Abstract
Protein translocation and folding in the endoplasmic reticulum of Saccharomyces cerevisiae involves two distinct Hsp70 chaperones, Lhs1p and Kar2p. Both proteins have the characteristic domain structure of the Hsp70 family consisting of a conserved N-terminal nucleotide binding domain and a C-terminal substrate binding domain. Kar2p is a canonical Hsp70 whose substrate binding activity is regulated by cochaperones that promote either ATP hydrolysis or nucleotide exchange. Lhs1p is a member of the Grp170/Lhs1p subfamily of Hsp70s and was previously shown to function as a nucleotide exchange factor (NEF) for Kar2p. Here we show that in addition to this NEF activity, Lhs1p can function as a holdase that prevents protein aggregation in vitro. Analysis of the nucleotide requirement of these functions demonstrates that nucleotide binding to Lhs1p stimulates the interaction with Kar2p and is essential for NEF activity. In contrast, Lhs1p holdase activity is nucleotide-independent and unaffected by mutations that interfere with ATP binding and NEF activity. In vivo, these mutants show severe protein translocation defects and are unable to support growth despite the presence of a second Kar2p-specific NEF, Sil1p. Thus, Lhs1p-dependent nucleotide exchange activity is vital for ER protein biogenesis in vivo.
Introduction
The endoplasmic reticulum (ER)2 of Saccharomyces cerevisiae contains two Hsp70 chaperones, Lhs1p and Kar2p. Kar2p is a canonical Hsp70 that plays an important role in ER protein folding (1), the import of proteins into the ER membrane via the membrane-embedded Sec61p complex (2) and in the export of misfolded proteins via the endoplasmic reticulum-associated degradation (ERAD) pathway (3–5). It has the two-domain structure that is characteristic of Hsp70s, consisting of a highly conserved N-terminal nucleotide binding domain (NBD) and a C-terminal substrate binding domain. Substrate binding by Hsp70s is regulated by nucleotide-dependent conformational changes (6) that are in turn modulated by cochaperones. An Hsp40/DnaJ-like protein promotes ATP hydrolysis inducing strong substrate interaction, whereas a nucleotide exchange factor (NEF) resets the ATPase cycle leading to substrate release (7, 8). Kar2p has two known nucleotide exchange factors, Lhs1p and Sil1p (9, 10).
Lhs1p is a member of the Grp170/Lhs1p subfamily of Hsp70 chaperones, whose distinguishing features include an extended substrate binding domain (11, 12) and subfamily-specific motifs in the nucleotide binding domain (11). In the absence of Lhs1p, cells show defects in ER protein translocation and folding (13–17) and require the Ire1p-dependent induction of the unfolded protein response (UPR) for survival (14, 16). Although Lhs1p has NEF activity (10) it cannot be excluded that Lhs1p might play additional roles in the ER lumen. For example, in vitro studies have shown that the mammalian Lhs1p homologue, Grp170, not only functions as a NEF for the mammalian Kar2p homologue BiP (18), but is also able to prevent heat-induced aggregation of substrate proteins without the assistance of other chaperones or cofactors (19). This suggests that in addition to its NEF activity, Grp170 can function as a BiP-independent chaperone. So far, the relative contribution of these two functions to ER protein folding and translocation has not been determined.
Most sequence similarity between Lhs1p and other members of the Hsp70 superfamily is found in its NBD, but although the residues implicated in nucleotide binding and hydrolysis are highly conserved, Lhs1p does not have a detectable endogenous ATPase activity like canonical Hsp70s (10). Recent studies suggest that divergent Hsp70s can have different requirements for nucleotide binding and hydrolysis. The cytosolic yeast Hsp110 Sse1p is a member of a different Hsp70 subfamily than Lhs1p, but has a similar function as an Hsp70 NEF (20, 21). Sse1p has been reported to hydrolyze ATP (22), but ATP hydrolysis is not essential for NEF activity (20). For the activity of the ribosome-associated atypical Hsp70 Ssz1p, nucleotide binding is dispensable (23, 24). These observations suggest that the NBD might play diverse functions in the different Hsp70 subfamilies.
