Abstract
GLI2 (GLI-Kruppel family member 2), a zinc finger transcription factor that mediates Hedgehog signaling, is implicated in the progression of an ever-growing number of human malignancies, including prostate and pancreatic cancer, as well as basal cell carcinoma of the skin. Its expression is up-regulated by transforming growth factor-β (TGF-β) in a variety of cell types, both normal and transformed. We report herein that TGF-β-driven GLI2 expression is transcriptional and does not result from stabilization of GLI2 transcripts. We describe the characterization of the 5′-flanking sequence of human GLI2 mRNA, the identification of a transcription start site, the cloning of ∼1,600 bp of the regulatory promoter region and the identification and functional analysis of a TGF-β-responsive region mapped to a 91-bp sequence between nucleotides −119 and −29 of the promoter. This region harbors SMAD and lymphoid enhancer factor/T cell factor binding sites that allow functional cooperation between SMAD3 and β-catenin, recruited to the promoter in response to TGF-β to drive GLI2 gene transcription.
Introduction
During embryonic development, several key signaling pathways such as Hedgehog (Hh)3, Wnt, and transforming growth factor-β (TGF-β) govern fundamental cell fate decisions that regulate proliferation and differentiation in a time- and position-dependent fashion. Deregulation of these pathways contributes to the onset or to the development of tumors (1–4). Constitutive activation of Hh signaling has been implicated in the growth of several human malignancies ranging from semimalignant tumors of the skin to highly aggressive cancers of the brain, pancreas, and prostate (5, 6). Cellular responses to Hh are initiated by ligand binding to the tumor suppressor transmembrane receptor PTCH-1 (Patched-1). This interaction releases the inhibitory effect of PTCH-1 on the seven-transmembrane signaling protein Smoothened (SMOH), thus initiating the Hh intracellular cascade, which ultimately leads to the activation and nuclear translocation of zinc finger transcription factors of the GLI family (7). While Hh signaling favors the stabilization of the GLI2 protein by inhibiting its proteasomal degradation, it also induces the expression of GLI1 and PTCH-1 in a GLI2-dependent manner (8).
There is broad evidence for a functional implication of GLI2 in the development of solid tumors. For example, GLI2 knockdown with specific small hairpin RNA or antisense oligonucleotides in prostate cancer cells reduces anchorage-independent colony formation, delays tumor xenograft growth in vivo, and enhances paclitaxel chemosensitivity (9, 10). Similarly, GLI2-specific antisense oligonucleotides inhibit the proliferation of hepatocellular carcinoma cell lines, through the regulation of genes implicated in cell cycle and apoptosis (11). Moreover, in a mouse tumor allograft model, Gli2 silencing in epithelial cells that constitutively express an active form of GLI2 (GLI2ΔN2) has unambiguously demonstrated the important role played by GLI2 in preventing apoptosis and promoting tumor microvascularization (12). Together, these data support the notion that suppression of GLI2 expression may represent a valuable therapeutic option for the treatment of several cancers (13).
Although GLI activation may result from Hh ligand- or Hh receptor-induced signaling, recent evidence has shown the possible implication of noncanonical, Hh-independent signaling pathways to regulate GLI expression and/or activity (14). Thus, the identification of pathways leading to GLI activation is critical for adequate therapeutic targeting. In this context, we have previously identified TGF-β as a potent and ubiquitous inducer of GLI1 and GLI2 expression in both normal and transformed cells (15).
TGF-βs encompass a large family of secreted proteins. To trigger their biological effects, TGF-βs bind to type I and II serine/threonine kinase receptor complexes. Ligand-dependent receptor activation leads to the recruitment and phosphorylation of intracellular mediators of TGF-β signal transduction, namely the SMAD proteins (16, 17). In most cell types, TGF-β1 induces the activation of SMAD2 and SMAD3 and their association with SMAD4. These complexes relay signals from the cell membrane to the nucleus where SMAD3·SMAD4 complexes bind to specific cis-elements, either alone or in cooperation with other transcription factors and co-activators that regulate the transcription of TGF-β target genes (18, 19).
In this report, we describe the cloning and functional characterization of the human GLI2 promoter. We also identify a 91-bp TGF-β responsive region whereby TGF-β recruits SMAD3 and β-catenin to induce GLI2 transcription.
EXPERIMENTAL PROCEDURES
Cell Cultures and Reagents
HaCaT immortalized human keratinocytes and HepG2 hepatocarcinoma cell lines were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum and antibiotics (Invitrogen, Cergy-Pontoise, France). When indicated, cells were serum-starved for 16 h and treated with 5 ng/ml human recombinant TGF-β1 (R&D Systems, Lille, France). Control cultures received corresponding TGF-β vehicle buffer (4 mm HCl, 0.1% bovine serum albumin). Cycloheximide and actinomycin D were obtained from Euromedex (Strasbourg, France). Small interfering RNAs (siRNAs) were purchased from Ambion/Applied Biosystems (Courtabœuf, France).
Multiplex and Real-time PCR
Total RNA was prepared using a column-based commercial kit (Macherey-Nagel, Hoerdt, France). Genomic DNA-free RNA was then converted into cDNA using the ThermoScript RT-PCR system (Invitrogen). cDNAs were used for multiplex PCR according to the manufacturer's recommendations (Qiagen, Courtabœuf, France), real-time PCR using a Power SYBR Green mixture on an AB7300 apparatus (Applied Biosystems), or for 5′-rapid amplification of cDNA ends (5′-RACE). PCR primer sequences and specific PCR conditions are available upon request.
