Abstract
Together with seven ADAMTS-like proteins, the 19 mammalian ADAMTS proteases constitute a superfamily. ADAMTS proteases are secreted zinc metalloproteases whose hallmark is an ancillary domain containing one or more thrombospondin type 1 repeats. ADAMTS-like proteins resemble ADAMTS ancillary domains and lack proteolytic activity. Vertebrate expansion of the superfamily reflects emergence of new substrates, duplication of proteolytic activities in new contexts, and cooperative functions of the duplicated genes. ADAMTS proteases are involved in maturation of procollagen and von Willebrand factor, as well as in extracellular matrix proteolysis relating to morphogenesis, angiogenesis, ovulation, cancer, and arthritis. New insights into ADAMTS mechanisms indicate significant regulatory roles for ADAMTS ancillary domains, propeptide processing, and glycosylation. ADAMTS-like proteins appear to have regulatory roles in the extracellular matrix.
Evolutionary Perspective
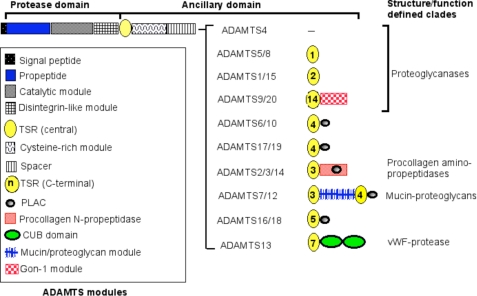
Comparison of the human ADAMTS repertoire with invertebrates (1, 2) suggests that at least five human ADAMTS clades arose early in deuterostome evolution, whereas the largest human clade, which contains proteoglycanases, arose later in chordates. The six Ciona intestinalis ADAMTS proteases represent core evolutionary branches in chordates, and the mammalian ADAMTS clades they root are distinctive, implying neofunctionalization (i.e. distinct substrate specificities) of each clade (1). ADAMTS proteases in each mammalian clade have, however, a similar primary structure (Fig. 1), implying subfunctionalization, i.e. maintenance of substrate specificity, but specialization in different biological contexts (1). As an example, ADAMTS2, ADAMTS3, and ADAMTS14 each process the major fibrillar procollagen types I–III in vitro, but their tissue-specific expression manifests each activity in discrete tissues (3). ADAMTS2 and ADAMTS3 are coexpressed with procollagen III (a vascular collagen) and procollagen II (a cartilage collagen), respectively, and may have dominant roles in their maturation, whereas procollagen I processing may be undertaken by both enzymes in a tissue-specific or overlapping manner (3).
FIGURE 1.
Mammalian ADAMTS proteases. The domain backbone shared by each ADAMTS protease is shown at the top. The unique structure of each ADAMTS protease C-terminal to the backbone is indicated on the right, and the key to these modules is located on the left. Some clades are named according to structural or functional characteristics that best define them; clades without a known function or a defining characteristic are not named. The proteoglycanases constitute a superclade comprising ADAMTS proteases with different domain structure. The figure is based on reference sequences obtained from GenBankTM. PLAC, protease and lacunin module.
Functions of ADAMTS Proteases
This minireview primarily discusses functions for which unequivocal genetic evidence exists and substrates whose deficient cleavage was implicated in a phenotype. ADAMTS13 mutations cause inherited TTP2(4, 5), whereas acquired TTP is caused by ADAMTS13 autoantibodies. ADAMTS13 processes vWF at a single peptide bond (Tyr1605–Met1606) and has no other known substrates (4, 5). TTP results from failure to process ultra-large vWF to an optimal size for normal coagulation. ADAMTS13 deficiency leads to occlusion of the circulation by platelet thrombi, with serious consequences for vital organs.
ADAMTS2 mutations cause Ehlers-Danlos syndrome (dermatosparactic type) (6), a connective tissue disorder first identified in cattle (7) and a consequence of failure to excise the amino propeptide of procollagen I in the dermis of skin. The retained propeptide hinders collagen fibril assembly, and the resulting thin branched fibrils are weak, leading to severe skin fragility (7). Other collagen-rich tissues (e.g. bone) are not fragile probably because of the compensating activity of ADAMTS3 (8) and/or ADAMTS14 (9). Procollagen processing is a specialized task, but unlike vWF processing, it is shared among three similar enzymes.
ADAMTS10 mutations cause a rare connective tissue disorder, recessive WMS (10). Several clinical features of WMS are the opposite of Marfan syndrome, caused by mutations of FBN1, which encodes the matrix glycoprotein fibrillin-1. FBN1 mutations lead to TGFβ dysregulation (11). Fibrillin-1 mutations can cause dominant WMS (12), suggesting that ADAMTS10 is functionally coupled to fibrillin-1. Indeed, we have found that ADAMTS10 specifically bound to fibrillin-1.3 This places ADAMTS10 squarely within a TGFβ-regulating extracellular network centered in fibrillin microfibrils (13).