In this study we analyzed the requirement of nucleotide binding for Lhs1p function. Although Lhs1p holdase activity is nucleotide-independent, nucleotide binding to wild-type Lhs1p stimulates interaction with Kar2p. Mutations that interfere with nucleotide binding abolish NEF activity in vitro, cause severe protein translocation defects in vivo, and are lethal in a Δire1 strain. The data demonstrate that nucleotide binding to Lhs1p is essential for its nucleotide exchange activity.
EXPERIMENTAL PROCEDURES
Strains and Plasmids
In vivo activity of the Lhs1p mutants was tested in Δlhs1 strain JTY33 (13) and Δire1Δlhs1 strain JTY44 (MATα ade2–1 can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 Δire1::KanMX4 Δlhs1::KanMX4). pRC42 (LHS1, HIS3) and pRC43 (LHS1, URA3) have been described previously (13, 14). To create yeast plasmids carrying lhs1 D26A or G28L the mutations were introduced into pJRT9 (10) by site-directed mutagenesis. The 3799-bp ApaI/SalI fragments of these vectors were cloned into the corresponding sites of pRS313 (25) resulting in pD26A and pG28L, respectively. For expression in Escherichia coli, the mutations were introduced into pETLhs1p (10) to create pET30a-Lhs1p(D26A) and pET30a-Lhs1p(G28L).
Purification of Recombinant Proteins and GdnHCl Treatment of Lhs1p
His6-tagged Kar2p, Lhs1p, and Lhs1p mutants were expressed in E. coli BL21 from pJT46 (His-Kar2p (13)), pETLhs1p (His-Lhs1p (10)), or the plasmids described above. Extracts were prepared as previously described (10) and loaded onto Hi-Trap chelating columns (GE Healthcare). The proteins were purified following several stringent wash steps to prevent contamination with E. coli DnaK (26). GST-tagged Kar2p, Kar2p(G246D), or GST fusion of the Sec63p J-domain were expressed in E. coli BL21 from pDF1 (10), pDF7 (27), or pGST/63J (28), respectively, and purified on a GST-Trap column (GE Healthcare) after stringent wash steps to prevent contamination with E. coli DnaK (26). All proteins were dialyzed against 40 mm Tris-HCl, pH 7.5, 80 mm NaCl, 0.8 mm dithiothreitol, 10% glycerol. To dissociate nucleotides bound to Lhs1p, purified Lhs1p was incubated in 5 m guanidine hydrochloride, 2 mm EDTA for 1 h at room temperature followed by dialysis against 40 mm Tris-HCl, pH 7.5, 80 mm NaCl, 0.8 mm dithiothreitol, 10% glycerol.
Nucleotide Analysis
The ATP Bioluminescence Assay Kit CLS II (Roche Applied Science) was used for analysis of the nucleotide content of purified Lhs1p. 200 pmol of Lhs1p was added to 1 ml of methanol followed by snap freezing in liquid nitrogen. After thawing, the sample was centrifuged for 20 min at 16,000 × g. The supernatant was removed, lyophilized, and resuspended in 200 μl of MilliQ water. For analysis of the ATP content 50-μl samples were mixed with an equal volume of luciferase reagent, followed by luminescence measurements in a luminometer (Turner Designs). For analysis of the ADP content, ADP was converted into ATP by mixing a 50-μl sample with 50 μl of 200 mm triethanolamine, pH 7.8, 400 mm KCl, 60 mm MgSO4, 3 mm phosphoenolpyruvate, and 10 units of pyruvate kinase followed by a 3-h incubation at 30 °C for 3 h. The ATP content was subsequently determined as described.
Analysis of ATP Binding, Nucleotide Exchange Activity, and Steady State ATPase
To analyze ATP binding, 5 μm Lhs1p or Kar2p in 50 mm Tris-HCl, pH 7.4, 50 mm KCl, 5 mm MgCl2 was incubated with 25 μm [α-32P]ATP (6.24 Ci/mmol) for 30 min at room temperature. After isolation of nucleotide-bound protein complexes on a PD SpinTrap G-25 column (GE Healthcare), samples were analyzed by thin layer chromatography (TLC) as described (10). Alternatively, Lhs1p with bound nucleotide was captured onto polyvinylidene difluoride filters (Millipore) by filtration. Unbound nucleotide was removed by three washes with ice-cold buffer and the amount of bound [α-32P]ATP was analyzed by scintillation counting. For ATP saturation binding experiments 3.5 μm Lhs1p was incubated for 1 h at room temperature with 1.5 μCi of [α-32P]ATP in the presence of increasing concentrations of cold ATP (0–200 μm). The amount of Lhs1p-bound nucleotides was determined by filter binding as described above. Binding curves were generated using SigmaPlot and fitted to a one-site saturation binding model. The steady state ATPase activity of Kar2p and the nucleotide exchange activity of Lhs1p were determined as described previously (10).