5′-RACE
5′-RACE was performed using a commercial kit (Invitrogen). cDNA synthesis was performed using total RNA from HaCaT cells primed with the following gene-specific primer, 5′-AGAGATCATGGAGAGCGTGG located in exon 4 of the GLI2 gene. PCR of the dC-tailed cDNA was performed with the AAP primer included in the kit, together with a primer corresponding to a sequence located in exon 2 of human GLI2: 5′-ATCTCAGCCGCTCATCGTC. Finally, nested PCR was performed using the AUAP primer of the kit and the exon 1 primer: 5′-TTGGGGAAGCTTCTGACCTTGCTCTTTGATGTG, which contains a HindIII site (underlined). The indicated exonic information refers to the published NM_005270.3 GenBankTM sequence. The PCR products containing the unknown 5′ upstream region of GLI2 transcripts were digested with MluI/HindIII and inserted into the MluI/HindIII sites of pGL3-Basic for sequencing.
Reporter Gene Constructs
Genomic DNA was extracted from HaCaT cells using a commercial kit (Qiagen). Various fragments of the human GLI2 promoter were amplified from genomic DNA, using BglII-containing forward primers and a HindIII-containing reverse primer. PCR products were subcloned into pGL3-Basic vector (Promega, Charbonnieres, France) and sequenced. The sequence of the constructs is identical to the sequence of human chromosome 2, GenbankTM NW_001838848.1. Site-directed mutagenesis to inactivate the SMAD and lymphoid enhancer factor/T cell factor (TCF/LEF) binding sites, respectively, at positions −33 and −60 of the promoter was carried out according to the DpnI-based QuikChange site-directed mutagenesis methodology (Stratagene, La Jolla, CA) with the following primers: mutant SMAD site (mS) forward, 5′-GTGGCGGGAGGGTATGTGGGATTTCAGGTTTCAGG; mS reverse, 5′-CCTGAAACCTGAAATCCCACATACCCTCCCGCCAC; mutant TCF/LEF site (mT) forward, 5′-CTCGTTAGAGGAGGCCGAAGAAACCAGGTGGCGGG; mT reverse, 5′-CCCGCCACCTGGTTTCTTCGGCCTCCTCTAACGAG. Putative binding sites are shown in bold, and mutated bases are underlined. After amplification, promoter fragments were subcloned into the BglII/HindIII sites of pGL3-Basic. TS constructs containing 1 (TS), 2 (TS2), or 4 (TS4) concatamerized copies of the −66/−25 region of the human GLI2 promoter, were obtained by inserting the following double-stranded oligonucleotides into the pGL3-Basic XhoI site (italic): 5′-TCGAGGAGGAGTTCAAAGAAACCAGGTGGCGGGAGGGTGTCTGGGATC and its complementary strand: 5′-TCGAGATCCCAGACACCCTCCCGCCACCTGGTTTCTTTGAACTCCTCC. TmS4, in which the SMAD binding element (bold) is mutated to ATGT (bold underlined), was obtained by subcloning four copies of the following oligonucleotide: 5′-TCGAGGAGGAGTTCAAAGAAACCAGGTGGCGGGAGGGTATGTGGGATC and its complementary strand: 5′-TCGAGATCCCACATACCCTCCCGCCACCTGGTTTCTTTGAACTCCTCC.
Transfections and Luciferase Reporter Assays
24 h prior to transfection, HaCaT and HepG2 cells were plated in 24-well plates and transfected with siRNA (Ambion/Applied Biosystems) using HiPerfectTM (Qiagen) when indicated. The following day, cells at ∼40% confluency were transfected with 200 ng of firefly luciferase promoter reporter construct together with 15 ng of expression vector (when necessary) using JetPEI (PolyPlus Transfection, Strasbourg, France). 40 ng of phRL-MLP Renilla luciferase expression vector was co-transfected to estimate transfection efficiencies. phRL-MLP was generated by cloning the adenovirus major late promoter (MLP) upstream of the Renilla luciferase gene as a BgIII/HindIII insert within phRL-null (Promega). For each protocol, at least three independent experiments were performed. Sixteen h post-transfection, cells were serum-starved in 0.5% serum-containing medium and treated 8 h later with TGF-β1 for 16 h. Cells were lysed, and luciferase activity was determined with the Dual-Luciferase reporter assay system (Promega) using a Fluoroskan Ascent FL (Thermo LabSystems). In each experiment, promoter activity is expressed as the firefly/Renilla ratio relative to internal control conditions. Expression vectors for GLI2, SMAD3, TCF4, and constitutively active β-catenin (B23) have been described previously (20–23).
Western Analyses
Cells were lysed in 1% SDS lysis buffer (10 mm Tris, pH 7.4, 1% SDS, and 1 mm sodium orthovanadate). Equal amounts of protein were boiled with Laemmli buffer and subjected to 10% SDS-PAGE. Proteins were then transferred onto nitrocellulose membranes (Amersham Biosciences). Membranes were blocked with 5% nonfat milk in phosphate-buffered saline containing 0.1% Tween 20 for 1 h at room temperature and incubated overnight at 4 °C with the primary antibody. After several washes, membranes were incubated in blocking buffer with a secondary antibody coupled to horseradish peroxidase (Santa Cruz Biotechnology) for 2 h at room temperature. Antigen·antibody complexes were detected using ECLplus (Amersham Biosciences/GE Healthcare, Velizy, France) and revealed with a Storm PhosphorImager (Amersham Biosciences/GE Healthcare). Anti-Myc, anti-SMAD3, and anti-HSP60 were purchased from Santa Cruz Biotechnology, whereas anti-β-catenin was from BD Biosciences. Anti-β-actin was obtained from Sigma-Aldrich.