Several ADAMTS proteases collectively referred to as proteoglycanases (Fig. 1) participate in proteolysis of the large aggregating proteoglycans aggrecan, versican, and brevican and have been the subject of intense study because aggrecan loss is a major feature of osteoarthritis. Mice with deletion of the ADAMTS5 catalytic module (Adamts5ΔCat) are resistant to induced arthritis (14, 15). Thus, ADAMTS5 is regarded as a major aggrecanase in mice and is a target for drug development in osteoarthritis.
Specific physiological roles of the proteoglycanases were sought using genetically engineered or naturally occurring mouse mutants. Although Adamts4ΔCat and Adamts5ΔCat mice and the doubly deleted mice are reportedly normal, 45% of Adamts1 null mice die perinatally (16). Surviving mice are small, with low female fertility and urogenital anomalies (16–18). Adamts1 is hormonally regulated and is essential for proteolysis of periovular versican during ovulation and for ovarian lymphangiogenesis (19, 20). ADAMTS1 is anti-angiogenic via several mechanisms. It sequesters VEGF165 (21), binds fibroblast growth factor-2 via a heparin bridge (22), and releases anti-angiogenic fragments from thrombospondin-1 and thrombospondin-2 (23). In Xenopus laevis, XADAMTS1 was found to regulate embryonic fibroblast growth factor signaling (24). ADAMTS1 is essential for versican turnover during mouse cardiac development and is regulated coordinately with myocardial trabecular growth and compaction (25).
Among the proteoglycanases, ADAMTS9 and ADAMTS20 constitute a highly conserved subset given their strong resemblance to the GON-1 protease in Caenorhabditis elegans. Adamts20 mutations occur in the white spotting mutant, Belted (bt) (26). Expression of Adamts20 mRNA in dermal mesenchyme coordinately with melanoblast migration during embryogenesis initially suggested that ADAMTS20 facilitated melanoblast migration (26). However, new evidence suggests that Adamts20 promotes survival of melanoblasts via versican proteolysis and Kit signaling (27). Adamts9 is widely expressed in mesoderm derivatives (28). Adamts9 null mice die by 7.5 days of gestation (27), currently precluding full analysis of its developmental role. ADAMTS9 was shown to be a tumor suppressor in esophageal and nasopharyngeal cancer using a functional complementation approach (29), and another proteoglycanase, ADAMTS15, was identified as a tumor suppressor gene in colorectal cancer (30). In addition to its tumor suppressor role, ADAMTS9 is a product of microvascular endothelial cells that acts cell-autonomously to suppress angiogenesis.4
Mutations of the C. elegans ADAMTS proteases GON-1 and MIG-17 have offered several novel insights. gon-1 mutants have gonad defects due to failed migration of pathfinding DTCs along the ventral body wall (31, 32), whereas mig-17 mutants have defects in gonadal arm migration along the dorsal body wall (33). gon-1 defects may result from lack of basement membrane degradation or release of migratory cues and can be rescued by transgenic overexpression of ADAMTS9 or ADAMTS4. RNA interference and engineered deletions of fibulin, but not of several other extracellular matrix proteins, suppressed the gon-1 phenotype (34). The mig-17 phenotype was suppressed only by dominant fibulin mutations, but not by fibulin inactivation (35). Fibulin mutations in the worm lead to expansion of the gonad and loss of integrity of the basement membrane separating it from the gut lumen (35); the protease mutations suppressed, in turn, the fibulin phenotype (34). Thus, fibulin and the two C. elegans proteases appear to interact in the gonadal basement membrane (34, 35), but neither GON-1 nor MIG-17 cleaves fibulin (34, 35). Intriguingly, MIG-17 controls cell migration by recruitment of nidogen to the basement membrane (36).
Cooperative Functions of ADAMTS Proteases
Evolutionary subfunctionalization suggests that closely related ADAMTS proteases may cooperatively maintain proteolysis above a functionally critical threshold in specific pathways. In support of a cooperative proteolysis model, hemizygosity of Adamts9 increased the depigmented area in bt mice (27). Furthermore, 100% of bt/bt:Adamts9+/− mice develop cleft palate, whereas bt and Adamts9+/− mice have delayed palatal closure but a very low incidence of cleft palate.5 ADAMTS5, ADAMTS9, and ADAMTS20 work cooperatively in resorption of interdigital webs in mice.6 In interdigital webs, the cardiac outflow tract, embryonic myocardium, and ovarian follicles, several proteoglycanases are coexpressed in versican-rich contexts and may work cooperatively to degrade this proteoglycan (25, 37–39). Cooperation of ADAMTS2 with ADAMTS3 and/or ADAMTS14 is suggested by the presence of varying amounts of completely processed procollagen I and III in dermatosparactic tissues (3). The contribution of GON-1 and MIG-17 to gonadogenesis is also a cooperative role, albeit at discrete stages of DTC migration.