Protease Protection Assay
5 μg of Lhs1p or Kar2p in 50 mm Tris-HCl, pH 7.4, 50 mm KCl, 5 mm MgCl2 was incubated for 30 min at room temperature in the presence of 2 mm ADP or ATP. Proteinase K was added to a final concentration of 3 μg/ml and after a 5-min incubation at room temperature the reaction was quenched by trichloroacetic acid precipitation. Trichloroacetic acid-precipitated proteins were resuspended in SDS-PAGE sample buffer and analyzed by SDS-PAGE.
GST Pulldown Assay
GST-Kar2p-Lhs1p pulldown experiments were done essentially as described (13). GST-Kar2p was immobilized on gluthatione-agarose (Sigma) by incubating the beads with cell extract of E. coli BL21 expressing GST-Kar2p from pDF1 (10) or pDF7 (27). The beads were washed with GST-binding buffer (13) and incubated for 1 h at 4 °C with 10 μg of purified Lhs1p. After three washes, GST-Kar2p-bound Lhs1p was eluted from the beads with SDS-PAGE sample buffer and analyzed by Western blotting.
Luciferase Aggregation Assay
The holdase activity of Lhs1p was determined essentially as described by Oh et al. (29). 0.3 μm Firefly Luciferase (Sigma) in buffer H (25 mm Hepes KOH, pH 7.9, 50 mm KCl, 5 mm MgOAc, 5 mm β-mercaptoethanol) was incubated at 43 °C in the presence or absence of the indicated amounts of Lhs1p. The optical density at 320 nm was followed on a Cary 300 Biospectrophotometer (Varian, Palo Alto, CA).
Other
Yeast transformations, cell growth, and preparations of total protein extracts were carried out as described (13, 30). Lhs1p and prepro-α-factor antibodies are described in Ref. 13.
RESULTS
Recombinant Lhs1p Is Purified in an ATP Bound State
The nucleotide binding domain of Lhs1p contains all the residues implicated in nucleotide binding and hydrolysis in canonical Hsp70s, suggesting that nucleotide binding plays a role in Lhs1p function. However, binding of radiolabeled nucleotide to purified Lhs1p proved surprisingly inefficient (see below). The apparent failure of Lhs1p to bind nucleotide under these conditions might reflect the intrinsic properties of the protein but could also be explained if Lhs1p had been purified in a nucleotide bound state. Indeed, when the nucleotide content of purified Lhs1p was analyzed in a bioluminescence assay, a significant amount of ATP was detected, corresponding to at least 28.4 ± 0.2% occupancy. The actual occupancy is probably higher, as later findings demonstrate this method significantly underestimates the percentage of bound nucleotides (see below).