Chromatin Immunoprecipitation
HaCaT cells were grown to 60–70% confluency on 15-cm plates then serum-starved in 0.5% serum-containing medium for 16 h and stimulated with TGF-β for the indicated time. Chromatin immunoprecipitation was carried out using the ChIP-IT express kit (Active Motif, Rixensart, Belgium). Briefly, 4 μg of enzymatically sheared chromatin were immunoprecipitated using 4 μg of antibody against IgG (Santa Cruz Biotechnology), SMAD3, or β-catenin overnight at 4 °C, then incubated for another 2 h at 4 °C with protein G beads. Precipitated DNA was used for PCR analysis using human GLI2 promoter-specific primers: forward, 5′-GAAGATCTCTGTGACTTTAATGCGGTGTG; reverse, 5′-TTGGGGAAGCTTCACCCTGAAACCTGAAATCCC. Amplimers were visualized by agarose gel electrophoresis.
RESULTS
TGF-β1 Increases GLI2 Expression but Does Not Stabilize Either GLI2 Transcripts or GLI2 Protein
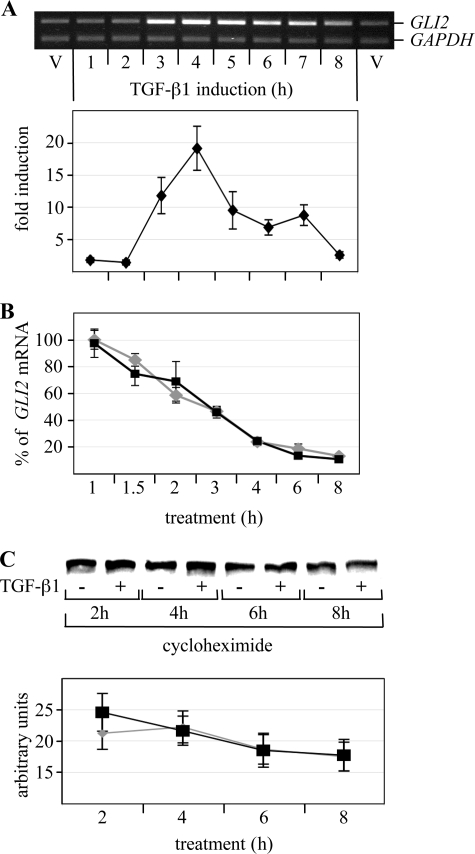
Treatment of human immortalized HaCaT keratinocytes with TGF-β1 led to rapid elevation of GLI2 mRNA, peaking 4 h after TGF-β1 induction and remaining significantly higher than basal expression up to 8 h poststimulation (Fig. 1A). Similar results were obtained in the human hepatocarcinoma cell line HepG2, in which basal levels of GLI2 transcripts are very low (see Fig. 2C). These data are consistent with our initial report (15). To determine whether TGF-β stabilizes GLI2 transcripts, GLI2 mRNA half-life was estimated in HaCaT cells using the transcription inhibitor actinomycin D. As shown in Fig. 1B, TGF-β1 did not modify the rate of GLI2 mRNA decay (half-life: 2.69 ± 0.02 versus 2.67 ± 0.02 h, respectively), suggesting that TGF-β-dependent elevation of GLI2 expression is not due to stabilization of GLI2 transcripts.
FIGURE 1.
TGF-β induced GLI2 expression occurs without stabilization of either GLI2 mRNA or protein. A, subconfluent HaCaT keratinocytes were serum-starved for 16 h and treated with human recombinant TGF-β1 for the indicated time periods. GLI2 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression was measured by multiplex reverse transcription-PCR (upper panel) or quantitative reverse transcription-PCR (lower panel) as described previously (15). Results are the mean ± S.D. of four quantitative PCR results. B, subconfluent HaCaT keratinocytes were serum-starved for 16 h, at which point 1 μg/ml actinomycin D was added to the culture medium to block transcription. 30 min later, cells were incubated in the absence or presence of TGF-β1. RNA was extracted at various time points, and GLI2 mRNA decay was measured by quantitative reverse transcription-PCR. Results are the mean ± S.D. of four quantitative PCR results. C, subconfluent HaCaT keratinocytes were transfected with a Myc-tagged full-length GLI2 expression vector and serum-starved for 16 h, at which point 10 μg/ml cycloheximide was added to the culture medium to block protein neosynthesis. TGF-β1 was added 30 min later. Myc-GLI2 protein content in control and TGF-β1-treated cultures was detected at the indicated time points by Western blotting using an anti-Myc antibody (upper panel). Myc-GLI2 protein decay was analyzed by scanning densitometry (lower panel). Results are the mean ± S.D. of two independent densitometric analyses.
FIGURE 2.
Characterization of the 5′ regulatory region of the human GLI2 gene. A, 5′-RACE was used to identify the transcription start site of human GLI2 transcripts. MW, molecular weight markers. B, human GLI2 gene promoter sequence obtained after PCR amplification of a ∼1,600-bp fragment upstream of the initiator site (Inr) identified by 5-RACE. DPE, downstream core promoter element. Residues in bold, exon I; residues in italic, intron I. C, −1624pHGLI2luc was transfected in HaCaT and HepG2 together with phRL-MLP to estimate transfection efficiency. RNA was extracted from parallel cultures. GLI2 expression was measured by multiplex PCR (right panel). D, a battery of human GLI2 promoter constructs was transfected in parallel into HaCaT and HepG2. The light gray box corresponds to exon 1; the dark box corresponds to intron I (B). Following TGF-β treatment (see under “Experimental Procedures” for experimental conditions), reporter activity was determined. Activity of each construct is expressed as fold activity relative to pGL3-Basic (pGL3b) arbitrarily set to 1. Results are expressed as mean ± S.D. of triplicate samples from one (representative) of three experiments. Fold induction by TGF-β (black bars) above controls (vehicle-treated) (gray bars) is indicated. E, nucleotide sequence of the 91-bp TGF-β responsive unit between nucleotides −119 and −29 relative to the transcription start site.
