fluorolog 3 spectrofluorometer (HORIBA Jobin Yvon, Edison, NJ), with the excitation wavelength set at 290 nm. Initial tag-free PDK2 concentrations (based on the dimer) were 0.5 μm in 50 mm potassium phosphate buffer (pH 7.5) containing 0.2 mm EDTA and 2 mm MgCl2. A, fluorescence spectra of WT and mutant PDK2 in the absence of DCA or ADP. B, quenching of PDK2 fluorescence by increasing concentrations of DCA. C, quenching of PDK2 fluorescence by 50 μm ADP and increasing concentrations of DCA. The quenching data were fit as described (39) and plotted with the GraphPad Prism program.
TABLE 2.
Binding affinities of wild-type and mutant PDK2 for DCA measured by fluorescence quenching in the absence or presence of ADP
Fluorescence quenching by DCA of untagged wild-type and mutant PDK2 proteins was carried out with or without 50 μm ADP as described under “Experimental Procedures.” The L0.5 values are the DCA concentrations causing half-maximum fluorescence quenching. The L0.5 and Qmax values are expressed as averages ± S.D. (n = 3). NM, not measurable.
| PDK2 | ADP (50 μm) | L0.5 | Qmax |
|---|---|---|---|
| μm | % | ||
| WT | − | 132 ± 8 | 68.8 ± 2.8 |
| WT | + | 16.8 ± 0.9 | 52.3 ± 0.6 |
| Y145F | − | 143 ± 8 | 37.2 ± 0.4 |
| Y145F | + | 30.1 ± 1.5 | 20.6 ± 0.2 |
| R149A | − | 135 ± 7 | 32.5 ± 0.6 |
| R149A | + | 30.0 ± 0.9 | 21.5 ± 0.1 |
| Y145F/R149A | − | 98.1 ± 7.8 | 27.0 ± 1.6 |
| Y145F/R149A | + | 29.9 ± 0.6 | 16.1 ± 2.3 |
| D382A/W383A | − | NM | NM |
Effects of DCA on Nucleotide Binding in Wild-type and Mutant PDK2
To investigate the effects of DCA on nucleotide binding, affinities (Kd) of wild-type and mutant PDK2 for ADP and ATP were determined by ITC. Wild-type PDK2 exhibits lower affinity for ADP (Kd = 7.43 μm) than ATP (Kd = 3.88 μm) (Table 3). In the presence of 0.3 mm DCA, affinities of wild-type PDK2 for ADP and ATP are increased by 3.7- and 2.0-fold, respectively, resulting in similar binding affinities for both nucleotides (Table 3). Substitutions of Tyr-145 and Arg-149, which bind the DW-motif, result in higher than wild-type but also similar affinities for both ADP and ATP in the respective mutants. The presence of DCA results in further increases in affinities of the Y145F mutant for ADP and ATP by 3.0- and 2.2-fold, respectively. In contrast, affinities of the R149A variant for ADP and ATP are similar and remain unchanged despite the presence of DCA. Affinities of the Y145F/R149A double mutant for ADP and ATP are comparable with those of the R149A single variant and are not affected by DCA. The DW-motif double mutant, D382A/W383A, possesses slightly higher than wild-type affinities for ADP and ATP, which increase slightly in the presence of DCA. The above ITC measurements indicate that the disruption of Arg-149 that binds the DW-motif nullifies DCA-mediated affinity increases for nucleotides ADP and ATP observed with wild-type PDK2.
TABLE 3.
Binding affinities of wild-type and mutant PDK2 proteins for nucleotides (ADP or ATP) measured by ITC in the absence or presence of DCA
The 150 μm concentration of ADP or ATP in the syringe was injected into the reaction cell containing 15 μm SUMO-PDK2 (based on the monomer) at 15 °C in the absence or presence of 0.3 mm DCA. A one-site binding model was used to fit the binding isotherms with the Origin 7 program. All Kd and ΔH values are averaged from three independent titrations (n = 3).