Structure, Post-translational Modification, and Proteolytic Mechanisms
ADAMTS proteases consist of a protease domain and an ancillary domain with a characteristic modular structure containing at least one TSR (Fig. 1). In general, ADAMTS ancillary domains provide substrate-binding specificity, and the protease domain provides cleavage site specificity. ADAMTS catalytic modules have a reprolysin active-site motif (HEXXH + HD), but their cysteine signatures differ from ADAM proteins (40). Inhibition of ADAMTS4 and ADAMTS5 by TIMP3, which also inhibits ADAM proteins (41), suggests structural similarities between the active sites of ADAMTS proteases and ADAM proteins despite several specific differences in catalytic cleft topography (42, 43). The crystal structure of ADAMTS1 (42) suggests that the disintegrin-like module is functionally a part of the protease domain. This is supported by absence of the disintegrin-like domain in all ADAMTS-like proteins (Fig. 2) and its presence in MIG-17, which lacks an ancillary domain. Indeed, residues in the ADAMTS13 disintegrin-like domain comprise an exosite that interacts with vWF (44).
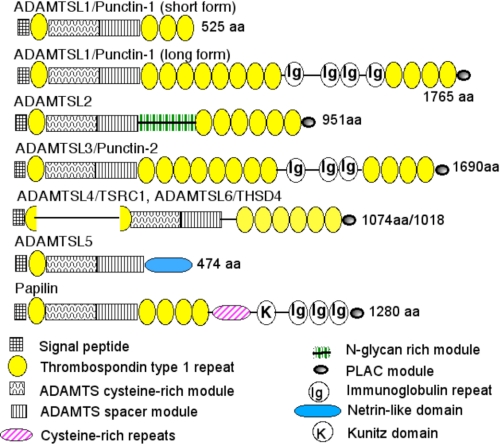
FIGURE 2.
Mammalian ADAMTSL proteins. The domain structure of each ADAMTSL is shown according to the key at the bottom. The two forms of ADAMTSL1 shown are splice variants, and the long form composes a clade with ADAMTSL3. ADAMTSL4 and ADAMTSL6 compose a distinct clade in which TSR1 is split by an insertion. The figure is based on reference sequences obtained from GenBankTM. aa, amino acids. PLAC, protease and lacunin module.
All ADAMTS proteases (except ADAMTS4) and ADAMTSL are modified by addition of N-linked carbohydrate. N-Glycosylation of the MIG-17 propeptide is critical for its spatial targeting during gonadal morphogenesis (45), and N-glycosylation of the ADAMTS9 propeptide is essential for its secretion (46). WXXW and CXX(S/T)CG motifs in TSRs are consensus sites for C-mannosylation (of Trp) and O-fucosylation (of Ser and Thr), respectively (47). Prevention of O-fucosylation of ADAMTSL1 and ADAMTS13 severely restricts their secretion (48, 49). Because O-fucosylation requires prior protein folding, it functions as a quality control mechanism in biosynthesis. C-Mannosylation was demonstrated in ADAMTSL1 and ADAMTS5, and it also appears to be a potential quality control mechanism (50). ADAMTS7 and ADAMTS12 are unusual in being mucins as well as chondroitin sulfate proteoglycans, with both modifications being present in a distinct module (Fig. 1) (51).
Excision of metalloprotease propeptides, a process termed “activation,” is a key post-translational step. ADAMTS propeptide excision is undertaken by proprotein convertases, e.g. furin. ADAMTS1 and ADAMTS4 are conventionally processed by furin because their processing occurs in the trans-Golgi and is required for proteolytic activity (52, 53). However, there are several idiosyncrasies in ADAMTS propeptide processing. For example, removal of the short ADAMTS13 propeptide is not essential for vWF cleavage (54). Furin processing of several ADAMTS proteases occurs extracellularly (51, 55, 56), and pro-ADAMTS9 specifically undergoes cell-surface furin processing (56). In contrast to most metalloproteases, cell-surface processing of the ADAMTS9 propeptide by furin reduces its proteolytic activity (46). MIG-17 undergoes a stage-specific extracellular activation during DTC migration (57). The furin-processing site in ADAMTS10 lacks a P4 or P6 Arg residue and is inefficiently processed by furin (55).
ADAMTS proteases require their ancillary domains for substrate recognition and correct tissue compartmentalization (58–63). The ancillary domain of several ADAMTS proteases is modified by C-terminal proteolysis (52, 55, 64), which potentially alters substrate recognition and enzyme localization. C-terminal fragments thus released may have independent biological activities, as exemplified by oxidative platelet fragmentation caused by a fragment of ADAMTS18 (65).
