Abstract
H2O2 acts as a signaling molecule by oxidizing critical thiol groups on redox-regulated target proteins. To explain the efficiency and selectivity of H2O2-based signaling, it has been proposed that oxidation of target proteins may be facilitated by H2O2-scavenging peroxidases. Recently, a peroxidase-based protein oxidation relay has been identified in yeast, namely the oxidation of the transcription factor Yap1 by the peroxidase Orp1. It has remained unclear whether the protein oxidase function of Orp1 is a singular adaptation or whether it may represent a more general principle. Here we show that Orp1 is in fact not restricted to oxidizing Yap1 but can also form a highly efficient redox relay with the oxidant target protein roGFP (redox-sensitive green fluorescent protein) in mammalian cells. Orp1 mediates near quantitative oxidation of roGFP2 by H2O2, and the Orp1-roGFP2 redox relay effectively converts physiological H2O2 signals into measurable fluorescent signals in living cells. Furthermore, the oxidant relay phenomenon is not restricted to Orp1 as the mammalian peroxidase Gpx4 also mediates oxidation of proximal roGFP2 in living cells. Together, these findings support the concept that certain peroxidases harbor an intrinsic and powerful capacity to act as H2O2-dependent protein thiol oxidases when they are recruited into proximity of oxidizable target proteins.
Introduction
By now, it is well accepted that hydrogen peroxide (H2O2) acts as a signaling molecule. In a variety of physiological situations, it is generated in a controlled manner and leads to the selective posttranslational modification of cysteine residues on target proteins (1). Reversible thiol oxidation, in particular disulfide bond formation, changes the functional properties of affected proteins. One prominent example is the transient inactivation of protein tyrosine phosphatases in receptor tyrosine kinase signaling (2).
It is less clear how H2O2 actually oxidizes its target proteins. It is frequently assumed that a low pKa is sufficient to make protein thiols directly reactive toward H2O2. However, it has been pointed out that a low pKa by itself will not lead to a high reaction rate as the rate-limiting step is still impeded by a high activation barrier (3). To overcome kinetic inhibition, thiol-based peroxidases are equipped with highly evolved catalytic mechanisms (triads or tetrads (4)) to bring about the reaction between the peroxidatic thiol and H2O2, thus achieving high rate constants (e.g. 2 × 107 m−1s−1 for Prx22 (5)). Specialized catalytic mechanisms for “self-oxidation” are unlikely to exist in redox-regulated proteins, thus explaining their slow reaction with H2O2 when probed in vitro (e.g. 20 m−1s−1 for PTP1B) (3). It has been argued recently that under the intracellular conditions of kinetic competition, given the high reactivity and abundance of peroxiredoxins, oxidant-sensitive redox-regulated proteins like PTP1B are not likely to be oxidized directly by H2O2 (6). A plausible alternative to direct oxidation is a mechanism whereby a peroxidase acts as a primary oxidant acceptor and then passes on the oxidation to a target protein. Oxidative redox relays may also explain the observed selectivity of H2O2 in redox signaling. In support of this idea, a few specific observations of peroxidase-based redox relays have been made in recent years.
The best known example is the Orp1-Yap1 redox relay in yeast (7). Orp1 (also known as Gpx3) is a seleno-independent member of the glutathione peroxidase (GPx) family. Orp1 is thioredoxin (Trx)-dependent and therefore functionally classified as a peroxiredoxin (8). Upon encountering H2O2, Orp1 forms a sulfenic acid (Cys-SOH) at its peroxidatic cysteine (Cys36). In the conventional catalytic cycle, Cys36-SOH rapidly condenses with the resolving cysteine of Orp1 (Cys82) to generate an intramolecular disulfide bond that is subject to reduction by Trx. In its redox relay mode, however, Orp1 mediates oxidation of the transcription factor Yap1 rather than oxidation of Trx. In a first step, Orp1 forms an intermolecular mixed disulfide bond with Yap1. It is thought that Cys36-SOH, instead of condensing with Cys82, directly reacts with the target thiol of Yap1 (Cys598). In a second step, the Orp1-Yap1 intermolecular disulfide is attacked by Cys303 of Yap1 and thus exchanged into a Yap1 intramolecular disulfide bond. The disulfide form of Yap1 accumulates in the nucleus and initiates a transcriptional response.
The case of Orp1-Yap1 clearly demonstrates the possibility of peroxidase-mediated protein oxidation. However, the Orp1 redox relay may be a unique adaptation, restricted to a specific pairing of proteins in the context of the yeast cell. Therefore, in this study, we asked whether Orp1, as well as other GPx-type peroxidases, may harbor a general intrinsic ability to promote the oxidation of other proteins.
Overall, two mechanisms of peroxidase-mediated protein oxidation are conceivable. Firstly, as suggested for Yap1, a target protein thiol may react directly with Cys-SOH (or Sec-SeOH in selenocysteine-based peroxidases), creating an intermolecular disulfide, which is then rearranged into a target intramolecular disulfide by virtue of a second target thiol. Secondly, following the conversion of Cys-SOH into a disulfide bond (either within or between peroxidase subunits or between peroxidase and glutathione), the target protein may be oxidized by a conventional thiol-disulfide exchange reaction. Either scenario implies that proximity and spatial orientation are important. Only if peroxidase and target protein are close to each other can the target protein thiol be expected to intercept either the Cys-SOH or the disulfide state of the peroxidase cycle, both of which are highly transient. Inside the cell, proximity and alignment may be established in a specific manner by adaptor or scaffolding proteins. In fact, the redox relay between Orp1 and Yap1 is known to depend on an additional protein, Ybp1 (9), which appears to act as a scaffold.