| PDK2 | DCA (0.3 mm) | Kd (ADP) | ΔH (ADP) | Kd (ATP) | ΔH (ATP) |
|---|---|---|---|---|---|
| μm | kcal/mol | μm | kcal/mol | ||
| WT | − | 7.43 ± 0.69 | −12.8 ± 0.7 | 3.88 ± 0.16 | −12.2 ± 0.2 |
| WT | + | 2.03 ± 0.08 | −18.7 ± 0.5 | 1.91 ± 0.26 | −15.1 ± 0.7 |
| Y145F | − | 2.75 ± 0.11 | −18.5 ± 0.5 | 3.18 ± 0.34 | −14.5 ± 0.9 |
| Y145F | + | 0.93 ± 0.07 | −23.7 ± 0.5 | 1.45 ± 0.01 | −21.0 ± 0.3 |
| R149A | − | 1.62 ± 0.17 | −17.4 ± 0.9 | 1.66 ± 0.29 | −14.2 ± 1.1 |
| R149A | + | 1.63 ± 0.27 | −21.8 ± 0.9 | 1.63 ± 0.04 | −19.4 ± 1.5 |
| Y145F/R149A | − | 1.82 ± 0.13 | −20.2 ± 1.6 | 1.86 ± 0.13 | −16.4 ± 0.8 |
| Y145F/R149A | + | 1.84 ± 0.07 | −17.8 ± 0.5 | 1.80 ± 0.17 | −19.2 ± 1.7 |
| D382A/W383A | − | 4.03 ± 0.07 | −19.4 ± 2.3 | 1.95 ± 0.11 | −15.4 ± 1.5 |
| D382A/W383A | + | 2.89 ± 0.12 | −24.2 ± 2.8 | 1.67 ± 0.13 | −22.7 ± 1.3 |
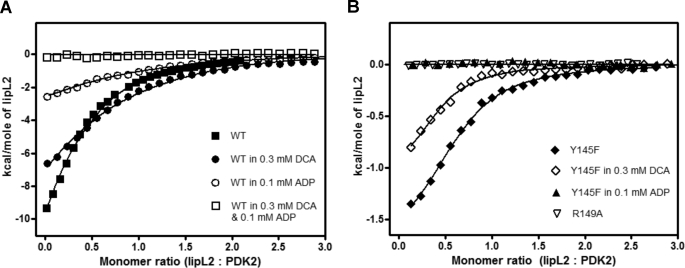
Effects of DCA and Nucleotides on lipL2 Binding to PDK2
It was shown previously that the presence of both DCA and ADP prevents binding of lipL2 to wild-type PDK2 (39). In this study, the effects of mutations in the DW-motif and its interacting residues at the DW-motif binding site on lipL2 binding were dissected by ITC. As shown in Fig. 5A and Table 4, the presence of 0.3 mm DCA, 0.1 mm ADP, or 0.1 mm ATP alone is without measurable effect on the binding affinity of lipL2 to wild-type PDK2 with Kd value ranging from 14.3 to 16.3 μm. However, enthalpy changes (ΔH) in the presence of ADP or ATP are decreased by 3.6- and 5.6-fold, respectively. The combined presence of DCA and ADP or DCA and ATP completely abolishes the binding of lipL2 to wild-type PDK2. For the Y145F mutant, the presence of 0.3 mm DCA alone does not change the binding affinity of the mutant for lipL2 with Kd = 5.2 μm with or without DCA (Fig. 5B and Table 4). However, the presence of ADP or ATP alone negates the binding of lipL2 to the Y145F variant. Significantly, with the R149A mutant, the ability to bind lipL2 is completely nullified in the absence of DCA and/or nucleotides. The loss of lipL2 binding in the absence of DCA or nucleotides is also observed with the Y145F/R149A and D382A/W383A double mutants. The above results, in combination, strongly suggest that the perturbation of the DW-motif or its binding site either by DCA together with ADP or ATP or by site-directed mutagenesis renders PDK2 unable to bind lipL2.
FIGURE 5.
Distinct binding properties of lipL2 to wild-type and mutant PDK2 in the absence or presence of different ligands measured by ITC. Binding affinities of wild-type and mutant PDK2 for lipL2 were determined by injecting 500 μm lipL2 in 10-μl increments into the cell containing 1.8 ml of 30 μm PDK2 (based on the monomer). Ligands (ADP, DCA, or ADP and DCA), alone or in combination, were included in both the syringe and the cell in equal concentrations. Titration data were initially processed by the Origin 7 program and subsequently presented with the GraphPad Prism program. A, WT-PDK2, WT-PDK2 in 0.3 mm DCA, WT-PDK2 in 0.1 mm ADP, WT-PDK2 in 0.3 mm DCA, and 0.1 mm ADP were titrated with lipL2 in the absence or presence of corresponding ligands. B, Y145F-PDK2, Y145F in 0.3 mm DCA, Y145F-PDK2 in 0.1 mm ADP, and R149A-PDK2 were titrated in the absence or presence of the corresponding ligands.
TABLE 4.