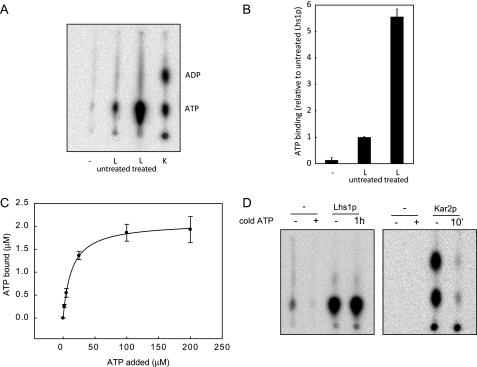
Several studies have reported on the presence of tightly bound nucleotides in the nucleotide binding pocket of members of the Hsp70 and Hsp110 subfamily (31, 32). Methods to remove these nucleotides include incubation with excess AMP-PNP followed by dialysis (31) or dialysis in the presence of alkaline phosphatase (33). These approaches, which both depend on spontaneous release of bound nucleotides, did not improve binding of radiolabeled ATP to Lhs1p (data not shown). We therefore examined the potential of the reversible denaturant, 5 m guanidine HCl, to release bound nucleotide. Purified Lhs1p was incubated in 5 m guanidine HCl (GdnHCl), 2 mm EDTA for 1 h at room temperature and then dialyzed overnight to remove the GdnHCl and any released nucleotides. After GdnHCl treatment, the ATP content of Lhs1p, as detected in our bioluminescence assay, was reduced to 4.2 ± 1.3%, demonstrating that GdnHCl treatment of Lhs1p dissociated a substantial proportion of the bound ATP. GdnHCl treatment did not affect the activity of Lhs1p, as stimulation of ATPase activity upon addition of limiting concentrations of GdnHCl-treated Lhs1p to Kar2p and its Sec63-J partner was indistinguishable from untreated Lhs1p (Fig. 1). However, the GdnHCl-treated Lhs1p exhibited a striking increase in [α-32P]ATP binding activity as shown by TLC analysis (Fig. 2A) and filter binding experiments (Fig. 2B). To determine the efficiency of nucleotide binding, Lhs1p was incubated for 1 h at room temperature with increasing concentrations of [α-32P]ATP. Lhs1p [α-32P]ATP complexes were captured on polyvinylidene difluoride filters, the amount of bound [α-32P]ATP was determined by scintillation counting, and the data were fitting to a one-site saturation binding model (SigmaPlot) (Fig. 2C). Using 3.5 μm apoLhs1p we observed a maximal binding of 2.1 ± 0.02 μm ATP with an affinity of 13.6 ± 0.5 μm. However, as the Lhs1p·ATP complex is essentially non-dissociating (see below), the true equilibrium affinity of Lhs1p for ATP is probably much higher. The maximal binding corresponds to ∼60% occupancy, which is considerably higher than the amount of nucleotide found associated with purified Lhs1p in our original bioluminescence assay. The bioluminescence assay measures only the ATP that is released from Lhs1p following methanol precipitation. When [α-32P]ATP·Lhs1p complexes were subject to the same precipitation conditions we regularly found only 50% of the available counts to be released into the supernatant (data not shown). These results suggest that the proportion of GdnHCl-treated Lhs1p, which binds ATP appears similar to that of the original purified protein. The observation that a substantial fraction of Lhs1p is purified in a nucleotide bound form suggests that dissociation is rather slow. To explore this possibility we analyzed the dissociation of Lhs1p-bound [α-32P]ATP. GdnHCl-treated Lhs1p was preloaded with [α-32P]ATP as before, followed by incubation in the presence of a greater than 1,000-fold excess of non-radioactive ATP. TLC analysis of Lhs1p-bound nucleotides demonstrated that the level of bound [α-32P]ATP was not significantly reduced even after 60 min (Fig. 2D). When the same assay was performed with Kar2p, the majority of [α-32P]ATP·Kar2p complexes were already dissociated after 10 min (Fig. 2D). These data confirm that, once formed, the Lhs1p·ATP complex is extremely stable. We therefore conclude that Lhs1p is purified in a nucleotide-bound form that can be dissociated by reversible denaturation with GdnHCl. The resultant “apoLhs1p” exhibits an efficient ATP binding activity the importance of which we investigated further.
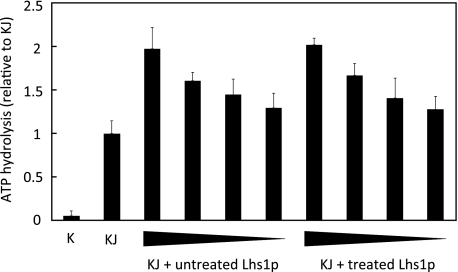
FIGURE 1.
GdnHCl-EDTA treatment does not affect Lhs1p activity. Steady state ATPase activity of 0.25 μm Kar2p (K) in the absence or presence of 1 μm of the soluble J-domain of Sec63p (J) and 1, 0.5, 0.25, or 0.13 μm untreated Lhs1p or GdnHCl-treated Lhs1p. ATP hydrolysis is plotted relative to hydrolysis in the presence of the Sec63 J domain (KJ). The mean ± S.D. (error bars) from three experiments is shown.