We first addressed the question whether Orp1 is generally able to promote oxidation of thiol proteins when they come into close proximity. As a model target protein, we used redox-sensitive green fluorescent protein (roGFP), which is equipped with an oxidizable dithiol pair on its surface and allows fluorometric real-time measurements of its redox state (10, 11). We demonstrate that Orp1 does indeed promote the oxidation of roGFP2 in a proximity-dependent manner, both in vitro and inside living cells. In vitro, close proximity of the two proteins in a fusion protein leads to near quantitative conversion of H2O2 molecules into roGFP disulfide bridges. Inside living cells, expression of a roGFP2-Orp1 fusion protein enables specific and sensitive real-time measurements of intracellular H2O2 under physiologically relevant conditions, thus demonstrating the extraordinary efficiency of proximity-based peroxidase redox relays. The mechanism of the Orp1-roGFP2 redox relay in living cells was found to depend on the resolving cysteine of Orp1 and to operate on the basis of conventional thiol-disulfide exchange. We then addressed the question whether mammalian peroxidases also have the ability to transfer oxidizing equivalents to roGFP. We found that the glutathione peroxidase Gpx4, but not the peroxiredoxin Prx6, promotes roGFP oxidation in living cells, suggesting that only certain peroxidases harbor the capacity to play pro-oxidative roles in mammalian redox signaling.
EXPERIMENTAL PROCEDURES
Cloning and Expression of roGFP2-Orp1, roGFP2-Gpx4, and Prx6-roGFP2
roGFP2-Orp1(wt) and roGFP2-Orp1(CS) were expressed with the 30-amino acid linker (GGSGG)6 between the two entities. Briefly, the coding sequence of roGFP2 was amplified by PCR (primers: 5′-CCCTCTAGACTCGAGATGGTGAGCAAGGGCG-3′ and 5′-CCCACTAGTCTTGTACAGCTCGTCCA-3′) and inserted into pBluescript II KS+ using restriction sites XbaI and SpeI. The linker was generated by the annealing of four oligonucleotides (5′-CTAGTGGTGGTTCAGGTGGTGGTGGTTCAGGTGGTGGTGGTTCAGGTGGA-3′, 5′-GGAGGATCAGGAGGAGGAGGATCAGGAGGAGGAGGATCAGGAGGAG-3′, 5′-TGATCCTCCTCCACCTGAACCACCACCACCTGAACCACCACCACCTGAACCACCA-3′, 5′-AATTCTCCTCCTGATCCTCCTCCTCCTGATCCTCCTCCTCC-3′) and inserted downstream of roGFP2 (restriction sites SpeI and EcoRI). Finally, the coding sequences of Orp1(wt) or Orp1(CS) were amplified (primers: 5′-CCCGATATCTCAGAATTCTATAAGCTAG-3′ and 5′-CCCATCGATCTATTCCACCTCTTTCAA-3′) and inserted downstream of the linker element using restriction sites EcoRV and ClaI. For expression in mammalian cells, constructs were inserted into the pLPCX retroviral vector (Clontech). For recombinant expression, constructs were equipped with a C-terminal hexahistidine tag by subcloning into the pQE-60 vector (Qiagen). Recombinant proteins (roGFP2-Orp1(wt), roGFP2-Orp1(CS), and roGFP2) were expressed in Escherichia coli strain BL21 (Stratagene), purified by nickel affinity chromatography (His-Trap HP columns, GE Healthcare), desalted (Slide-A-Lyzer dialysis cassettes, Pierce), and stored at −80 °C.
roGFP2-Gpx4 was generated by exchanging Orp1 with Gpx4-SECIS, using restriction sites EcoRV and ClaI (PCR primer: 5′-CCCGATATCTGTGCATCCCGCGATG-3′ and 5′-CCCATCGATTGGACCTTCCTCCGG-3′). Prx6-roGFP2 was generated from the Grx1-roGFP2 construct (11) by replacing Grx1 with Prx6, using restriction sites XhoI and SpeI (PCR primers: 5′-CCCCTCGAGCCATGGGATCCGCATGCGAG-3′ and 5′-CCCACTAGTGGTGGCAACCCTTTTTT-3′).
In Vitro Fluorescence-based Assays
Recombinant proteins were diluted into standard reaction buffer (100 mm potassium phosphate, 5 mm EDTA, pH 7.0, degassed and then saturated with N2) to a final concentration of 1 μm. The fluorescence emission of roGFP2 (510–530 nm) after excitation at 390 and 480 nm was measured in a fluorescence plate reader (FLUOstar Omega; BMG Labtech) equipped with built-in injectors. The ratio of the emissions was calculated and plotted against time. For oxidation experiments, proteins were first reduced with 20 mm dithiothreitol (DTT) for 20 min on ice and afterward desalted with Zeba spin desalting columns (Pierce).
Cell Culture and Imaging of Living Cells
HeLa cells were grown in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum, 2 mm l-glutamine, 100 units/ml penicillin, and 100 mg/ml streptomycin (Invitrogen). For transient transfection, the calcium phosphate precipitation method was used as described (12). HeLa cells stably expressing roGFP2-Orp1(wt) were generated by retroviral transduction and selection with 0.5 mg/ml puromycin. Cells were seeded and imaged in FD-35 Fluorodish cell culture dishes (World Precision Instruments) using a PerkinElmer Life Sciences Ultra View ERS confocal system on a Nikon TE2000 U inverted microscope equipped with a Plan Apo VC 60× WI objective (numerical aperture 1.2, water immersion). roGFP2 was excited by the 405- and 488-nm laser lines followed by detection of the emission through a 500–554-nm band-pass filter on an electron-multiplying charge-coupled device (EM-CCD camera C9100-02; Hamamatsu). Raw data were exported to ImageJ software (National Institutes of Health) as 16-bit TIFF for analysis. The background of the images was subtracted, and a threshold was set to avoid ratio-created artifacts. The ratio image was created by dividing the 405-nm image by the 488-nm image pixel by pixel.
Flow Cytometry
HeLa cells expressing roGFP2-Orp1(wt) were seeded into cell culture flasks and grown overnight. Adherent cells were washed twice with PBS and supplied with fresh medium with or without fetal calf serum. After 20 h, cells were washed once with PBS and then treated with PBS/20 mm N-ethyl maleimide for 5 min at room temperature. Cells were detached with trypsin, washed twice with PBS, and fixed with 3% paraformaldehyde in PBS for 7 min at room temperature. Cells were washed three times with PBS and analyzed using a FACSAria cell sorting system (BD Biosciences). The ratio of emission (510/10 nm) after excitation at 407 and 488 nm was calculated. As control, starved or non-starved HeLa cells were loaded with 10 μm 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA, Molecular Probes) for 60 min, detached from the culture dish, washed twice with PBS, and analyzed for dichlorofluorescein (DCF) fluorescence (excitation laser 488 nm, emission filter 515–545 nm) using a FACSCalibur flow cytometer (BD Biosciences). Data were analyzed using the FlowJo software.