Some ADAMTS proteases are operational cell-surface proteases via the affinity of their ancillary domain or propeptides for cell-surface molecules (55, 59, 66) and may participate in cell-surface ectodomain shedding; indeed, ADAMTS1 sheds syndecan-4 (67) and increases the shedding of two heparin-binding epidermal growth factors, amphiregulin and heparin-binding epidermal growth factor (68). Processing of the ADAMTS4 ancillary domain is initiated at the chondrocyte cell surface within a complex comprising a heparan sulfate proteoglycan (syndecan-1) and a membrane-anchored metalloprotease (66).
ADAMTS activity may be influenced by cofactors or substrate modification. ADAMTS1 associates with fibulin-1 and, like GON-1, does not cut it, but the interaction enhances its aggrecanase activity by 10-fold (69). ADAMTS1 and ADAMTS4 bind fibronectin, but whereas the aggrecanase activity of ADAMTS4 is inhibited, that of ADAMTS1 is enhanced 2-fold by fibronectin (69, 70). The heparin-binding property of some ADAMTS proteases such as ADAMTS1 provides a basis for cell binding and matrix association. Therefore, intermolecular interactions may define niches for ADAMTS proteases, bring them into defined networks, and act as cofactors for their activity. As shown by MIG-17 recruitment of nidogen (36), ADAMTS proteases may also be instrumental in forming networks.
Post-translational modification of some ADAMTS substrates is essential for their cleavage. ADAMTS2 processes heat-denatured procollagen inefficiently, requiring the triple-helical conformation of procollagen or the hairpin conformation of the propeptide folded back onto the triple helix (71). ADAMTS4 requires the glycosaminoglycan side chains on aggrecan for efficient processing (62). ADAMTS13 processing is enhanced by hydrodynamic shear-mediated stretching of the vWF A1 and A3 domains in circulating blood, which exposes the scissile bond (72); thus, in test tube assays, vWF processing by ADAMTS13 is inefficient and requires mild denaturation for optimization.
ADAMTS-like Proteins
ADAMTS-like proteins (Fig. 2) are products of distinct genes, not alternatively spliced products of ADAMTS genes, and they appear to be components of the extracellular matrix (73, 74). Their resemblance to ADAMTS proteases and matrix-binding properties suggest a potential role in ADAMTS regulation. Indeed, bovine ADAMTS2 is inhibited non-competitively by Drosophila papilin (74), and isoforms of MIG-6, a C. elegans papilin ortholog, influence DTC migration and act in the same pathway as MIG-17 (75). Alternatively, ADAMTSL may have architectural or regulatory functions in the matrix that are independent of ADAMTS proteases. Human ADAMTSL2 mutations cause geleophysic dysplasia, a rare disorder that resembles WMS and that is characterized by high levels of TGFβ activity (76). ADAMTSL2 binds to latent TGFβ-binding protein-1 (76), a fibrillin-1 ligand, as well as to fibrillin-1,7 suggesting that ADAMTSL2 and ADAMTS10 operate in the same pathway. A homozygous ADAMTSL4 mutation was recently identified in isolated ectopia lentis (dislocation of the ocular lens) (77). The suspensory ligament of the lens, which centers the lens in the path of light, is composed primarily of fibrillin-1. Intriguingly, ectopia lentis is a major clinical feature of both WMS and Marfan syndrome, and dominant ectopia lentis is caused by FBN1 mutations (11). Taken together, the role of ADAMTSL2 and ADAMTSL4 in TGFβ regulation and maintenance of the lens, respectively, suggests that these ADAMTSL proteins may regulate fibrillin supramolecular assembly and/or architecture.
Conclusions
Beginning with the discovery of ADAMTS1 only a dozen years ago (78), the ADAMTS field has advanced considerably. We now appreciate that some of these molecules work in highly specialized contexts (vWF and procollagen processing), whereas broad subsets of the superfamily may operate within fibrillin networks and in proteoglycan cleavage. The concept of cooperative proteolysis is likely to apply to several biological contexts. The concepts and knowledge that have been developed will be invaluable for investigating several superfamily members (e.g. ADAMTS17, ADAMTS19, ADAMTSL5, and ADAMTSL6) of whom little is currently known beyond their primary structure.
Supplementary Material
Acknowledgments
I am indebted to my colleagues in the ADAMTS field whose work has shaped the ideas presented here. I thank J. Evan Sadler and Kiyoji Nishiwaki for helpful comments.
This work was supported by National Institutes of Health Grants AR43890 and AR49930. This work was also supported by the National Marfan Foundation, the Arthritis Foundation, and the Yamanouchi USA Foundation. This is the ninth article in the Thematic Minireview Series on Proteolytic Enzymes. The first article was published in the November 7, 2008 issue; the second and third articles were published in the May 22, 2009 issue; the fourth and fifth articles were published in the July 31, 2009 issue; the sixth and seventh articles were published in the August 14, 2009 issue; and the eighth article was published in the August 28, 2009 issue. This minireview will be reprinted in the 2009 Minireview Compendium, which will be available in January, 2010.