Hh is known to stabilize the GLI2 protein (24). In an attempt to determine whether TGF-β may also favor GLI2 protein accumulation, HaCaT cells were transfected with a Myc-tagged GLI2 expression vector (20), and the stability of the expressed fusion protein was determined by Western blotting. As shown in Fig. 1C, there was no detectable effect of TGF-β on Myc-GLI2 stability when protein neosynthesis was blocked by cycloheximide. It is thus likely that TGF-β exerts its effect on GLI2 expression primarily at the transcriptional level, although we cannot exclude a stabilization of endogenous GLI2 protein by TGF-β; its low abundance or our detection capability with currently available commercial antibodies did not allow us to determine its half-life (data not shown).
Identification of the 5′-regulatory Region of the Human GLI2 Gene
As a first step toward cloning the human GLI2 gene promoter region, it was necessary to characterize the gene transcription start site(s). For this purpose, 5′-RACE by PCR followed by nested amplification with a primer localized on exon 1 according to the human GLI2 GenBankTM sequence NM_005270.3, was performed using total human RNA from HaCaT cells as a template (Fig. 2A). Sequencing of the ∼320-bp PCR product resulting from the 5′-RACE reaction identified a canonical initiator site (Fig. 2B): TCATTCT, with an adenosine as transcription start site. This initiator is located 85 bp upstream of the described exon 1 from GenBankTM sequence NM_005270.3. Initiator sites are normally associated with a downstream core promoter element (25). Such sequence was found in position +35/+39 of the GLI2 promoter (see Fig. 2B).
We were next able to PCR-amplify a ∼2-kb fragment containing 1,624 bp of the human GLI2 gene upstream of the initiator site (Fig. 2B), identical to a region of chromosome 2 located 5′ of the GLI2 coding sequence (GenbankTM MW_001838848.1). We failed to identify any canonical TATA box near the transcription start site. Yet, other features typical of a promoter were found, such as two potential CAAT boxes and numerous putative cis-acting elements upstream of the initiator element (for details, see supplemental Table 1), as determined using the MatInspectorTM software (Genomatix).
To address the question of whether this 2-kb GLI2 genomic region exhibits transcriptional activity, the fragment was cloned upstream of the luciferase gene into pGL3-Basic to generate −1624pHGLI2luc. Parallel transient cell transfection experiments in HaCaT keratinocytes and HepG2 cells comparing the activity of −1624pHGLI2luc to that of pGL3-Basic identified robust transcriptional activity for this GLI2 genomic fragment in HaCaT cells (3-fold above pGL3-Basic activity), not in HepG2 cells (Fig. 2C, left panel), consistent with the subdetectable expression of GLI2 transcripts in the latter cell type as opposed to HaCaT cells (Fig. 2C, right panel). These data attest for a promoter function for this region of the GLI2 gene.
Delineation of a TGF-β-responsive Region within the Human GLI2 Promoter
A battery of deletion reporter constructs of the human GLI2 promoter was assessed for transcriptional activity in transient cell transfection experiments, in both HaCaT and HepG2 cells, treated or not with TGF-β. Consistent with the data obtained with −1624pHGLI2luc, all shorter constructs had clearly detectable basal transcriptional activity in HaCaT cells (2–3-fold above pGL3-luc activity) but were inactive in HepG2 cells (Fig. 2D). A stimulatory effect of TGF-β was observed with all constructs containing between 119 and 1305 bp of the promoter in both cell types, except for −1624pHGLI2luc, suggesting the existence of a potential repressor element within the −1624/−1305 region of the promoter.
Whereas −119pHGLI2luc was efficiently transactivated in response to TGF-β, the −29-bp construct was not, thus identifying the 91-bp region between nucleotides −119 and −29 as required for activation by TGF-β1 (TGF-β-responsive unit; TβRU). Sequence examination identified putative LEF/TCF (5′-TTCAAAGA) and SMAD3/4 (5′-GTCT) binding elements (TBE and SBE, respectively) within this short fragment of the GLI2 promoter (Fig. 2E).
The Human GLI2 Promoter TGF-β-responsive Unit Drives SMAD and β-Catenin/TCF4 Responses
We initially demonstrated that siRNA-mediated SMAD3 knockdown reduces TGF-β1-driven GLI2 expression (15). In accordance with these findings, SMAD3 knockdown effectively prevented TGF-β1-induced GLI2 expression and −119pHGLI2luc transactivation in HaCaT (Fig. 3A) and HepG2 cells (data not shown). Conversely, overexpression of SMAD3 activated the proximal promoter, increased GLI2 expression, and enhanced TGF-β effect in both HaCaT and HepG2 cells (Fig. 3B).
FIGURE 3.
SMAD3 and TCF4/β-catenin contribute to GLI2 promoter transactivation by TGF-β. A, SMAD3 expression was reduced in HaCaT by specific siRNA knockdown. Efficacy of the knockdown was verified by Western blotting with an anti-SMAD3 antibody (left panel). An HSP60 antibody was used as control. GLI2 expression was determined by quantitative PCR (center panel). Right panel, HaCaT cells were cotransfected with control or SMAD3 siRNA together with −119pHGLI2luc and incubated with or without TGF-β for 16 h. For both RNA and promoter data, results are expressed as fold induction by TGF-β above vehicle-treated cultures. B, HaCaT (left panel) and HepG2 (center panel) cells were transfected with −119pHGLI2luc in the presence of either empty pcDNA3.1 or SMAD3 expression vector and treated with TGF-β. SMAD3 expression was verified by Western blotting (upper panels). An anti-HSP60 antibody was used as control. Right panel, HepG2 cells were transfected with either pcDNA3.1 or a SMAD3 expression vector. 24 h later, cells were serum-starved for 16 h and incubated with or without TGF-β. RNA was extracted 7 h later, and GLI2 expression was measured by quantitative PCR. Results are expressed as relative activity compared with vehicle-treated pcDNA3.1-transfected cells. C, HepG2 (left panel) cells were transfected with −119pHGLI2luc together with either TCF4 or SMAD3 expression vectors either alone or in combination, whereas HaCaT cells (right panel) were transfected with −119pHGLI2luc together with expression vectors for either SMAD3 or a constitutively active form of β-catenin, alone or in combination, and then treated with TGF-β. Luciferase activity was determined 16 h later. D, β-catenin expression was knocked down with specific siRNAs. Efficacy was verified by Western blotting (left panel). HepG2 cells were transfected with −119pHGLI2luc together with control or β-catenin siRNAs and treated with TGF-β.