Binding affinities of wild-type and mutant PDK2 proteins for lipL2 measured by ITC in the absence or presence of different ligands
The 500 μm concentration of lipL2 in the syringe was injected into the reaction cell containing 20 μm SUMO-PDK2 (based on the monomer) at 15 °C in the absence or presence of the indicated ligands (DCA, ADP, and/or ATP). The concentrations of these ligands are DCA at 0.3 mm and both ADP and ATP at 0.1 mm. All Kd values are expressed as average ± S.D. (n = 3). NM, not measurable.
| PDK2 | DCA (0.3 mm) | ADP (0.1 mm) | ATP (0.1 mm) | Kd | ΔH |
|---|---|---|---|---|---|
| μm | kcal/mol | ||||
| WT | − | − | − | 14.4 ± 2.7 | −16.1 ± 2.0 |
| WT | + | − | − | 14.3 ± 1.8 | −12.8 ± 2.2 |
| WT | − | + | − | 16.3 ± 3.5 | −4.5 ± 0.5 |
| WT | − | − | + | 16.1 ± 1.5 | −2.9 ± 0.5 |
| WT | + | + | − | NM | NM |
| WT | + | − | + | NM | NM |
| Y145F | − | − | − | 5.2 ± 0.1 | −1.5 ± 0.3 |
| Y145F | + | − | − | 5.2 ± 0.4 | −1.4 ± 0.4 |
| Y145F | − | + | − | NM | NM |
| Y145F | − | − | + | NM | NM |
| R149A | − | − | − | NM | NM |
| Y145F/R149A | − | − | − | NM | NM |
| D382A/W383A | − | − | − | NM | NM |
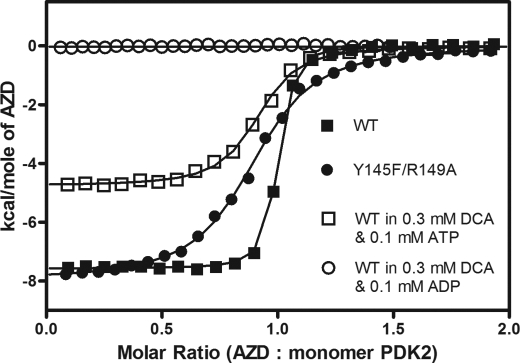
Effects of DCA and Nucleotides on AZD7545 Binding to Wild-type and Mutant PDK2
Dihydrolipoamide mimetics AZD7545 and AZ12 were shown to bind to the lipoyl domain-binding sites of PDK1 (22) and PDK2 (21), respectively. Effects of DCA and nucleotide on the binding of AZD7545, which lacks the protein moiety of the lipoyl domain, to wild-type and mutant PDK2 were also dissected by ITC. The presence of 0.3 mm DCA or 0.1 mm ATP alone has no effect on the binding affinity of wild-type PDK2 for AZD7545, as indicated by similar Kd values in the range of 18.4–24.0 nm (Table 5). The presence of DCA and ATP significantly lowers the affinity of wild-type PDK2 for AZD7545 by 3.5-fold. Notably, the combined presence of DCA and ADP eliminates the binding of AZD7545 to wild-type PDK2, similar to that observed with lipL2 (Fig. 6).
TABLE 5.
Binding affinities of wild-type and mutant PDK2 for AZD7545 determined by ITC in the absence or presence of different ligands
The 100 μm concentration of AZD7545 in the syringe was injected into the ITC reaction cell containing 10 μm SUMO-PDK2 (based on the monomer) at 15 °C in the absence or presence of the indicated ligands (DCA alone, DCA combined with ADP, or DCA combined with ATP). The concentrations of these ligands are DCA at 0.3 mm and both ADP and ATP at 0.1 mm. Values are expressed as average ± S.D. (n = 3). NM, not measurable.
| PDK2 | DCA (0.3 mm) | ADP (0.1 mm) | ATP (0.1 mm) | Kd | ΔH |
|---|---|---|---|---|---|
| nm | kcal/mol | ||||
| WT | − | − | − | 19.2 ± 0.6 | −7.9 ± 0.6 |
| WT | + | − | − | 18.4 ± 0.6 | −11.7 ± 1.2 |
| WT | − | + | − | 50.2 ± 4.6 | −5.0 ± 0.1 |
| WT | − | − | + | 24.0 ± 0.3 | −6.2 ± 0.1 |
| WT | + | + | − | NM | NM |
| WT | + | − | + | 66.5 ± 4.2 | −4.8 ± 0.2 |
| Y145F | − | − | − | 17.7 ± 0.7 | −9.4 ± 0.8 |
| Y145F | + | − | − | 19.1 ± 1.2 | −9.6 ± 0.6 |
| Y145F | − | + | − | 40.7 ± 5.5 | −6.8 ± 0.5 |
| Y145F | − | − | + | 21.1 ± 1.7 | −6.3 ± 0.3 |
| R149A | − | − | − | 36.7 ± 0.7 | −6.2 ± 0.2 |
| Y145F/R149A | − | − | − | 220 ± 11 | −7.1 ± 1.0 |
| D382A/W383A | − | − | − | 16.7 ± 1.8 | −9.3 ± 0.3 |
FIGURE 6.