FIGURE 2.
GdnHCl-EDTA-treated Lhs1p exhibits efficient nucleotide binding activity. A, untreated Lhs1p (L), GdnHCl-treated Lhs1p or Kar2p (K) were incubated with [α-32P]ATP for 30 min at room temperature. After removal of free nucleotides by gel filtration binding, bound nucleotides were analyzed by thin layer chromatography. Consistent with previous observations (10), Lhs1p does not hydrolyze [α-32P]ATP like the canonical Hsp70 Kar2p. B, [α-32P]ATP·Lhs1p complexes were captured onto polyvinylidene difluoride filters (Millipore), followed by removal of unbound nucleotides by three washes with ice-cold ATPase assay buffer. The amount of bound [α-32P]ATP was analyzed by scintillation counting. The mean ± S.D. (error bars) from three experiments is shown. C, 3.5 μm Lhs1p was incubated for 1 h at room temperature with 0–200 μm ATP, supplemented with [α-32P]ATP. Lhs1p with bound nucleotide was captured onto polyvinylidene difluoride filters as described in B, the amount of bound [α-32P]ATP was analyzed by scintillation counting, and binding curves were generated using a one-site saturation binding model. Error bars represent the mean of two experiments that were each performed in triplicate. D, GdnHCl-treated Lhs1p or Kar2p were preincubated with [α-32P]ATP for 3 h or 30 min, respectively, a more than 1000-fold excess of cold ATP was added and the proteins were incubated for an additional hour (Lhs1p) or 10 min (Kar2p). After removal of free nucleotides by gel filtration, bound nucleotides were analyzed by thin layer chromatography.
Nucleotide Binding to Lhs1p Causes a Conformational Change
Canonical Hsp70 chaperones adopt specific conformations with different protease sensitivities upon binding of ADP or ATP (34–36). To determine whether Lhs1p undergoes a conformational change upon nucleotide binding, GdnHCl-treated Lhs1p was preincubated with or without nucleotide and subsequently treated with proteinase K. In the absence of nucleotides, incubation with proteinase K resulted in almost complete degradation of Lhs1p. However, after preincubation with either ADP or ATP, a substantial amount of Lhs1p remained protected from proteolysis (Fig. 3), demonstrating that nucleotide binding results in a conformational change that alters the susceptibility to proteinase K. Consistent with our finding that Lhs1p was originally purified in an ATP-bound form we found that untreated Lhs1p displayed a similar protease protection pattern to that observed for nucleotide bound GdnHCl-treated Lhs1p and that this was independent of any added nucleotide.
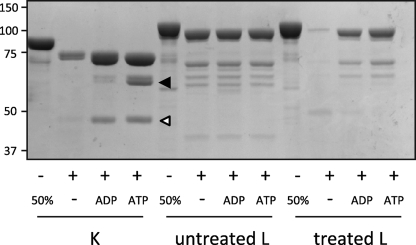
FIGURE 3.
Nucleotide binding alters the proteinase K susceptibility of Lhs1p. Untreated Lhs1p (L), GdnHCl-treated Lhs1p and Kar2p (K) were preincubated for 30 min at room temperature in the presence of nucleotides before addition of proteinase K. Cleavage products were separated by SDS-PAGE and visualized by Coomassie Brilliant Blue staining. The ∼44 and ∼60 kDa cleavage products of Kar2p are indicated by a white and black arrows, respectively.
The proteolytic pattern of nucleotide-bound Lhs1p differed from canonical Hsp70s, where ADP and ATP have different effects on protease susceptibility. ADP binding protects a ∼44-kDa NBD fragment, whereas ATP protects a ∼60-kDa fragment that includes both the NBD and the substrate binding domain (Fig. 3, Kar2p, white and black arrows, respectively) (34–36). These results demonstrate that, like canonical Hsp70 chaperones, Lhs1p undergoes a conformational change upon nucleotide binding, although the nature of these conformational changes appears to differ between the two subfamilies.