W. E. Kutz, L. W. Wang, and S. S. Apte, manuscript in preparation.
B.-H. Koo and S. S. Apte, manuscript in preparation.
H. Enomoto, C. Nelson, and S. S. Apte, unpublished data.
McCulloch, D. R., Nelson, C. M., Dixon, L. J., Silver, D. L., Wylie, J. D., Lindner, V., Sasaki, T., Cooley, M. A., Argraves, W. S., and Apte, S. S. (2009) Dev. Cell, in press.
W. Wang and S. S. Apte, unpublished data.
- TTP
- thrombotic thrombocytopenic purpura
- vWF
- von Willebrand factor
- WMS
- Weill-Marchesani syndrome
- TGFβ
- transforming growth factor-β
- DTC
- distal tip cell
- TSR
- thrombospondin type 1 repeat.
REFERENCES
- 1.Huxley-Jones J., Apte S. S., Robertson D. L., Boot-Handford R. P. (2005) Int. J. Biochem. Cell Biol. 37, 1838–1845 [DOI] [PubMed] [Google Scholar]
- 2.Angerer L., Hussain S., Wei Z., Livingston B. T. (2006) Dev. Biol. 300, 267–281 [DOI] [PubMed] [Google Scholar]
- 3.Le Goff C., Somerville R. P., Kesteloot F., Powell K., Birk D. E., Colige A. C., Apte S. S. (2006) Development 133, 1587–1596 [DOI] [PubMed] [Google Scholar]
- 4.Levy G. G., Nichols W. C., Lian E. C., Foroud T., McClintick J. N., McGee B. M., Yang A. Y., Siemieniak D. R., Stark K. R., Gruppo R., Sarode R., Shurin S. B., Chandrasekaran V., Stabler S. P., Sabio H., Bouhassira E. E., Upshaw J. D., Jr., Ginsburg D., Tsai H. M. (2001) Nature 413, 488–494 [DOI] [PubMed] [Google Scholar]
- 5.Zheng X., Chung D., Takayama T. K., Majerus E. M., Sadler J. E., Fujikawa K. (2001) J. Biol. Chem. 276, 41059–41063 [DOI] [PubMed] [Google Scholar]
- 6.Colige A., Sieron A. L., Li S. W., Schwarze U., Petty E., Wertelecki W., Wilcox W., Krakow D., Cohn D. H., Reardon W., Byers P. H., Lapière C. M., Prockop D. J., Nusgens B. V. (1999) Am. J. Hum. Genet. 65, 308–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lapière C. M., Nusgens B. V. (1993) Arch. Dermatol. 129, 1316–1319 [DOI] [PubMed] [Google Scholar]
- 8.Fernandes R. J., Hirohata S., Engle J. M., Colige A., Cohn D. H., Eyre D. R., Apte S. S. (2001) J. Biol. Chem. 276, 31502–31509 [DOI] [PubMed] [Google Scholar]
- 9.Colige A., Vandenberghe I., Thiry M., Lambert C. A., Van Beeumen J., Li S. W., Prockop D. J., Lapiere C. M., Nusgens B. V. (2002) J. Biol. Chem. 277, 5756–5766 [DOI] [PubMed] [Google Scholar]
- 10.Dagoneau N., Benoist-Lasselin C., Huber C., Faivre L., Mégarbané A., Alswaid A., Dollfus H., Alembik Y., Munnich A., Legeai-Mallet L., Cormier-Daire V. (2004) Am. J. Hum. Genet. 75, 801–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinson P. N., Arteaga-Solis E., Baldock C., Collod-Béroud G., Booms P., De Paepe A., Dietz H. C., Guo G., Handford P. A., Judge D. P., Kielty C. M., Loeys B., Milewicz D. M., Ney A., Ramirez F., Reinhardt D. P., Tiedemann K., Whiteman P., Godfrey M. (2006) J. Med. Genet 43, 769–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faivre L., Gorlin R. J., Wirtz M. K., Godfrey M., Dagoneau N., Samples J. R., Le Merrer M., Collod-Beroud G., Boileau C., Munnich A., Cormier-Daire V. (2003) J. Med. Genet. 40, 34–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charbonneau N. L., Ono R. N., Corson G. M., Keene D. R., Sakai L. Y. (2004) Birth Defects Res. Part C Embryo Today Rev. 72, 37–50 [DOI] [PubMed] [Google Scholar]
- 14.Glasson S. S., Askew R., Sheppard B., Carito B., Blanchet T., Ma H. L., Flannery C. R., Peluso D., Kanki K., Yang Z., Majumdar M. K., Morris E. A. (2005) Nature 434, 644–648 [DOI] [PubMed] [Google Scholar]