Isolation, Transfection, and Flow Cytometry of Primary T Cells
Primary human peripheral blood mononuclear cells were obtained by Ficoll-Hypaque (Linaris) density gradient centrifugation of heparinized blood from healthy volunteers. T cells were purified from the peripheral blood mononuclear cells with negative magnetic bead selection using the Pan T cell isolation kit (Miltenyi Biotech), as described in the manufacturer's instructions. For transfection into primary blood T cells, the human T cell nucleofector kit (Amaxa Biosystems) was used as described (13). Cells were treated with 0.1 μg/ml anti-CD3 (OKT-3, ATCC) or IgG2a isotype control antibody (Pharmingen) together with 1 μg/ml goat-anti-mouse antibody (Dianova) as cross-linker. Data were acquired with an LSRII flow cytometer (BD Biosciences) using 405- and 488-nm laser lines for excitation and 500–550- as well as 515–545-nm filter sets for emission. Data were analyzed using the FlowJo software.
RESULTS
Orp1 Promotes the Oxidation of roGFP2
To determine whether Orp1 is able to promote oxidation of roGFP2 in principle, a 5-fold molar excess of reduced recombinant Orp1 was mixed with reduced recombinant roGFP2 in vitro, and the reaction was started by the injection of 1 mm H2O2. The redox state of roGFP2 was monitored in real time by ratiometric fluorescence measurements (10). Two different variants of the Orp1 protein were compared: wild type Orp1 (Orp1(wt)), capable of forming the intramolecular disulfide bond (Cys36–Cys82), and the Orp1 C82S mutant (Orp1(CS)), lacking the resolving cysteine and thus maintaining a stabilized Cys36-SOH (14, 15). Orp1(wt) promoted roGFP2 oxidation at a slow but significant rate, suggesting that thiol-disulfide exchange between the two proteins is possible but not efficient under the given conditions. In contrast, Orp1(CS) mediated pronounced oxidation of roGFP2, indicating that roGFP2 is more efficiently oxidized by the Cys36-SOH form of Orp1 (Fig. 1A). Based on the observation that both Orp1 proteins, wild type and mutant, are generally able to mediate H2O2-dependent oxidation of roGFP2, we decided to study fusion proteins that bring the two interacting proteins into close spatial proximity.
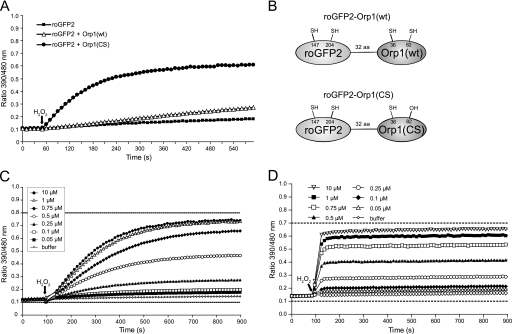
FIGURE 1.
Orp1 mediates the oxidation of roGFP2. A, reduced roGFP2 (1 μm) was incubated with either reduced Orp1(wt) or Orp1(CS) (5 μm). H2O2 (1 mm) was injected after 50 s. The response of roGFP2 in the absence of Orp1 was used as a negative control. B, schematic representation of roGFP2-Orp1 fusion proteins. C, response of reduced roGFP2-Orp1(wt) to increasing concentrations of H2O2 (0.1–10 μm). D, response of reduced roGFP2-Orp1(CS) to increasing concentrations of H2O2 (0.1–10 μm). In C and D, dotted horizontal lines indicate the dynamic range as defined by complete reduction (10 mm DTT, lower dotted line, set to 0.1) and oxidation (1 mm diamide, upper dotted line). The ratiometric dynamic range of roGFP2-Orp1(CS) is lower (7-fold rather than 8-fold) because of its susceptibility to overoxidation. In each experiment, the ratio of roGFP2 emissions (510–530 nm) after excitation at 390 and 480 nm was calculated and plotted against time.
A Fusion Protein of Orp1 and roGFP2 Is a Sensitive Redox Relay for H2O2
Wild type and mutant Orp1 were fused to the C terminus of roGFP2 using a 32-amino acid linker peptide to allow for flexible interactions (Fig. 1B). Following the addition of H2O2, both roGFP2-Orp1(wt) (Fig. 1C) and roGFP2-Orp1(CS) (Fig. 1D) were oxidized almost stoichiometrically and reached near maximal oxidation at a 1:1 molar ratio of H2O2 to protein (1 μm each). This result demonstrated that proximity affords a highly efficient conversion of oxidizing equivalents into roGFP2 disulfide bridges. Although reaching the same end-point oxidation state in response to H2O2, the reaction rate of the two fusion proteins was markedly different. roGFP2-Orp1(wt) required several minutes to complete oxidation (Fig. 1C), whereas roGFP2-Orp1(CS) responded almost instantly (in less than 1 min) (Fig. 1D).
A Fusion Protein of Orp1 and roGFP2 Is a Specific Redox Relay for H2O2
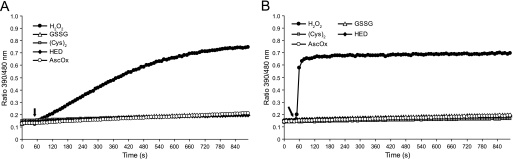
Orp1 is known to respond to H2O2 with high specificity (14, 16). To investigate the specificity of the fusion proteins, roGFP2-Orp1(wt) and roGFP2-Orp1(CS) were exposed to several alternative oxidants, GSSG, cystine (Cys2), hydroxyethyl disulfide (HED), or dehydroascorbic acid (AscOx). A significant response could only be elicited by H2O2 (Fig. 2, A and B), confirming that the established specificity of Orp1 also holds true for the roGFP2 fusion proteins. Again a significant kinetic difference between roGFP2-Orp1(wt) (Fig. 2A) and roGFP2-Orp1(CS) (Fig. 2B) was evident, prompting us to investigate more closely the mechanism of oxidant transfer between Orp1 and roGFP2.