Binding of AZD7545 to wild-type and mutant PDK2 in the absence or presence of different ligands measured by ITC. Binding affinities of PDK2 and AZD7545 were determined by injecting 100 μm AZD7545 in 10-μl increments into the cell containing 1.8 ml of 10 μm PDK2 (based on the monomer). Ligands (DCA and ATP or ADP) were included in both the syringe and the cell in equal concentrations. Titration data were initially processed by the Origin 7 program and subsequently presented with the GraphPad Prism program.
On the other hand, the Y145F PDK2 mutant shows binding affinity for AZD7545 comparable with wild-type PDK2 (Kd = 17.7–21.1 nm, respectively). The binding of AZD7545 to this mutant is not affected by the presence of DCA or ATP alone. The presence of 0.1 mm ADP alone decreases the affinity of the Y145F variant by 2.3-fold. Interestingly, both the R149A mutant and the Y145F/R149A double mutant, which do not bind lipL2, are able to bind AZD7545, with binding affinities reduced by 1.9- and 11.5-fold, respectively, compared with wild-type PDK2. Likewise, the DW-motif double mutant D382A/W383A, which fails to bind lipL2, shows wild-type binding affinity for AZD7545 with Kd of 16.7 nm.
DISCUSSION
In the present study, PDK2 was chosen to investigate the mechanism of DCA inhibition because it has the highest solubility among the four PDK isoforms, which facilitates in vitro biophysical measurements. PDK3 has the lowest binding affinity for DCA (40) and is therefore not suitable for studying the mechanism of DCA inhibition. The inability of lipL2 and the E2p/E3BP core to stimulate PDK1 and PDK4 activities (18) precludes investigations into interactions between the bound DCA and the lipoyl domain-binding sites in these PDK isoforms. The markedly reduced basal activities in the PDK2 mutants affecting the DW-motif and its interacting residues Tyr-145 and Arg-149 corroborate the hypothesis that the DW-motif is critical for kinase activity, similar to that observed with the DW-motif double mutant of PDK4 (18). Nonetheless, the presence of residual basal activity in the PDK2 mutants permitted the measurements of apparent IC50 for DCA inhibition (Fig. 3 and Table 1). The R149A mutant shows a far more robust increase in apparent IC50 for basal activity than the Y145F variant when compared with wild-type PDK2. The higher apparent IC50 value and the lower residual basal activity of the R149A mutant than the Y145F is explained by the fact that Arg-149 has more extensive interactions with the DW-motif than Tyr-145, as revealed by the PDK2 structure (Fig. 1). The possibility can also be entertained that the more drastic amino acid substitution of R149A than Y145F may contribute, in part, to the more pronounced conformational changes at the DW-motif binding interface, resulting in lower basal activity in the former PDK2 variant.
We showed that the presence of ADP significantly increases affinity of wild-type PDK2 for DCA (Table 2), which is in good agreement with the findings reported previously (39, 41). Perturbations in the DW-motif anchoring site do not have appreciable effects on the affinities of Y145F, R149A, and Y145F/R149A mutants for DCA either in the absence or presence of ADP. Thus, the DW-motif is pivotal in the transmission of information from the nucleotide-binding site to the DCA binding site (Fig. 7). Nucleotide binding promotes a disorder-to-order transition of the ATP lid in the nucleotide-binding site of the PDK3-L2 complex (19) and PDK2 (21). The results suggest that the nucleotide-induced ordering of the ATP lid promotes the high affinity binding of DCA to its binding site. However, the preferential positive effects of ADP over ATP on DCA binding reported previously (39) are presently not understood. A recent report showed that the T302A mutation in the ATP lid significantly mitigates DCA inhibition in PDK2 (41). This result can be explained by the fact that the T302A mutation impedes the binding of nucleotides to PDK2 as measured by ITC (supplemental Fig. S2A) and fluorescence quenching (41). The absence of bound ADP in the T302A mutant hampers ADP-induced high affinity DCA binding and therefore abolishes the inhibition of kinase activity by DCA (41). The abrogation of ADP binding also explains the similar-to-wild-type binding affinity of the T302A variant for lipL2, either in the absence or presence of DCA and ADP (supplemental Fig. S2B). In contrast, the presence of both DCA and ADP completely prevents the binding of lipL2 to wild-type PDK2.
FIGURE 7.
The signaling network between different ligand-binding sites in human PDK2. This figure originates from Fig. 1A with minor modifications. As a signal axis, helices α6, α7, and α8 are highlighted as solid schematic models with the rest of the protein shown as a transparency. The red arrows indicate the directional communications between the different ligand binding sites. See “Discussion” for details of site-to-site communications mediated by the DW-motif.