- 15.Stanton H., Rogerson F. M., East C. J., Golub S. B., Lawlor K. E., Meeker C. T., Little C. B., Last K., Farmer P. J., Campbell I. K., Fourie A. M., Fosang A. J. (2005) Nature 434, 648–652 [DOI] [PubMed] [Google Scholar]
- 16.Mittaz L., Russell D. L., Wilson T., Brasted M., Tkalcevic J., Salamonsen L. A., Hertzog P. J., Pritchard M. A. (2004) Biol. Reprod. 70, 1096–1105 [DOI] [PubMed] [Google Scholar]
- 17.Shindo T., Kurihara H., Kuno K., Yokoyama H., Wada T., Kurihara Y., Imai T., Wang Y., Ogata M., Nishimatsu H., Moriyama N., Oh-hashi Y., Morita H., Ishikawa T., Nagai R., Yazaki Y., Matsushima K. (2000) J. Clin. Invest. 105, 1345–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mittaz L., Ricardo S., Martinez G., Kola I., Kelly D. J., Little M. H., Hertzog P. J., Pritchard M. A. (2005) Nephrol. Dial. Transplant. 20, 419–423 [DOI] [PubMed] [Google Scholar]
- 19.Brown H. M., Dunning K. R., Robker R. L., Pritchard M., Russell D. L. (2006) Dev. Biol. 300, 699–709 [DOI] [PubMed] [Google Scholar]
- 20.Shozu M., Minami N., Yokoyama H., Inoue M., Kurihara H., Matsushima K., Kuno K. (2005) J. Mol. Endocrinol. 35, 343–355 [DOI] [PubMed] [Google Scholar]
- 21.Luque A., Carpizo D. R., Iruela-Arispe M. L. (2003) J. Biol. Chem. 278, 23656–23665 [DOI] [PubMed] [Google Scholar]
- 22.Krampert M., Kuenzle S., Thai S. N., Lee N., Iruela-Arispe M. L., Werner S. (2005) J. Biol. Chem. 280, 23844–23852 [DOI] [PubMed] [Google Scholar]
- 23.Lee N. V., Sato M., Annis D. S., Loo J. A., Wu L., Mosher D. F., Iruela-Arispe M. L. (2006) EMBO J. 25, 5270–5283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suga A., Hikasa H., Taira M. (2006) Dev. Biol. 295, 26–39 [DOI] [PubMed] [Google Scholar]
- 25.Stankunas K., Hang C. T., Tsun Z. Y., Chen H., Lee N. V., Wu J. I., Shang C., Bayle J. H., Shou W., Iruela-Arispe M. L., Chang C. P. (2008) Dev. Cell 14, 298–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rao C., Foernzler D., Loftus S. K., Liu S., McPherson J. D., Jungers K. A., Apte S. S., Pavan W. J., Beier D. R. (2003) Development 130, 4665–4672 [DOI] [PubMed] [Google Scholar]
- 27.Silver D. L., Hou L., Somerville R., Young M. E., Apte S. S., Pavan W. J. (2008) PLoS. Genet. 4, e1000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jungers K. A., Le Goff C., Somerville R. P., Apte S. S. (2005) Gene Expr. Patterns 5, 609–617 [DOI] [PubMed] [Google Scholar]
- 29.Lo P. H., Leung A. C., Kwok C. Y., Cheung W. S., Ko J. M., Yang L. C., Law S., Wang L. D., Li J., Stanbridge E. J., Srivastava G., Tang J. C., Tsao S. W., Lung M. L. (2007) Oncogene 26, 148–157 [DOI] [PubMed] [Google Scholar]
- 30.Viloria C. G., Obaya A. J., Moncada-Pazos A., Llamazares M., Astudillo A., Capellá G., Cal S., López-Otín C. (2009) Cancer Res. 69, 4926–4934 [DOI] [PubMed] [Google Scholar]
- 31.Blelloch R., Anna-Arriola S. S., Gao D., Li Y., Hodgkin J., Kimble J. (1999) Dev. Biol. 216, 382–393 [DOI] [PubMed] [Google Scholar]
- 32.Blelloch R., Kimble J. (1999) Nature 399, 586–590 [DOI] [PubMed] [Google Scholar]
- 33.Nishiwaki K., Hisamoto N., Matsumoto K. (2000) Science 288, 2205–2208 [DOI] [PubMed] [Google Scholar]
- 34.Hesselson D., Newman C., Kim K. W., Kimble J. (2004) Curr. Biol. 14, 2005–2010 [DOI] [PubMed] [Google Scholar]
- 35.Kubota Y., Kuroki R., Nishiwaki K. (2004) Curr. Biol. 14, 2011–2018 [DOI] [PubMed] [Google Scholar]