FIGURE 2.
roGFP2-Orp1 specifically responds to H2O2. A and B, reduced roGFP2-Orp1(wt) (A) and roGFP2-Orp1(CS) (B) were exposed to the oxidants H2O2, GSSG, cystine (Cys2), hydroxyethyl disulfide (HED), and dehydroascorbic acid (AscOx) (10 μm each) 50 s after starting the measurement (arrow).
Wild Type Orp1 Oxidizes roGFP2 by Thiol-Disulfide Exchange
Cys36-SOH of Orp1 can be blocked irreversibly with 5,5-dimethyl-1,3-cyclohexadione (dimedone) and thus be prevented from reacting with thiols (15). The presence of dimedone in the reaction buffer substantially attenuated H2O2-dependent roGFP oxidation in both fusion proteins (Fig. 3A), thus verifying the requirement for a sulfenic acid intermediate in the redox relay mechanism of both roGFP2-Orp1(wt) and roGFP2-Orp1(CS).
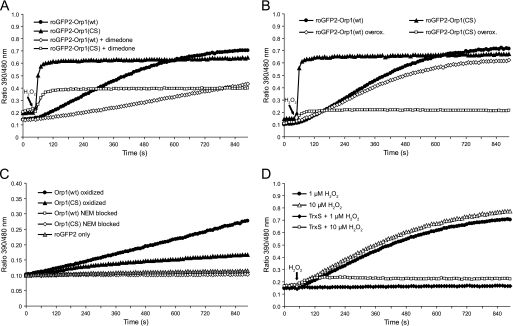
FIGURE 3.
Mechanism of Orp1-mediated roGFP2 oxidation. A, dimedone attenuates the response of both roGFP2-Orp1(wt) and roGFP2-Orp1(CS). Reduced fusion proteins were treated with 1 μm H2O2 in the presence or absence of 50 mm dimedone. B, roGFP2-Orp1(CS), but not roGFP2-Orp1(wt), is susceptible to overoxidation (overox.). Reduced fusion proteins were exposed to 5 mm H2O2 for 30 min, desalted, exposed to DTT (10 mm, 20 min), and desalted again. The fusion proteins (1 μm) were then treated with 10 μm H2O2. Untreated fusion proteins served as controls. C, the disulfide form of Orp1 is capable of mediating roGFP2 oxidation. Wild type and mutant Orp1 proteins were oxidized with a 5-fold molar excess of H2O2 or blocked with N-ethyl maleimide (NEM) followed by desalting. 1 μm reduced roGFP2 was incubated with 50 μm pretreated Orp1(wt) or Orp1(CS). D, thioredoxin competes thiol-disulfide exchange between roGFP2 and wild type Orp1. roGFP2-Orp1(wt) was exposed to 1 or 10 μm H2O2 in the presence or absence of a functional TrxS consisting of Trx1, Trx reductase, and NADPH. In A, B, and D, H2O2 was injected after 50 s.
If the roGFP2 moiety of roGFP2-Orp1(CS) is directly oxidized via Cys36-SOH, its response should be susceptible to overoxidation. In contrast, if the roGFP2 moiety of roGFP2-Orp1(wt) is oxidized by the Cys36–Cys82 disulfide bond, it should be largely resistant to overoxidation. To determine susceptibility to overoxidation, we exposed both fusion proteins to millimolar H2O2. The fusion proteins were then treated with DTT and tested for their responsiveness to micromolar H2O2. roGFP2-Orp1(CS) was almost completely inactivated (Fig. 3B), consistent with the fact that DTT does not regenerate the peroxidatic thiol from the sulfinic or sulfonic acid modification. In contrast, roGFP2-Orp1(wt) retained almost full activity. The observed protection against overoxidation in the presence of Cys82 confirms the rapid formation of the Orp1 intramolecular disulfide bond and the thiol-disulfide exchange mechanism of roGFP2 oxidation.
To demonstrate more directly that wild type Orp1 is capable of oxidizing roGFP2 by thiol-disulfide exchange, solitary Orp1(wt) was completely oxidized to the disulfide form, separated from residual H2O2 by gel filtration, and then incubated with roGFP2 in the absence of additional oxidants. To compensate for the lack of proximity, Orp1 was used in 50-fold molar excess over roGFP2. The disulfide form of Orp1(wt) was found to mediate oxidation of roGFP2 (Fig. 3C), thus confirming that Orp1(wt) can oxidize roGFP2 by thiol-disulfide exchange.
Thioredoxin Competes Thiol-Disulfide Exchange between roGFP2 and Orp1(wt)
Previous experiments established that Trx does not act on the roGFP2 disulfide bond (11). However, the thioredoxin system (TrxS) is the natural reductant of the Orp1 intramolecular disulfide (7). Hence, Trx is expected to compete with roGFP2 for the Orp1 intramolecular disulfide and to disrupt the redox relay. We therefore exposed reduced roGFP2-Orp1(wt) to different concentrations of H2O2 in the presence or absence of a TrxS. The stoichiometric response to 1 μm H2O2 was fully neutralized and a 10-fold molar excess of H2O2 (10 μm) was mostly neutralized in the presence of TrxS (Fig. 3D). Again, the susceptibility of roGFP2-Orp1(wt) to Trx supports the notion that the redox relay is based on thiol-disulfide exchange between the disulfide form of Orp1(wt) and the dithiol form of roGFP2.