Gene transcription in response to β-catenin requires the association of nuclear β-catenin to transcription factors of the LEF/TCF family bound to cognate cis-elements (3). It has been shown previously that TGF-β1 induces the accumulation of both cytoplasmic and nuclear β-catenin in HaCaT cells (26). Functional cooperation between SMAD and LEF/TCF transcription factors has been described to mediate TGF-β signaling and interactions with the Wnt pathway (27–31). We thus investigated a possible cooperation between β-catenin/TCF4 and SMAD3 to regulate GLI2 promoter activity. Firstly, we observed that coexpression of TCF4 with SMAD3 in HepG2 cells, in which the β-catenin pathway is constitutively activated (32), resulted in synergistic transactivation of −119pHGLI2luc, indicative of efficient cooperation of these transcription factors to transactivate the GLI2 promoter (Fig. 3C, left panel). TCF4 was also found to enhance TGF-β-driven gene transcription. Secondly, TCF4 was ectopically expressed in HaCaT cells, without or with a constitutively active form of β-catenin, B23, known to interact with and favor the transcriptional activity of LEF/TCF transcription factors. TCF4 alone had a slight repressor effect on −119pHGLI2luc activity. Such a result is somewhat expected, given the low levels of active β-catenin in HaCaT cells as compared with HepG2 cells, which probably allows the formation of inhibitory TCF4 homodimers (3). Consistent with this hypothesis, overexpression of active β-catenin increased the basal activity and amplified the TGF-β response of −119pHGLI2luc and reversed the effect of TCF4 to that of a transcriptional activator (Fig. 3C, right panel). This experimental approach somewhat mimics the endogenous situation in HepG2 cells (that contain constitutively active β-catenin) and indicates a cooperation of SMAD3 with TCF4/β-catenin to drive GLI2 transcription.
To validate the implication of β-catenin in TGF-β-driven GLI2 promoter transactivation in HepG2 cells, endogenous β-catenin expression was knocked down with specific siRNAs. Western analysis (Fig. 3D, left panel) revealed that siRNA treatment efficiently reduced the expression of both short (constitutively active) and long (wild-type) forms of β-catenin protein found in HepG2 cells (32). Subsequently, we found that β-catenin siRNA largely abolished TGF-β effect on −119pHGLI2luc activity (Fig. 3D, right panel). These findings establish that the β-catenin pathway is involved in the regulation of GLI2 promoter activation by TGF-β in HepG2 cells.
Functional Analysis of the Putative SMAD3/4 and LEF/TCF Cis-Elements Identified within the TGF-β-responsive Unit of the Human GLI2 Promoter
To determine whether the putative TBE and SBE cooperate and are sufficient to mediate a TGF-β-dependent transcriptional response, a short 42-bp sequence spanning both SBE and TBE sites was concatamerized upstream of a minimal promoter in MLP-luc, itself unresponsive to TGF-β (33). As shown in Fig. 4A (left panel), TGF-β induced a robust transcriptional response of these constructs in HepG2 cells, proportional to the number of sequences inserted in the reporter vectors. Mutation of the SBE site within each monomer of the 4× construct (TmS4) abolished TGF-β responsiveness (Fig. 4A, right panel), indicating (a), that the SBE is critical for TGF-β response and (b), that the LEF/TCF site is not sufficient to drive TGF-β responsiveness in the context of a heterologous promoter.
FIGURE 4.
Contribution of the SBE and TBE sites within the HGLI2 promoter TβRU for TGF-β responsiveness. A, left panel: HepG2 cells were transfected with TS1luc, TS2luc, and TS4luc, respectively, containing one, two, or four copies of the −66/−25 GLI2 promoter fragment and then treated with TGF-β. Right panel: HepG2 cells were transfected in parallel with TS4luc and TmS4luc and then treated with TGF-β. Luciferase activity measured 16 h after TGF-β addition is expressed relative to empty MLP-luc activity, arbitrarily set to 1. B, HaCaT (left panel) and HepG2 (right panel) cells were transfected in parallel with either −119/+104pHGLI2luc (WT), or the corresponding mS, mT, or mTmS mutants, and then treated with TGF-β. Luciferase activity measured 16 h after TGF-β addition is expressed relative to WT −119/+104pHGLI2luc activity, arbitrarily set to 1. C, HaCaT cells were cotransfected in parallel with either WT −119/+104pHGLI2luc or the corresponding mS and mT mutants, together with either empty pcDNA3.1 (pcDNA), SMAD3 (S3), or constitutively active β-catenin (B23) expression vectors and then treated with TGF-β. Luciferase activity measured 16 h after TGF-β addition is expressed relative to −119/+104pHGLI2luc activity cotransfected with pcDNA3.1, arbitrarily set to 1. Schematic representations of the SBE and TBE mutation status within each GLI2 promoter construct are shown as insets. D, HepG2 cells were cotransfected in parallel with either WT −119/+104pHGLI2luc or the corresponding mS and mT mutants, together with either empty pcDNA3.1, SMAD3, or TCF4 (T4) expression vectors and then treated with TGF-β. Luciferase activity measured 16 h after TGF-β addition is expressed relative to −119/+104pHGLI2luc activity cotransfected with pcDNA3.1, arbitrarily set to 1. E, digested chromatin from control or TGF-β-treated (1 and 3 h) HaCaT cells was immuno-precipitated with either anti-IgG, anti-SMAD3, or anti-β-catenin antibodies, as indicated. Results show representative PCR reactions with primers spanning the TGF-β responsive unit. Input lanes are PCR reactions using chromatin as template, without immunoprecipitation (IP).