Nucleotide Binding to Lhs1p Stimulates the Interaction with Kar2p
Previously we have shown that Lhs1p interacts with GST-tagged Kar2p (10). The interaction between purified (untreated) Lhs1p and GST-Kar2p occurs both in the presence or absence of ADP but is substantially inhibited by the addition of ATP (Fig. 4A). Given that the purified Lhs1p is largely ATP bound, then it might appear probable that the observed inhibitory effect of ATP on the interaction is mediated via its binding to Kar2p. To test this possibility we created a G246D mutant form of Kar2p that is equivalent to the nucleotide binding G226D mutation in the mammalian Kar2p homologue BiP (34). As predicted, the G246D mutant form of Kar2p was defective in ATP binding and its interaction with Lhs1p was now found to be unaffected by the addition of ATP (Fig. 4, A and B). These data suggest that Lhs1p binds preferentially to the apo- and ADP-bound forms of Kar2p but not to its ATP-bound form.
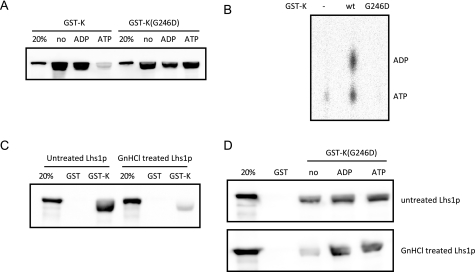
FIGURE 4.
Nucleotide binding to Lhs1p is required for efficient interaction with Kar2p. A, purified untreated Lhs1p was incubated with immobilized GST-Kar2p (GST-K) or GST-Kar2p(G246D) (GST-K(G246D)) in the presence or absence of 2 mm ADP or ATP. Samples of bound proteins were examined by Western blotting and detected with antibodies against Lhs1p. The altered migration of Lhs1p is caused by overloading with GST-Kar2p (data not shown). B, GST-Kar2p(G246D) does not bind ATP. GST-Kar2p or GST-Kar2p(G246D) were incubated with [α-32P]ATP for 30 min at room temperature. After removal of free nucleotides by gel filtration binding, bound nucleotides were analyzed by thin layer chromatography. C, interaction between untreated and GdnHCl-treated Lhs1p with GST or GST-Kar2p analyzed, as described in A. D, interaction between untreated or GdnHCl-treated Lhs1p and GST-Kar2p(G246D) (GST-K(G246D)) analyzed in the presence or absence of 2 mm ADP or ATP, as described in A. wt, wild-type.
Next we examined the requirement for nucleotide binding to Lhs1p in the Kar2p interaction. Strikingly, we found that removal of bound ATP from Lhs1p by GdnHCl treatment led to a substantial reduction in the interaction with GST-Kar2p (Fig. 4C). Whereas the interaction of untreated Lhs1p with GST-Kar2p(G246D) was unaffected by the presence of nucleotides, ADP and ATP dramatically stimulated the GST-Kar2p(G246D) binding of GdnHCl-treated Lhs1p (Fig. 4D). These data demonstrate that nucleotide binding to Lhs1p stimulates its interaction with Kar2p. Interestingly, the interaction seems to be promoted to a similar extent by ADP and ATP.
Lhs1p Nucleotide Binding Mutations Interfere with NEF Activity
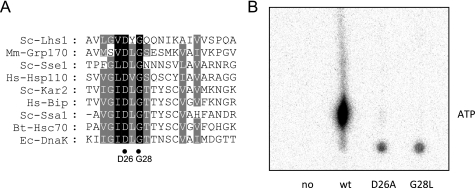
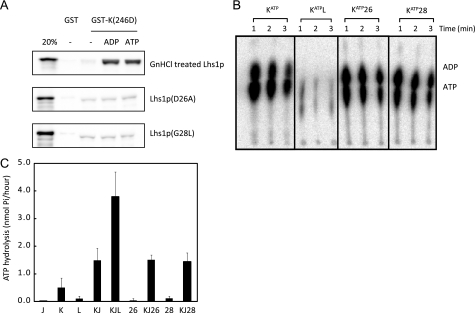
To further analyze the importance of nucleotide binding for Lhs1p function we examined two novel mutations in the nucleotide binding domain. Lhs1p Asp-26 and Gly-28 are identical throughout the NBDs of the Hsp70 superfamily (Fig. 5A) and their equivalents in bovine Hsc70 (37) and Sse1p (38–40) are localized in the nucleotide binding pocket where the conserved aspartate residue coordinates the Mg2+ATP (38–41). Substitution of Asp-26 or Gly-28 with alanine or leucine, respectively, severely affected [α-32P]ATP binding to GdnHCl-treated Lhs1p (Fig. 5B), demonstrating that the NBD mutations resulted in nucleotide binding defects. The observation that