Glutathione Quenches Stabilized Cys-SOH but Does Not Interfere with Disulfide Exchange
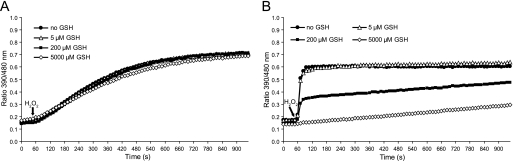
Cys-SOH residues are highly reactive and easily condense with otherwise unreactive thiols (17). We reasoned that the Cys-SOH-initiated response of roGFP2-Orp1(CS) may therefore be highly susceptible to quenching by GSH. When exposed to increasing concentrations of GSH, the response of roGFP2-Orp1(wt) was not significantly compromised (Fig. 4A). In contrast, the response of roGFP2-Orp1(CS) was strongly inhibited in a dose-dependent manner and completely abolished by GSH in the millimolar range (Fig. 4B). These results suggested that only roGFP2-Orp1(wt) would be expected to work as a redox relay under physiological glutathione concentrations.
FIGURE 4.
Glutathione interferes with the sulfenic acid but not with the disulfide exchange mechanism. A and B, 1 μm roGFP2-Orp1(wt) (A) or 1 μm roGFP2-Orp1(CS) (B) was exposed to 1 μm H2O2 after 50 s in the presence of increasing GSH concentrations (0, 5, 200, 5000 μm).
roGFP2-Orp1(wt), but Not roGFP2-Orp1(CS), Responds to H2O2 in Living Cells
Having characterized both fusion proteins in vitro, we tested their behavior in intact cells. In an initial experiment, HeLa cells were transiently transfected with corresponding expression constructs. The response to exogenously applied H2O2 was quantified by live cell microscopy. Consistently, roGFP2-Orp1(wt), but not roGFP2-Orp1(CS), exhibited an Orp1-dependent response in living cells (Fig. 5A). As suggested by the previous in vitro experiment (Fig. 4), the lack of response by roGFP2-Orp1(CS) is most likely due to the quenching influence of intracellular GSH.
FIGURE 5.
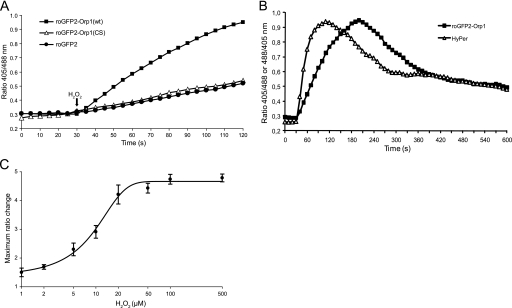
roGFP2-Orp1 responds to H2O2 in living cells. A, roGFP2-Orp1(wt), but not roGFP2-Orp1(CS), facilitates monitoring of H2O2 in living cells. HeLa cells were transiently transfected with roGFP2-Orp1(wt), roGFP2-Orp1(CS), or roGFP2 and treated with 50 μm H2O2 after 30 s. Cells were excited with 405- and 488- nm lasers, and the ratio of the emissions (500–554 nm) was calculated. B, roGFP2-Orp1 and HyPer detect H2O2 with similar sensitivity. HeLa cells stably expressing roGFP2-Orp1(wt) or HyPer were exposed to 20 μm H2O2 after 30 s. The ratio (405/488 nm for roGFP2-Orp1(wt) and 488/405 nm for HyPer) of individual cells was plotted against time. C, based on experiments similar to B, the maximal ratiometric change of roGFP2-Orp1(wt)-positive cells was plotted against H2O2 concentration. A value of 1 indicates a fully reduced probe (calibration by treatment with 1 mm DTT). For each H2O2 concentration, measurements on six cells from two different experiments were averaged. Error bars represent S.D.
Comparison between roGFP2-Orp1 and the OxyR-based Redox Probe HyPer
HyPer, a recently described genetically encoded probe for H2O2, is based on the formation of a single disulfide bond in the bacterial H2O2-sensing transcription factor OxyR (18). In contrast to roGFP2-Orp1, HyPer does not involve a redox relay mechanism. A direct comparison between roGFP2-Orp1 and HyPer, both transfected into HeLa cells, revealed a very similar dynamic response to H2O2 in live cell imaging (Fig. 5B). HyPer appeared to respond faster, but otherwise both probes demonstrate a very similar sensitivity and course of recovery. roGFP2-Orp1 responds to low micromolar concentrations of exogenously applied H2O2 (likely leading to intracellular concentrations in the middle to upper nanomolar range) and exhibits a favorable dynamic range of 4.8 in live cell imaging (Fig. 5C).
roGFP2-Orp1 Responds to Oxidative Changes Caused by Growth Factor Deprivation
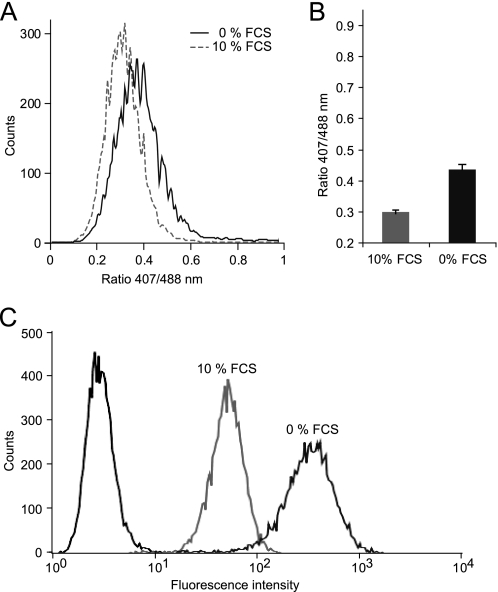
As shown previously with redox-sensitive dyes (19) and using Grx1-roGFP2 (11), growth factor deprivation increases intracellular oxidation. To demonstrate that roGFP2-Orp1 responds to endogenous oxidative processes, we cultured roGFP2-Orp1-expressing HeLa cells with or without serum. Flow cytometry revealed a significant and reproducible ratiometric shift induced by serum deprivation, as shown by a representative histogram (Fig. 6A) and the quantification of multiple experiments (Fig. 6B). The generation of oxidants in serum-starved cells was independently confirmed in the same experiment using the oxidant-sensitive dye H2DCFDA (Fig. 6C).