Wild-type PDK2 shows higher affinity for ATP than ADP, as indicated by Kd values as determined by ITC (Table 3). The disparate affinities for adenine nucleotides are significant because it promotes the ATP/ADP exchange in favor of substrate ATP, allowing cycles of the phosphoryl transfer reaction to proceed. The preferential increase in affinities for ADP over ATP in the presence of DCA (Table 3) supports the previous finding that the binding of pyruvate or its analog DCA slows the rate of ADP dissociation from PDK2, resulting in a PDK2-ADP inhibitor dead-end complex (42). It is also significant that, in the presence of DCA, wild-type PDK2 exhibits similar affinities for both ADP and ATP, which prevents the efficient removal of product ADP during phosphorylation reaction cycles. Because ADP is a potent feedback inhibitor for PDK during the phosphoryl transfer reaction (43), the considerably slower release of ADP from PDK2 is a plausible mechanism for the inhibition of PDK activity by pyruvate or DCA. On the other hand, the R149A mutation, which disrupts the DW-motif anchoring site, increases affinities of the PDK2 variant to new similar levels for both ADP and ATP, mimicking the effects of DCA (Table 3). Binding affinities of this mutant for ADP and ATP remain unchanged despite the presence of 0.3 mm DCA. This is reflected by the ∼340-fold increase of apparent IC50 for DCA in the R149A mutant (Table 1). Compared with the R149A variant, smaller increases in affinities for ADP and apparent IC50 for DCA are obtained with the Y145F and the D382A/W383A mutants, which is consistent with the lower degree of disturbance at the DW-motif interface caused by the latter mutations. Alanine substitutions of Asp-382 and Trp-383 in the DW-motif do not result in a complete disordering of the C-terminal cross-tails in the mutant PDK2 structure (data not shown). The alternate cross-tail conformation may account for the less robust increase in affinity for ADP in the D382A/W383A double mutant compared with the R149A variant. Collectively, the above results corroborate the central role of the DW-motif in fostering the increased affinity for ADP, leading to the significant inhibition of PDK2 activity by DCA. The PDK2-ADP-DCA complex with disordered C-terminal tails locks PDK2 in the inactive closed conformation, which, in the absence of ligands, is in equilibrium with the active intermediate open conformation (supplemental Fig. S1). Thus, the direction of signal transmission originates from the DCA binding site → the DW-motif → the nucleotide-binding site (Fig. 7).
The presence of ADP alone (20) or DCA combined with ADP (21) results in the disruption of the DW-motif anchors and complete disordering of C-terminal tails in wild-type PDK2. The binding of DCA itself does not promote the collapse of the DW-motif anchors and the cross-tail conformation in PDK1 (22). However, DCA binding induces a flip in the His-149 (the leader sequence included) side chain, resulting in the unwinding of a helical segment between α6 and α7 helices (22) (Fig. 7). These conformational changes probably interfere with the communication between the ATP lid in the nucleotide-binding site and the α7 helix that forms a side wall of the active site cleft. The α8 helix, on the other hand, harbors the DW-motif-interacting residues Tyr-145 and Arg-149. Thus, it is conceivable that helices α6/α7 and α8 form a “signaling axis” that transmits signals from nucleotide binding site → DCA-binding site → the DW-motif anchor, leading to the collapse of the DW-motif anchors in wild-type PDK2. Residues between helices α7 and α8 (e.g. Tyr-298, Tyr-151 and Asp-148) may be important in maintaining the above signaling axis for communications between the three ligand-binding sites.
We have shown previously that the binding of lipL2 to PDK3 produces a completely ordered close-tail conformation buttressed by the DW-motif anchors, which fosters a widening of the active site cleft (i.e. the open conformation), resulting in the stimulation of PDK3 activity (19). The truncations of the C-terminal tail segment containing the DW-motif in PDK3 (44) or mutations in the DW-motif in PDK2 (Table 4) forestall lipL2 binding. These findings establish unequivocally that the DW-motif is indispensable for lipL2 binding and the stimulation of kinase activity by the E2p/E3BP core. The disordering of C-terminal tails induced by the bound DCA and ADP in the PDK2 structure eliminates amino acid determinants on the C-terminal tail, which interact with the protein moiety from lipL2 (19, 44); this mechanism accounts for the loss of lipL2 binding, as determined by ITC (Table 4) and by analytical ultracentrifugation (39). The critical role of the DW-motif in mediating lipL2 binding is further supported by the absence of lipL2 binding in the R149A, Y145F/R149A, and D382A/W383A mutants. In these mutants, the DW-motif interface is intrinsically altered to the extent that the bound DCA and/or ADP are no longer required to prevent PDK2 from binding to lipL2. The Y145F variant, in which the DW-motif interface is only partially impaired, requires the presence of nucleotide ATP or ADP to halt lipL2 binding. Thus, the present results reinforce the thesis that the DW-motif is the key mediator for lipL2 binding to PDK2, with a signaling route transmitted from DCA binding site → the DW-motif → lipoyl domain binding site.