The dramatic induction of −119pHGLI2luc upon TCF4 expression led us to analyze sequences downstream of the TβRU. A putative LEF/TCF site (5′-ATCAAAGA) was found at position +236 within exon I. 3′-End deletion of −119pHGLI2luc was performed to generate −119/+104pHGLI2luc that contains no other SBE and TBE besides those found in the TβRU. Regulation of −119/+104pHGLI2luc by TGF-β and/or SMAD3 overexpression was similar to that of −119pHGLI2luc whereas its response to TCF4 and/or β-catenin overexpression was reduced (see below and compare with Fig. 3C).
To determine the respective contribution of the putative SBE (5′-GTCT) and TBE (5′-TTCAAAGA) sites of the TβRU for GLI2 promoter responsiveness to TGF-β, GLI2 promoter/reporter constructs were generated in which the SBE or the TBE were mutated, either alone (mS and mT, respectively) or in combination (mTmS) within −119/+104pHGLI2luc. Both mutations efficiently prevent binding of their cognate transcription factors, as demonstrated using in vitro-translated TCF4 and SMAD3 protein in electrophoretic mobility shift assay experiments with corresponding labeled oligonucleotides as probes (data not shown). In both HaCaT and HepG2 cells, wild-type −119/+104pHGLI2luc responded to TGF-β with a 3–8-fold transactivation (Fig. 4B). Mutation of the SBE abolished TGF-β responsiveness, as shown by the lack of transactivation of the mS and mTmS constructs. Thus, the SBE site is critical for TGF-β responsiveness of the GLI2 promoter. On the other hand, TGF-β-driven promoter transactivation was only partially affected by mutation of the TBE. Thus, integrity of both SBE and TBE is required for full TGF-β response.
The functionality of the SBE and TBE sites was next examined by overexpressing SMAD3 and/or β-catenin in HaCaT keratinocytes, in the absence or presence of exogenous TGF-β stimulation. Although SMAD3 and β-catenin both transactivated −119/+104pHGLI2luc and enhanced TGF-β response, mutation of the SBE fully abolished SMAD3- and TGF-β-driven transactivation. Despite the mS mutation, β-catenin still exerted its stimulatory effect on basal promoter activity but did not rescue TGF-β responsiveness. Mutation of the TBE site (mT construct) only slightly affected SMAD3 and TGF-β responsiveness but prevented both promoter transactivation by β-catenin and cooperation of β-catenin with SMAD3 to enhance the TGF-β response. Thus, integrity of the TBE, although not essential for SMAD3/TGF-β-driven promoter activation, is required for full cooperation of overexpressed SMAD3 and β-catenin.
We then used a similar experimental approach in HepG2 cells. TCF4 was found to transactivate the WT promoter and to cooperate with SMAD3 to enhance GLI2 promoter activity and TGF-β-driven transactivation. Mutation of the SBE essentially abolished GLI2 promoter by overexpressed TCF4 and blocked SMAD3 and TGF-β effects. Interestingly, in the context of a promoter in which the TBE is inactivated (mT construct), TCF4 still drove promoter activity and enhanced TGF-β-induced transcription. Thus, in a cellular context with constitutively active β-catenin, transactivation of the GLI2 promoter by TCF4 requires a functional SBE site, suggesting a direct cooperation of SMAD3 and TCF4 via the SBE site. In this context, integrity of the nearby TBE potentiates TGF-β response by allowing a stronger TCF4 effect.
We finally examined the possible recruitment of SMAD3 and β-catenin to the TβRU by chromatin immunoprecipitation in HaCaT keratinocytes. As shown in Fig. 4C, neither SMAD3 nor β-catenin bound the human GLI2 promoter TβRU in the absence of TGF-β stimulation. However, a rapid and sustained recruitment of SMAD3 was observed as early as 1 h following TGF-β addition to the culture medium and was still present after 3 h. At this later time point, β-catenin was also recruited to the TβRU in response to TGF-β. Taken together, these results demonstrate that both SMAD3 and β-catenin are recruited to the GLI2 promoter in response to TGF-β.
DISCUSSION
In this report, we have addressed three essential issues pertaining to the transcriptional mechanisms controlling the human GLI2 gene. First, we have identified the transcription start site. Second, we have characterized the functional GLI2 core promoter. Third, we have demonstrated the cooperation between SMAD3 and LEF/TCF transcription factors as well as β-catenin in TGF-β-induced up-regulation of GLI2 promoter activity.
Abnormal activation of the Hh pathway has been implicated in a variety of cancers (6), and targeted anti-cancer therapy focused on Hh signaling is trying to address unmet needs for efficient cancer treatment. In this context, small molecule Hh antagonists have been developed that either block SMOH function or interfere with GLI binding to DNA (13). However, it becomes more and more clear that noncanonical signaling events are often responsible for the expression of Hh mediators of the GLI family (15, 34, 35), indicating that therapeutic intervention against deleterious GLI expression may also be possible by targeting upstream inducers such as TGF-β or other pathways. In this context, we previously demonstrated that pancreatic adenocarcinoma cell lines resistant to the Hh inhibitor cyclopamine, are growth-inhibited by a small molecule inhibitor of TGF-β receptor 1 (15). In the context of breast cancer, it was recently found that progression from ductal carcinoma in situ to invasive carcinoma implicates TGF-β signaling and elevated GLI2 expression (35). Consistent with our initial observations (15), the authors found that TGF-β elevates GLI2 levels and GLI-dependent transcription, influencing myoepithelial cell differentiation and progression to invasion.