FIGURE 6.
roGFP2-Orp1 responds to endogenous oxidative changes. HeLa cells expressing roGFP2-Orp1(wt) were cultured for 20 h in medium containing either 0% (solid line) or 10% (dotted line) fetal calf serum (FCS) and analyzed by flow cytometry. A, histogram of one exemplary experiment (10,000 cells). B, quantification of three independent experiments. Error bars represent S.D. C, serum-starved (0% FCS) and non-starved (10% FCS) HeLa cells were loaded with H2DCFDA or left untreated. A histogram of DCF fluorescence (10,000 cells) is shown.
roGFP2-Orp1 Senses H2O2 Signals Triggered by Receptor Engagement on Primary Cells
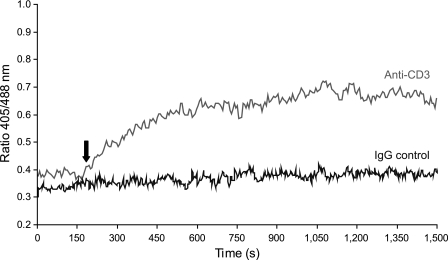
T cell receptor stimulation induces the endogenous generation of H2O2, which appears to play important roles in T cell signaling (20, 21). To test whether roGFP2-Orp1 can be used to detect a weak oxidative signal in a physiologically relevant situation, as shown previously for Grx1-roGFP2 (11), primary peripheral human T cells were transiently transfected with roGFP2-Orp1, stimulated by T cell receptor cross-linking, and analyzed by time-resolved flow cytometry. Cells treated with a cross-linking antibody responded by an increase in the fluorescent ratio, whereas cells treated with control antibody did not (Fig. 7). In conclusion, roGFP2-Orp1 can be used to specifically detect H2O2-based redox signaling events in living cells.
FIGURE 7.
roGFP2-Orp1 visualizes H2O2 signals in primary human T cells. Primary human T cells were isolated, transiently transfected with roGFP2-Orp1(wt), and analyzed by time-resolved flow cytometry. After 200 s (arrow), cells were either stimulated with an anti-CD3 antibody (grey line) or stimulated with an IgG control antibody (black line) together with a cross-linking secondary antibody in both cases. Cells were excited with 405- and 488-nm lasers, and the ratio of the emissions was plotted against time. Approximately 40–60 cells were averaged per second.
Mammalian Peroxidase Gpx4, but Not Prx6, Oxidizes roGFP2 in Living Cells
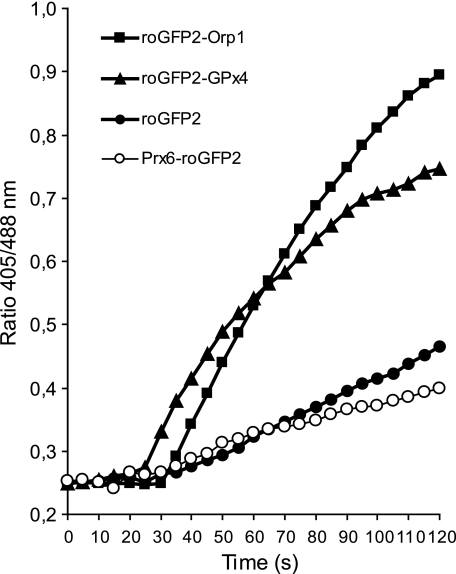
Having found that Orp1 can promote oxidation of proteins other than Yap1, we asked whether a protein thiol oxidase function is also a common property of mammalian peroxidases. Exemplarily, we expressed roGFP2 fusion proteins of glutathione peroxidase Gpx4 and peroxiredoxin Prx6 in murine embryonic fibroblasts and HeLa cells, respectively, and observed their oxidant relay behavior in response to H2O2. roGFP2-Gpx4, but not Prx6-roGFP2, showed a response comparable with that of roGFP2-Orp1 (Fig. 8), suggesting that certain mammalian peroxidases have an intrinsic ability to oxidize co-localized target proteins.
FIGURE 8.
Mammalian peroxidase Gpx4, but not Prx6, oxidizes fused roGFP2. roGFP2 fusion proteins of glutathione peroxidase 4 (roGFP2-Gpx4) and peroxiredoxin-6 (Prx6-roGFP2) were transiently expressed in HeLa cells. Oxidation in response to 50 μm H2O2 was monitored by live cell microscopy.
DISCUSSION
Peroxiredoxins and glutathione peroxidases have exceptionally high reaction rates with H2O2 (22) and are likely to trap most of the H2O2 generated under physiological conditions. Assuming that typical oxidant target proteins are in kinetic competition with “professional” H2O2 scavengers, it is not obvious how they become oxidized (3). One possible resolution of the paradox is that certain peroxidases serve as primary oxidant receptors to convey oxidation to secondary oxidant target proteins. The best studied example for such a peroxidase redox relay is the interaction of the oxidant receptor Orp1 with the oxidant target Yap1. It has not been known whether the Orp1-Yap1 relay represents a unique adaptation, whether Orp1 is more generally able to pass on oxidative equivalents to other proteins, or whether GPx-like peroxidases commonly fulfill relay functions.
We have found that Orp1 efficiently mediates electron flow between H2O2 and roGFP in intact cells. Therefore, Orp1 is not mechanistically restricted to use Yap1 as the recipient of oxidative equivalents and thus harbors a more general ability to act as a thiol oxidase for other proteins. This observation suggests the further reaching possibility that other peroxidases, especially those of the GPx family, have a similar intrinsic capacity to promote the oxidation of other proteins. In fact, the human selenocysteine-based glutathione peroxidase Gpx4 was already implicated in the disulfide cross-linking of protamines (23) and, as shown here, is also capable of mediating the oxidation of roGFP2 in living cells.
Coupling peroxidase redox activity to a fluorescent real-time probe offered the opportunity to characterize the conditions required to establish and maintain a functional peroxidase-based redox relay. As expected, it turned out that proximity between the peroxidase and the target protein is a key factor for directing the flow of oxidative equivalents toward the target protein. The efficiency by which wild type Orp1 oxidized roGFP2 was dramatically improved by the co-localization of both proteins within the same fusion protein. Likewise, the natural Orp1-Yap1 redox relay depends on a dedicated adapter protein (9). It is therefore conceivable that repositioning of peroxidases relative to other proteins, by adapters or otherwise, allows them to switch between oxidant scavenging (i.e. the transfer of oxidative equivalents to Trx or GSH) and oxidant signaling (i.e. the diversion of oxidative equivalents toward redox-regulated target proteins).