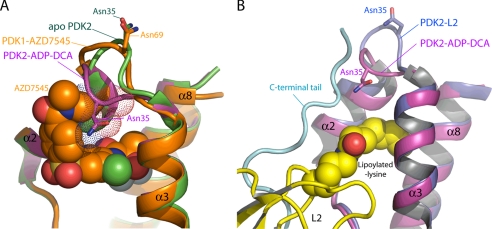
The non-proteinaceous lipoamide mimetic AZD7545 binds with high affinity to the lipoyl domain-binding site of wild-type PDK2 (Table 5). AZD7545 has been shown to robustly stimulate PDK3 basal activity, to a level similar to that obtained with lipL2 (22). However, the C-terminal end segment downstream of the DW-motif is not ordered by AZD7545 and is absent from the lipoyl binding site in the PDK1-AZD7545 structure (22) (Fig. 8A). When the lipoyl domain-binding site in the PDK2-DCA-ADP structure is superimposed with that in apo-PDK2, the Cα backbone of Asn-35 in PDK2 (Asn-69 in PDK1) located in a loop region moves by 6.5 Å, with a concomitant flip of the loop region (Fig. 8A). The movement of the loop region induced by DCA and ADP binding poses a steric hindrance between the Asn-35 side chain and the tail portion of AZD7545, thereby occluding the binding of AZD7545 to PDK2, as indicated by ITC measurements (Fig. 6). The loop region movement in the lipoyl domain-binding site induced by DCA and ADP does not appear to cause steric clashes with the linear dihydrolipoamide moiety when superimposed with the PDK2-L2-AMP-PNP structure (Fig. 8B). However, the complete disordering of the C-terminal cross-tails in the presence of DCA and ADP precludes the binding of lipL2 to wild-type PDK2. The C-terminal tail region contains key determinants that interact with both the protein and dihydrolipoamide moieties of lipL2 (19). In contrast, mutations in the DW-motif or its interacting residues result in essentially wild-type binding affinities for AZD7545 in the Y145F, R149A, and D382A/W383A mutants. The only exception is the Y145F/R149A double mutant, which shows substantially reduced binding affinity for AZD7545. These results suggest that, unlike the lipL2 binding, an intact DW-motif is not essential for the binding of non-proteinaceous AZD7545 to PDK2.
FIGURE 8.
Conformational changes in the lipoyl domain-binding site induced by DCA and ADP binding. A, a steric clash between AZD7545 and the loop region at the lipoyl-binding site of PDK2. The structures of apo-PDK2 (green, Protein Data Bank code 2BTZ) (21) and PDK2-ADP-DCA (Protein Data Bank code 2BU8 magenta) are superimposed on that of PDK1-AZD7545 (orange, Protein Data Bank code 2Q8G) (22). PDK residue numbers include the leader sequence. Asn-35 in the flipped loop between helices α2 and α3 of PDK2-ADP-DCA could cause a steric clash with the bound AZD7545 in PDK1, which may explain the failure of WT PDK2 to bind AZD7545 in the presence of both ADP and DCA (Table 5). AZD7545 is shown as a solid sphere model. The van der Waals radius of Asn-35 is depicted with dotted spheres. B, comparison of the lipoyl-binding site in the PDK2-L2 and PDK2-ADP-DCA structures. The PDK2-L2 complex structure (Protein Data Bank code 3CRL) (48) (PDK2 in blue and L2 in yellow) is superimposed on the structure of the PDK2-ADP-DCA ternary complex (21) in magenta. The ordered C-terminal tail in the PDK2-L2 is shown in cyan. The inversion of the loop between helices α2 and α3 induced by ADP and DCA binding would not interfere with the binding of lipoyl group of L2 to PDK2.