- 36.Kubota Y., Ohkura K., Tamai K. K., Nagata K., Nishiwaki K. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 20804–20809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kern C. B., Twal W. O., Mjaatvedt C. H., Fairey S. E., Toole B. P., Iruela-Arispe M. L., Argraves W. S. (2006) Dev. Dyn. 235, 2238–2247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCulloch D. R., Le Goff C., Bhatt S., Dixon L. J., Sandy J. D., Apte S. S. (2009) Gene Expr. Patterns 9, 314–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richards J. S., Hernandez-Gonzalez I., Gonzalez-Robayna I., Teuling E., Lo Y., Boerboom D., Falender A. E., Doyle K. H., LeBaron R. G., Thompson V., Sandy J. D. (2005) Biol. Reprod. 72, 1241–1255 [DOI] [PubMed] [Google Scholar]
- 40.Hurskainen T. L., Hirohata S., Seldin M. F., Apte S. S. (1999) J. Biol. Chem. 274, 25555–25563 [DOI] [PubMed] [Google Scholar]
- 41.Kashiwagi M., Tortorella M., Nagase H., Brew K. (2001) J. Biol. Chem. 276, 12501–12504 [DOI] [PubMed] [Google Scholar]
- 42.Gerhardt S., Hassall G., Hawtin P., McCall E., Flavell L., Minshull C., Hargreaves D., Ting A., Pauptit R. A., Parker A. E., Abbott W. M. (2007) J. Mol. Biol. 373, 891–902 [DOI] [PubMed] [Google Scholar]
- 43.Tortorella M. D., Malfait F., Barve R. A., Shieh H. S., Malfait A. M. (2009) Curr. Pharm. Des. 15, 2359–2374 [DOI] [PubMed] [Google Scholar]
- 44.de Groot R., Bardhan A., Ramroop N., Lane D. A., Crawley J. T. (2009) Blood 113, 5609–5616 [DOI] [PubMed] [Google Scholar]
- 45.Ihara S., Nishiwaki K. (2007) EMBO J. 26, 2607–2620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koo B. H., Longpré J. M., Somerville R. P., Alexander J. P., Leduc R., Apte S. S. (2007) J. Biol. Chem. 282, 16146–16154 [DOI] [PubMed] [Google Scholar]
- 47.Hofsteenge J., Huwiler K. G., Macek B., Hess D., Lawler J., Mosher D. F., Peter-Katalinic J. (2001) J. Biol. Chem. 276, 6485–6498 [DOI] [PubMed] [Google Scholar]
- 48.Ricketts L. M., Dlugosz M., Luther K. B., Haltiwanger R. S., Majerus E. M. (2007) J. Biol. Chem. 282, 17014–17023 [DOI] [PubMed] [Google Scholar]
- 49.Wang L. W., Dlugosz M., Somerville R. P., Raed M., Haltiwanger R. S., Apte S. S. (2007) J. Biol. Chem. 282, 17024–17031 [DOI] [PubMed] [Google Scholar]
- 50.Wang L. W., Leonhard-Melief C., Haltiwanger R. S., Apte S. S. (2009) J. Biol. Chem. 284, 30004–30015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Somerville R. P., Longpré J. M., Apel E. D., Lewis R. M., Wang L. W., Sanes J. R., Leduc R., Apte S. S. (2004) J. Biol. Chem. 279, 35159–35175 [DOI] [PubMed] [Google Scholar]
- 52.Rodriguez-Manzaneque J. C., Milchanowski A. B., Dufour E. K., Leduc R., Iruela-Arispe M. L. (2000) J. Biol. Chem. 275, 33471–33479 [DOI] [PubMed] [Google Scholar]
- 53.Wang P., Tortorella M., England K., Malfait A. M., Thomas G., Arner E. C., Pei D. (2004) J. Biol. Chem. 279, 15434–15440 [DOI] [PubMed] [Google Scholar]
- 54.Majerus E. M., Zheng X., Tuley E. A., Sadler J. E. (2003) J. Biol. Chem. 278, 46643–46648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Somerville R. P., Jungers K. A., Apte S. S. (2004) J. Biol. Chem. 279, 51208–51217 [DOI] [PubMed] [Google Scholar]
- 56.Koo B. H., Longpré J. M., Somerville R. P., Alexander J. P., Leduc R., Apte S. S. (2006) J. Biol. Chem. 281, 12485–12494 [DOI] [PubMed] [Google Scholar]
- 57.Ihara S., Nishiwaki K. (2008) FEBS J 275, 4296–4305 [DOI] [PubMed] [Google Scholar]
- 58.Majerus E. M., Anderson P. J., Sadler J. E. (2005) J. Biol. Chem. 280, 21773–21778 [DOI] [PubMed] [Google Scholar]