By comparing roGFP2 fusion proteins based on wild type and mutant Orp1 in dynamic redox measurements, we addressed the reaction mechanism of the Orp1-roGFP2 redox relay. Generally, there seem to exist two possibilities for a target protein to pick up an oxidizing equivalent from a peroxidase. Either it is able to tap the Cys-SOH before the same is converted into a disulfide bridge, or it engages with the disulfide bridge before the same is eliminated by the responsible reducing system. The potential advantage of the first mechanism is that Cys-SOH are highly reactive and that their condensation with free thiols is thermodynamically favored and not significantly impeded by an activation barrier (24). However, due to the high driving force of the reaction, the target thiol would have to be prepositioned very close to the nascent Cys-SOH to preempt its reaction with other thiols.
We used an Orp1 mutant lacking the resolving cysteine (Cys82) to explore the consequences of stabilizing Cys36-SOH within the fusion protein. In the absence of other thiols, roGFP2-Orp1(CS) responded to H2O2 more rapidly than roGFP2-Orp1(wt). However, the stabilization of Cys36-SOH made the fusion protein prone to inactivation by overoxidation (Fig. 3B), and the whole redox relay became highly susceptible to quenching by glutathione (Fig. 4B). Accordingly, we confirmed that in living cells, Orp1(CS) does not form a working redox relay with roGFP2. The reason appears to be the competition by millimolar concentrations of GSH in the cytosolic environment.
Wild type Orp1 was found to mediate roGFP2 oxidation exclusively by thiol-disulfide exchange, in agreement with the previous observation that newly formed Cys36-SOH is rapidly converted into the intramolecular Cys36–Cys82 disulfide bond (16). Although thiol-disulfide exchange is slower than Cys-SOH-triggered roGFP oxidation, the degree of oxidation reached after equilibration was almost identical for both kinds of fusion proteins.
The Orp1-Yap1 redox relay is presumed to depend on the direct condensation between Cys36-SOH and Cys598 of Yap1 (7). The fact that in yeast cells the Orp1-Yap1 relay is fully operational in the absence of Cys82 demonstrates that the Orp1 intramolecular disulfide is dispensable for Yap1 oxidation (14). However, this observation does not strictly exclude the possibility that Yap1/Ybp1-associated wild type Orp1 forms the intramolecular disulfide and also uses thiol-disulfide exchange to oxidize Yap1. Nevertheless, it is possible that the Orp1-Yap1 relay exclusively uses the Cys-SOH pathway and, thus, actually differs from the Orp1-roGFP2 relay. As Orp1, Yap1, and Ybp1 co-evolved to interact with one another, they may create a special microenvironment for the optimal alignment between Cys-SOH and the target thiol. The same microenvironment may also protect Cys-SOH against competing thiol-based reductants, including GSH. It is furthermore possible that Ybp1, rather than being a simple adapter, actively suppresses formation of the Orp1 intramolecular disulfide to enforce the Cys-SOH pathway of target oxidation. In contrast, the flexible peptide linker that brings together Orp1 and roGFP2 in the fusion protein, although effective in creating proximity and facilitating oxidant transfer, apparently does not create a microenvironment suitably shielded to support Cys-SOH-mediated oxidant transfer. Thus, the interaction of Orp1 with Yap1 and roGFP2, respectively, may be representative of the two principal pathways by which peroxidases can oxidize target protein thiols. Of note, it has been reported that oxidation of the Saccharomyces pombe Yap1-homologue Pap1 by the peroxiredoxin Tpx1 depends on both the peroxidatic and the resolving cysteine (25), suggesting an underlying thiol-disulfide exchange mechanism similar to the Orp1-roGFP2 redox relay. In any case, the in vivo responsiveness of roGFP2-Orp1 demonstrates that a functional peroxidase redox relay does not have to be based on the Cys-SOH mechanism and does not necessarily require highly specialized adapter proteins.
Our observations support the idea that at least some peroxidases in the GPx family possess a general propensity to act as protein thiol oxidases when specifically recruited to oxidizable target proteins. Indirect protein thiol oxidation by scaffold-based redox relays potentially is more target-specific than direct protein oxidation by H2O2, thus suggesting a mechanistic basis for H2O2-based redox signaling.
Finally, our work establishes peroxidase-roGFP relays as a novel design principle for genetically encoded redox probes. roGFP-based fusion proteins now offer the possibility to study the intracellular behavior of individual peroxidases. We compared roGFP2-Orp1 with the recently described OxyR-based H2O2 probe HyPer. In live cell imaging, both probes showed a similar response and sensitivity. The somewhat slower response of roGFP2-Orp1, relative to HyPer (Fig. 5B), is not unexpected as it is based on the redox equilibration between thiol-disulfide pairs of two protein domains, whereas OxyR transmits a conformational change through the formation of a single intramolecular disulfide bond (26). Nevertheless, as we have made no attempts to further optimize the roGFP2-Orp1 construct, in terms of linker length or otherwise, the generation of kinetically enhanced versions is well conceivable.
Supplementary Material
Acknowledgments
We thank J. Remington (University of Oregon, Eugene, OR) for the original roGFP2 construct, M. Toledano (Commissariat à l'Energie Atomique-Saclay, Gif-sur-Yvette, France) for Orp1 protein samples and plasmids, V. Belousov (Shemiakin-Ovchinnikov Institute of Bioorganic Chemistry, Moscow, Russia) for the HyPer construct, M. Conrad (Helmholtz Zentrum München, Germany) for the Gpx4 expression plasmid, B. Knoops (Université catholique de Louvain, Louvain-la-Neuve, Belgium) for the Prx6 plasmid, G. Kuntz for technical assistance, and U. Engel and C. Ackermann (Nikon Imaging Center at the University of Heidelberg, Germany) for microscope access and assistance.