The present study demonstrates the central role of the DW-motif in mediating communications between the N-terminal and C-terminal domains, which dictate the equilibrium between the closed and the open conformations in PDK isoforms. A metastable DW-motif favoring the intermediate-open conformation was shown to be responsible for the robust basal activity of PDK4 in the absence of the E2p/E3BP core (18). Because DCA is an analog of pyruvate, the latter itself being a potent inhibitor of PDK2 (45), the present results with DCA provide mechanistic insight into the feed-forward activation of PDC activity by its own substrate pyruvate. This mechanism is important for the regulation of glucose oxidation by reversible phosphorylation of PDC under hormonal and nutritional stimuli (46, 47). More importantly, the present results establish the DW-motif anchoring site as a drug target for the development of small molecule or peptidomimetic inhibitors for PDK isoforms. This new generation of PDK inhibitors may prove useful for use in metabolic targeting as a new strategy against cancer and type 2 diabetes.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants DK62306 and DK26758. This work was also supported by Welch Foundation Grant I-1286.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- PDC
- pyruvate dehydrogenase complex
- E1p
- pyruvate dehydrogenase
- E2p
- dihydrolipoyl transacetylase
- E3
- dihydrolipoamide dehydrogenase
- E3BP
- E3-binding protein
- L1
- lipoyl domain 1 of E2p
- L2
- inner lipoyl domain 2 of E2p
- L3
- lipoyl domain 3 of E3BP
- PDK
- pyruvate dehydrogenase kinase
- lipL2
- lipoylated L2
- DW-motif
- C-terminal region containing the invariant Asp-382 and Trp-383 in PDK2
- DCA
- dichloroacetic acid
- SUMO
- small ubiquitin modifier
- ITC
- isothermal titration calorimetry
- WT
- wild-type
- AMP-PNP
- 5′-adenylylimido-diphosphate
- AZD7545
- 2R-N-{4-[4-(dimethylcarbamoyl)phenylsulfonyl]-2-chlorophenyl}-3,3,3-trifluoro-2- hydroxy-2-methylpropanamide.
REFERENCES
- 1.Reed L. J., Damuni Z., Merryfield M. L. (1985) Curr. Top. Cell Regul. 27, 41–49 [DOI] [PubMed] [Google Scholar]
- 2.Patel M. S., Roche T. E. (1990) FASEB J. 4, 3224–3233 [DOI] [PubMed] [Google Scholar]
- 3.Reed L. J. (2001) J. Biol. Chem. 276, 38329–38336 [DOI] [PubMed] [Google Scholar]
- 4.Harris R. A., Huang B., Wu P. (2001) Adv. Enzyme Regul. 41, 269–288 [DOI] [PubMed] [Google Scholar]
- 5.Holness M. J., Sugden M. C. (2003) Biochem. Soc. Trans. 31, 1143–1151 [DOI] [PubMed] [Google Scholar]
- 6.Yeaman S. J., Hutcheson E. T., Roche T. E., Pettit F. H., Brown J. R., Reed L. J., Watson D. C., Dixon G. H. (1978) Biochemistry 17, 2364–2370 [DOI] [PubMed] [Google Scholar]
- 7.Teague W. M., Pettit F. H., Yeaman S. J., Reed L. J. (1979) Biochem. Biophys. Res. Commun. 87, 244–252 [DOI] [PubMed] [Google Scholar]
- 8.Sale G. J., Randle P. J. (1981) Eur. J. Biochem. 120, 535–540 [DOI] [PubMed] [Google Scholar]
- 9.Korotchkina L. G., Patel M. S. (2001) J. Biol. Chem. 276, 5731–5738 [DOI] [PubMed] [Google Scholar]
- 10.Korotchkina L. G., Patel M. S. (2001) J. Biol. Chem. 276, 37223–37229 [DOI] [PubMed] [Google Scholar]
- 11.Kolobova E., Tuganova A., Boulatnikov I., Popov K. M. (2001) Biochem. J. 358, 69–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kato M., Wynn R. M., Chuang J. L., Tso S. C., Machius M., Li J., Chuang D. T. (2008) Structure 16, 1849–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu S., Baker J. C., Roche T. E. (1995) J. Biol. Chem. 270, 793–800 [DOI] [PubMed] [Google Scholar]
- 14.Radke G. A., Ono K., Ravindran S., Roche T. E. (1993) Biochem. Biophys. Res. Commun. 190, 982–991 [DOI] [PubMed] [Google Scholar]
- 15.Tuganova A., Boulatnikov I., Popov K. M. (2002) Biochem. J. 366, 129–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baker J. C., Yan X., Peng T., Kasten S., Roche T. E. (2000) J. Biol. Chem. 275, 15773–15781 [DOI] [PubMed] [Google Scholar]
- 17.Roche T. E., Hiromasa Y., Turkan A., Gong X., Peng T., Yan X., Kasten S. A., Bao H., Dong J. (2003) Eur. J. Biochem. 270, 1050–1056 [DOI] [PubMed] [Google Scholar]