- 59.Somerville R. P., Longpre J. M., Jungers K. A., Engle J. M., Ross M., Evanko S., Wight T. N., Leduc R., Apte S. S. (2003) J. Biol. Chem. 278, 9503–9513 [DOI] [PubMed] [Google Scholar]
- 60.Kuno K., Matsushima K. (1998) J. Biol. Chem. 273, 13912–13917 [DOI] [PubMed] [Google Scholar]
- 61.Kashiwagi M., Enghild J. J., Gendron C., Hughes C., Caterson B., Itoh Y., Nagase H. (2004) J. Biol. Chem. 279, 10109–10119 [DOI] [PubMed] [Google Scholar]
- 62.Tortorella M., Pratta M., Liu R. Q., Abbaszade I., Ross H., Burn T., Arner E. (2000) J. Biol. Chem. 275, 25791–25797 [DOI] [PubMed] [Google Scholar]
- 63.Colige A., Ruggiero F., Vandenberghe I., Dubail J., Kesteloot F., Van Beeumen J., Beschin A., Brys L., Lapière C. M., Nusgens B. (2005) J. Biol. Chem. 280, 34397–34408 [DOI] [PubMed] [Google Scholar]
- 64.Flannery C. R., Zeng W., Corcoran C., Collins-Racie L. A., Chockalingam P. S., Hebert T., Mackie S. A., McDonagh T., Crawford T. K., Tomkinson K. N., LaVallie E. R., Morris E. A. (2002) J. Biol. Chem. 277, 42775–42780 [DOI] [PubMed] [Google Scholar]
- 65.Li Z., Nardi M. A., Li Y. S., Zhang W., Pan R., Dang S., Yee H., Quartermain D., Jonas S., Karpatkin S. (2009) Blood 113, 6051–6060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gao G., Plaas A., Thompson V. P., Jin S., Zuo F., Sandy J. D. (2004) J. Biol. Chem. 279, 10042–10051 [DOI] [PubMed] [Google Scholar]
- 67.Rodríguez-Manzaneque J. C., Carpizo D., Plaza-Calonge Mdel C., Torres-Collado A. X., Thai S. N., Simons M., Horowitz A., Iruela-Arispe M. L. (2009) Int. J. Biochem. Cell Biol. 41, 800–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu Y. J., Xu Y., Yu Q. (2006) Oncogene 25, 2452–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee N. V., Rodriguez-Manzaneque J. C., Thai S. N., Twal W. O., Luque A., Lyons K. M., Argraves W. S., Iruela-Arispe M. L. (2005) J. Biol. Chem. 280, 34796–34804 [DOI] [PubMed] [Google Scholar]
- 70.Hashimoto G., Shimoda M., Okada Y. (2004) J. Biol. Chem. 279, 32483–32491 [DOI] [PubMed] [Google Scholar]
- 71.Tuderman L., Kivirikko K. I., Prockop D. J. (1978) Biochemistry 17, 2948–2954 [DOI] [PubMed] [Google Scholar]
- 72.Dong J. F., Moake J. L., Bernardo A., Fujikawa K., Ball C., Nolasco L., López J. A., Cruz M. A. (2003) J. Biol. Chem. 278, 29633–29639 [DOI] [PubMed] [Google Scholar]
- 73.Hirohata S., Wang L. W., Miyagi M., Yan L., Seldin M. F., Keene D. R., Crabb J. W., Apte S. S. (2002) J. Biol. Chem. 277, 12182–12189 [DOI] [PubMed] [Google Scholar]
- 74.Kramerova I. A., Kawaguchi N., Fessler L. I., Nelson R. E., Chen Y., Kramerov A. A., Kusche-Gullberg M., Kramer J. M., Ackley B. D., Sieron A. L., Prockop D. J., Fessler J. H. (2000) Development 127, 5475–5485 [DOI] [PubMed] [Google Scholar]
- 75.Kawano T., Zheng H., Merz D. C., Kohara Y., Tamai K. K., Nishiwaki K., Culotti J. G. (2009) Development 136, 1433–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Le Goff C., Morice-Picard F., Dagoneau N., Wang L. W., Perrot C., Crow Y. J., Bauer F., Flori E., Prost-Squarcioni C., Krakow D., Ge G., Greenspan D. S., Bonnet D., Le Merrer M., Munnich A., Apte S. S., Cormier-Daire V. (2008) Nat. Genet. 40, 1119–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ahram D., Sato T. S., Kohilan A., Tayeh M., Chen S., Leal S., Al-Salem M., El-Shanti H. (2009) Am. J. Hum. Genet. 84, 274–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kuno K., Kanada N., Nakashima E., Fujiki F., Ichimura F., Matsushima K. (1997) J. Biol. Chem. 272, 556–562 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.