This work was supported by European Commission Grant MEXT 2761 (to T. P. D.).
This article was selected as a Paper of the Week.
- Prx
- peroxiredoxin
- Trx
- thioredoxin
- TrxS
- thioredoxin system
- roGFP
- redox-sensitive green fluorescent protein
- Gpx
- glutathione peroxidase
- Cys-SOH
- cysteine sulfenic acid
- H2DCFDA
- 2′,7′-dichlorodihydrofluorescein diacetate
- DCF
- dichlorofluorescein
- DTT
- dithiothreitol
- PBS
- phosphate-buffered saline
- dimedone
- 5,5-dimethyl-1,3-cyclohexadione
- wt
- wild type.
REFERENCES
- 1.D'Autréaux B., Toledano M. B. (2007) Nat. Rev. Mol. Cell Biol. 8, 813–824 [DOI] [PubMed] [Google Scholar]
- 2.Tonks N. K. (2006) Nat. Rev. Mol. Cell Biol. 7, 833–846 [DOI] [PubMed] [Google Scholar]
- 3.Winterbourn C. C., Hampton M. B. (2008) Free Radic. Biol. Med. 45, 549–561 [DOI] [PubMed] [Google Scholar]
- 4.Tosatto S. C., Bosello V., Fogolari F., Mauri P., Roveri A., Toppo S., Flohé L., Ursini F., Maiorino M. (2008) Antioxid. Redox Signal. 10, 1515–1526 [DOI] [PubMed] [Google Scholar]
- 5.Peskin A. V., Low F. M., Paton L. N., Maghzal G. J., Hampton M. B., Winterbourn C. C. (2007) J. Biol. Chem. 282, 11885–11892 [DOI] [PubMed] [Google Scholar]
- 6.Winterbourn C. C. (2008) Nat. Chem. Biol. 4, 278–286 [DOI] [PubMed] [Google Scholar]
- 7.Toledano M. B., Delaunay A., Monceau L., Tacnet F. (2004) Trends Biochem. Sci. 29, 351–357 [DOI] [PubMed] [Google Scholar]
- 8.Maiorino M., Ursini F., Bosello V., Toppo S., Tosatto S. C., Mauri P., Becker K., Roveri A., Bulato C., Benazzi L., De Palma A., Flohé L. (2007) J. Mol. Biol. 365, 1033–1046 [DOI] [PubMed] [Google Scholar]
- 9.Veal E. A., Ross S. J., Malakasi P., Peacock E., Morgan B. A. (2003) J. Biol. Chem. 278, 30896–30904 [DOI] [PubMed] [Google Scholar]
- 10.Dooley C. T., Dore T. M., Hanson G. T., Jackson W. C., Remington S. J., Tsien R. Y. (2004) J. Biol. Chem. 279, 22284–22293 [DOI] [PubMed] [Google Scholar]
- 11.Gutscher M., Pauleau A. L., Marty L., Brach T., Wabnitz G. H., Samstag Y., Meyer A. J., Dick T. P. (2008) Nat. Methods 5, 553–559 [DOI] [PubMed] [Google Scholar]
- 12.Chen C., Okayama H. (1987) Mol. Cell. Biol. 7, 2745–2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wabnitz G. H., Nebl G., Klemke M., Schröder A. J., Samstag Y. (2006) J. Immunol. 176, 1668–1674 [DOI] [PubMed] [Google Scholar]
- 14.Delaunay A., Pflieger D., Barrault M. B., Vinh J., Toledano M. B. (2002) Cell 111, 471–481 [DOI] [PubMed] [Google Scholar]
- 15.Paulsen C. E., Carroll K. S. (2009) Chem. Biol. 16, 217–225 [DOI] [PubMed] [Google Scholar]
- 16.Ma L. H., Takanishi C. L., Wood M. J. (2007) J. Biol. Chem. 282, 31429–31436 [DOI] [PubMed] [Google Scholar]
- 17.Poole L. B., Karplus P. A., Claiborne A. (2004) Annu. Rev. Pharmacol. Toxicol. 44, 325–347 [DOI] [PubMed] [Google Scholar]
- 18.Belousov V. V., Fradkov A. F., Lukyanov K. A., Staroverov D. B., Shakhbazov K. S., Terskikh A. V., Lukyanov S. (2006) Nat. Methods 3, 281–286 [DOI] [PubMed] [Google Scholar]
- 19.Satoh T., Sakai N., Enokido Y., Uchiyama Y., Hatanaka H. (1996) Brain Res. 733, 9–14 [DOI] [PubMed] [Google Scholar]
- 20.Jackson S. H., Devadas S., Kwon J., Pinto L. A., Williams M. S. (2004) Nat. Immunol. 5, 818–827 [DOI] [PubMed] [Google Scholar]
- 21.Gülow K., Kaminski M., Darvas K., Süss D., Li-Weber M., Krammer P. H. (2005) J. Immunol. 174, 5249–5260 [DOI] [PubMed] [Google Scholar]
- 22.Fourquet S., Huang M. E., D'Autreaux B., Toledano M. B. (2008) Antioxid. Redox Signal. 10, 1565–1576 [DOI] [PubMed] [Google Scholar]
- 23.Conrad M., Moreno S. G., Sinowatz F., Ursini F., Kölle S., Roveri A., Brielmeier M., Wurst W., Maiorino M., Bornkamm G. W. (2005) Mol. Cell. Biol. 25, 7637–7644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Claiborne A., Yeh J. I., Mallett T. C., Luba J., Crane E. J., 3rd, Charrier V., Parsonage D. (1999) Biochemistry 38, 15407–15416 [DOI] [PubMed] [Google Scholar]
- 25.Bozonet S. M., Findlay V. J., Day A. M., Cameron J., Veal E. A., Morgan B. A. (2005) J. Biol. Chem. 280, 23319–23327 [DOI] [PubMed] [Google Scholar]
- 26.Lee C., Lee S. M., Mukhopadhyay P., Kim S. J., Lee S. C., Ahn W. S., Yu M. H., Storz G., Ryu S. E. (2004) Nat. Struct. Mol. Biol. 11, 1179–1185 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.