- 18.Wynn R. M., Kato M., Chuang J. L., Tso S. C., Li J., Chuang D. T. (2008) J. Biol. Chem. 283, 25305–25315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato M., Chuang J. L., Tso S. C., Wynn R. M., Chuang D. T. (2005) EMBO J. 24, 1763–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steussy C. N., Popov K. M., Bowker-Kinley M. M., Sloan R. B., Jr., Harris R. A., Hamilton J. A. (2001) J. Biol. Chem. 276, 37443–37450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knoechel T. R., Tucker A. D., Robinson C. M., Phillips C., Taylor W., Bungay P. J., Kasten S. A., Roche T. E., Brown D. G. (2006) Biochemistry 45, 402–415 [DOI] [PubMed] [Google Scholar]
- 22.Kato M., Li J., Chuang J. L., Chuang D. T. (2007) Structure 15, 992–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu P., Sato J., Zhao Y., Jaskiewicz J., Popov K. M., Harris R. A. (1998) Biochem. J. 329, 197–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitehouse S., Cooper R. H., Randle P. J. (1974) Biochem. J. 141, 761–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bersin R. M., Stacpoole P. W. (1997) Am. Heart J. 134, 841–855 [DOI] [PubMed] [Google Scholar]
- 26.Stacpoole P. (1989) Compr. Ther. 15, 28–44 [PubMed] [Google Scholar]
- 27.Henderson G. N., Curry S. H., Derendorf H., Wright E. C., Stacpoole P. W. (1997) J. Clin. Pharmacol. 37, 416–425 [DOI] [PubMed] [Google Scholar]
- 28.Kim J. W., Tchernyshyov I., Semenza G. L., Dang C. V. (2006) Cell Metab. 3, 177–185 [DOI] [PubMed] [Google Scholar]
- 29.Papandreou I., Cairns R. A., Fontana L., Lim A. L., Denko N. C. (2006) Cell Metab. 3, 187–197 [DOI] [PubMed] [Google Scholar]
- 30.Koukourakis M. I., Giatromanolaki A., Sivridis E., Gatter K. C., Harris A. L. (2005) Neoplasia 7, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu C. W., Lin S. C., Chen K. F., Lai Y. Y., Tsai S. J. (2008) J. Biol. Chem. 283, 28106–28114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Warburg O. (1956) Science 124, 269–270 [PubMed] [Google Scholar]
- 33.Bonnet S., Archer S. L., Allalunis-Turner J., Haromy A., Beaulieu C., Thompson R., Lee C. T., Lopaschuk G. D., Puttagunta L., Bonnet S., Harry G., Hashimoto K., Porter C. J., Andrade M. A., Thebaud B., Michelakis E. D. (2007) Cancer Cell 11, 37–51 [DOI] [PubMed] [Google Scholar]
- 34.Cairns R. A., Papandreou I., Sutphin P. D., Denko N. C. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 9445–9450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Michelakis E. D., Webster L., Mackey J. R. (2008) Br. J. Cancer 99, 989–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klyuyeva A., Tuganova A., Popov K. M. (2007) FEBS Lett. 581, 2988–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mayers R. M., Butlin R. J., Kilgour E., Leighton B., Martin D., Myatt J., Orme J. P., Holloway B. R. (2003) Biochem. Soc. Trans. 31, 1165–1167 [DOI] [PubMed] [Google Scholar]
- 38.Harris R. A., Bowker-Kinley M. M., Wu P., Jeng J., Popov K. M. (1997) J. Biol. Chem. 272, 19746–19751 [DOI] [PubMed] [Google Scholar]
- 39.Hiromasa Y., Hu L., Roche T. E. (2006) J. Biol. Chem. 281, 12568–12579 [DOI] [PubMed] [Google Scholar]
- 40.Bowker-Kinley M. M., Davis W. I., Wu P., Harris R. A., Popov K. M. (1998) Biochem. J. 329, 191–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klyuyeva A., Tuganova A., Popov K. M. (2008) Biochemistry 47, 8358–8366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bao H., Kasten S. A., Yan X., Roche T. E. (2004) Biochemistry 43, 13432–13441 [DOI] [PubMed] [Google Scholar]
- 43.Roche T. E., Reed L. J. (1974) Biochem. Biophys. Res. Commun. 59, 1341–1348 [DOI] [PubMed] [Google Scholar]
- 44.Tso S. C., Kato M., Chuang J. L., Chuang D. T. (2006) J. Biol. Chem. 281, 27197–27204 [DOI] [PubMed] [Google Scholar]
- 45.Pratt M. L., Roche T. E. (1979) J. Biol. Chem. 254, 7191–7196 [PubMed] [Google Scholar]
- 46.Harris R. A., Bowker-Kinley M. M., Huang B., Wu P. (2002) Adv. Enzyme Regul. 42, 249–259 [DOI] [PubMed] [Google Scholar]
- 47.Sugden M. C., Holness M. J. (2003) Am. J. Physiol. Endocrinol. Metab. 284, E855–E862 [DOI] [PubMed] [Google Scholar]
- 48.Green T., Grigorian A., Klyuyeva A., Tuganova A., Luo M., Popov K. M. (2008) J. Biol. Chem. 283, 15789–15798 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.