Lineage-independent activation of the Wnt/β-catenin pathway is often found during cancer progression (36). Remarkably, the Wnt/β-catenin pathway has been shown to enhance GLI1-dependent transcription in stomach, lung, and colon cancer cells (37). Also, various Wnt genes are Hh/GLI targets and mediate some of GLI functions during embryogenesis (38), suggesting intricate regulatory mechanisms between these pathways. The β-catenin pathway is constitutively active in the HepG2 cell line due to heterozygote mutation in the CTNNB1 gene leading to a large deletion in the protein that favors its nuclear accumulation and transcriptional activity (32). Why HepG2 cells do not express GLI2 unless stimulated by TGF-β remains an enigma, but this suggests that the constitutively active β-catenin pathway is not sufficient per se to allow GLI2 transcription. This may be either due to the existence of antagonistic repressory pathways, yet to be identified, or to the absolute requirement of transcriptional partner(s) such as activated SMADs for β-catenin to activate GLI2 expression.
Functional interactions between the SMAD and β-catenin machineries have been described. Their net outcome and mechanisms leading to target gene expression involves various mechanistic possibilities, as both pathways may function as cofactor for the other. It was initially found, using overexpressed proteins in 293T cells, that SMAD4 physically interacts with β-catenin and that LEF1 interacts with both SMAD4 and β-catenin to form a complex that activates the Xtwn gene, a Wnt/β-catenin target, via binding to the promoter region (31). A parallel report also identified SMAD2 and SMAD3 as a LEF1/β-catenin interactor in COS-1 cells, allowing synergistic Xtwn promoter activation by TGF-β and Wnt signaling, via distinct SMAD and LEF/TCF binding sites (27). In glutathione S-transferase pulldown studies, Lei et al. (28) were able to identify the interaction of both SMAD3 and SMAD4 to heteromeric TCF4·β-catenin complexes. SMADs and TCF4·β-catenin overexpression synergistically activated the human gastrin promoter in gastric adenocarcinoma cells. We showed that SMAD·TCF·β-catenin complexes were also able to regulate transcription at isolated SMAD or TCF sites. In the latter two studies, the cooperation of SMADs with LEF/TCF was shown to implicate the transcriptional coactivator p300/CREB-binding protein. It was recently demonstrated that the integrity of two TCF/LEF binding sites are necessary for vascular endothelial growth factor A regulation by TGF-β and involves down-regulation of glycogen synthase kinase 3β-dependent phosphorylation of β-catenin in response to TGF-β and recruitment of unphosphorylated β-catenin to TCF4·SMAD2/3 complexes (29). Our data, which demonstrates that TCF4 cooperates with SMADs to transactivate the GLI2 promoter in HepG2 cells even when the TBE is mutated, support the notion that β-catenin and SMADs act as cofactors to regulate SMAD-dependent gene transcription. The presence of a LEF/TCF site near the SBE further accentuates the cooperation of these transcription factors.
It is likely that signals leading to β-catenin activation such as those initiated by Wnts will cooperate with TGF-β to up-regulate GLI2 expression, thereby providing de novo substrate for enhanced Hh signaling. Such signal convergence leading to GLI2 expression is likely to occur in a number of cancers independent of lineage. In this context, our work represents critical groundwork for elucidating the mechanisms regulating GLI2 transcription, an important step to elaborate adequate targeted intervention against upstream signals responsible for GLI2 expression.
Supplementary Material
Acknowledgments
We thank Dr. A. Dlugosz (University of Michigan, Ann Arbor, MI) for providing reagents critical for experiments and Dr. D. Bernuau (INSERM U697) for helpful discussions.
This work was supported by the Donation Henriette et Emile Goutière (to A. M.), Cancéropôle Ile-de-France, and INSERM.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1.
Recipient of a postdoctoral fellowship from Institut National du Cancer and Région Ile-de-France.
- Hh
- Hedgehog
- siRNA
- small interfering RNA
- 5′-RACE
- 5′-rapid amplification of cDNA ends
- TβRU
- TGF-β-responsive unit
- WT
- wild-type
- CREB
- cAMP-responsive element-binding protein
- LEF/TCF
- lympoid enhancer factor/T cell factor
- TBE
- LEF/TCF binding element
- SBE
- SMAD3/4 binding element
- MLP
- major late promoter.
REFERENCES
- 1.Evangelista M., Tian H., de Sauvage F. J. (2006) Clin Cancer Res. 12, 5924–5928 [DOI] [PubMed] [Google Scholar]
- 2.Kelleher F. C., Fennelly D., Rafferty M. (2006) Acta Oncol. 45, 375–388 [DOI] [PubMed] [Google Scholar]
- 3.Hoppler S., Kavanagh C. L. (2007) J. Cell Sci. 120, 385–393 [DOI] [PubMed] [Google Scholar]
- 4.Massague J. (2008) Cell 134, 215–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pasca di Magliano M., Hebrok M. (2003) Nat. Rev. Cancer 3, 903–911 [DOI] [PubMed] [Google Scholar]
- 6.Ruiz i Altaba A., Sanchez P., Dahmane N. (2002) Nat. Rev. Cancer 2, 361–372 [DOI] [PubMed] [Google Scholar]
- 7.Varjosalo M., Taipale J. (2008) Genes Dev. 22, 2454–2472 [DOI] [PubMed] [Google Scholar]
- 8.Kasper M., Regl G., Frischauf A. M., Aberger F. (2006) Eur. J. Cancer 42, 437–445 [DOI] [PubMed] [Google Scholar]
- 9.Thiyagarajan S., Bhatia N., Reagan-Shaw S., Cozma D., Thomas-Tikhonenko A., Ahmad N., Spiegelman V. S. (2007) Cancer Res. 67, 10642–10646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Narita S., So A., Ettinger S., Hayashi N., Muramaki M., Fazli L., Kim Y., Gleave M. E. (2008) Clin. Cancer Res. 14, 5769–5777 [DOI] [PubMed] [Google Scholar]
- 11.Kim Y., Yoon J. W., Xiao X., Dean N. M., Monia B. P., Marcusson E. G. (2007) Cancer Res. 67, 3583–3593 [DOI] [PubMed] [Google Scholar]
- 12.Ji J., Kump E., Wernli M., Erb P. (2008) Int. J. Cancer 122, 50–56 [DOI] [PubMed] [Google Scholar]
- 13.Rubin L. L., de Sauvage F. J. (2006) Nat. Rev. Drug Discov. 5, 1026–1033 [DOI] [PubMed] [Google Scholar]
- 14.Lauth M., Toftgard R. (2007) Cell Cycle 6, 2458–2463 [DOI] [PubMed] [Google Scholar]
- 15.Dennler S., Andre J., Alexaki I., Li A., Magnaldo T., ten Dijke P., Wang X. J., Verrecchia F., Mauviel A. (2007) Cancer Res. 67, 6981–6986 [DOI] [PubMed] [Google Scholar]
- 16.Massague J., Seoane J., Wotton D. (2005) Genes Dev. 19, 2783–2810 [DOI] [PubMed] [Google Scholar]
- 17.Schmierer B., Hill C. S. (2007) Nat. Rev. Mol. Cell Biol. 8, 970–982 [DOI] [PubMed] [Google Scholar]
- 18.Javelaud D., Mauviel A. (2004) Int. J. Biochem. Cell Biol. 36, 1161–1165 [DOI] [PubMed] [Google Scholar]
- 19.ten Dijke P., Hill C. S. (2004) Trends Biochem. Sci. 29, 265–273 [DOI] [PubMed] [Google Scholar]
- 20.Roessler E., Ermilov A. N., Grange D. K., Wang A., Grachtchouk M., Dlugosz A. A., Muenke M. (2005) Hum. Mol. Genet. 14, 2181–2188 [DOI] [PubMed] [Google Scholar]
- 21.Dennler S., Huet S., Gauthier J. M. (1999) Oncogene 18, 1643–1648 [DOI] [PubMed] [Google Scholar]
- 22.Roose J., Huls G., van Beest M., Moerer P., van der Horn K., Goldschmeding R., Logtenberg T., Clevers H. (1999) Science 285, 1923–1926 [DOI] [PubMed] [Google Scholar]
- 23.Wei Y., Renard C. A., Labalette C., Wu Y., Levy L., Neuveut C., Prieur X., Flajolet M., Prigent S., Buendia M. A. (2003) J. Biol. Chem. 278, 5188–5194 [DOI] [PubMed] [Google Scholar]
- 24.Pan Y., Bai C. B., Joyner A. L., Wang B. (2006) Mol. Cell Biol. 26, 3365–3377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smale S. T., Kadonaga J. T. (2003) Annu. Rev. Biochem. 72, 449–479 [DOI] [PubMed] [Google Scholar]
- 26.Edlund S., Lee S. Y., Grimsby S., Zhang S., Aspenstrom P., Heldin C. H., Landstrom M. (2005) Mol. Cell Biol. 25, 1475–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Labbé E., Letamendia A., Attisano L. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 8358–8363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lei S., Dubeykovskiy A., Chakladar A., Wojtukiewicz L., Wang T. C. (2004) J. Biol. Chem. 279, 42492–42502 [DOI] [PubMed] [Google Scholar]
- 29.Clifford R. L., Deacon K., Knox A. J. (2008) J. Biol. Chem. 283, 35337–35353 [DOI] [PubMed] [Google Scholar]
- 30.Hirota M., Watanabe K., Hamada S., Sun Y., Strizzi L., Mancino M., Nagaoka T., Gonzales M., Seno M., Bianco C., Salomon D. S. (2008) Cell Signal 20, 1632–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishita M., Hashimoto M. K., Ogata S., Laurent M. N., Ueno N., Shibuya H., Cho K. W. (2000) Nature 403, 781–785 [DOI] [PubMed] [Google Scholar]
- 32.de La Coste A., Romagnolo B., Billuart P., Renard C. A., Buendia M. A., Soubrane O., Fabre M., Chelly J., Beldjord C., Kahn A., Perret C. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 8847–8851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dennler S., Itoh S., Vivien D., ten Dijke P., Huet S., Gauthier J. M. (1998) EMBO J. 17, 3091–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nolan-Stevaux O., Lau J., Truitt M. L., Chu G. C., Hebrok M., Fernandez-Zapico M. E., Hanahan D. (2009) Genes Dev. 23, 24–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu M., Yao J., Carroll D. K., Weremowicz S., Chen H., Carrasco D., Richardson A., Violette S., Nikolskaya T., Nikolsky Y., Bauerlein E. L., Hahn W. C., Gelman R. S., Allred C., Bissell M. J., Schnitt S., Polyak K. (2008) Cancer Cell 13, 394–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paul S., Dey A. (2008) Neoplasma 55, 165–176 [PubMed] [Google Scholar]
- 37.Maeda O., Kondo M., Fujita T., Usami N., Fukui T., Shimokata K., Ando T., Goto H., Sekido Y. (2006) Oncol. Rep. 16, 91–96 [PubMed] [Google Scholar]
- 38.Mullor J. L., Dahmane N., Sun T., Ruiz i Altaba A. (2001) Curr. Biol. 11, 769–773 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.