an efficient interaction between wild-type Lhs1p and Kar2p is nucleotide-dependent suggests that nucleotide binding mutations would affect the Lhs1p-Kar2p interaction. Indeed, when assayed in a GST-Kar2p(G246D) pulldown assay, both nucleotide binding mutants showed a reduced Kar2p interaction, which could not be stimulated by nucleotides (Fig. 6A). To determine whether the residual interaction observed in the pulldown assay was sufficient to stimulate nucleotide exchange on Kar2p, Kar2p was preloaded with [α-32P]ATP and incubated with wild-type Lhs1p or the Lhs1p mutants. The release of nucleotides from Kar2p was monitored by thin layer chromatography. As reported previously (10) incubation with wild-type Lhs1p led to a rapid reduction in the amount Kar2p-bound [α-32P]ADP and [α-32P]ATP (Fig. 6B). However, in the presence of the Lhs1p mutants, the amount of bound nucleotide remained similar to the control (Fig. 6B), demonstrating that both NBD mutations strongly interfere with the NEF activity of Lhs1p. This conclusion was further supported by the observation that, unlike wild-type Lhs1p, the NBD mutants were unable to stimulate the level of ATP hydrolysis when added to Kar2p and its Sec63-J partner (Fig. 6C). Taken together, these results demonstrate that nucleotide binding to Lhs1p is essential for its nucleotide exchange activity.
FIGURE 5.
Mutation of conserved residues in the nucleotide binding domain prevent ATP binding to Lhs1p. A, alignment of a conserved region in the ATPase domains of Hsp70 superfamily members. The position of the conserved Lhs1p residues Asp-26 and Gly-28 are indicated with black dots. B, NBD mutations affect ATP binding. GdnHCl-treated Lhs1p or Lhs1p carrying the D26A or G28L mutations was incubated with [α-32P]ATP for 30 min at room temperature. After removal of free nucleotides by gel filtration binding, bound nucleotides were analyzed by thin layer chromatography. wt, wild-type.
FIGURE 6.
The Lhs1p D26A and G28L mutations inhibit functional interaction with Kar2p. A, the Lhs1p D26A and G28L mutations severely affect the interaction with Kar2p. Interaction of untreated Lhs1p, GdnHCl-treated Lhs1p, Lhs1p(D26A), or Lhs1p(G28L) with GST-Kar2p(G246D) were analyzed in the presence or absence of 2 mm ADP or ATP as described in the legend to Fig. 3A. B, the Lhs1p D26A and G28L mutants do not stimulate nucleotide exchange on Kar2p. Kar2p·[α-32P]ATP complex was isolated and incubated with Lhs1p or Lhs1p mutants in the presence of 100 μm unlabeled ATP. At the indicated time points, proteins were reisolated, and the bound nucleotides were examined by TLC. C, steady state ATPase activity of Kar2p (K) in the absence or presence of the soluble J-domain of Sec63p (J), Lhs1p (L), Lhs1p(D26A) (26), or Lhs1p(G28L) (28). The mean ± S.D. from a minimum of three independent experiments is shown.
Lhs1p Holdase Activity Is Independent of Nucleotides
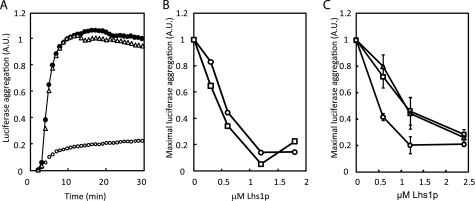
The mammalian Lhs1p homologue Grp170 not only functions as a nucleotide exchange factor (18), but can also act as a holdase that prevents heat-induced aggregation of proteins in vitro (19). To verify that holdase activity is a general feature of the Grp170/Lhs1p subfamily, the model substrate firefly luciferase was incubated at 43 °C in the presence or absence of untreated Lhs1p and its thermal aggregation was followed by an increase in optical density. As reported for Grp170, wild-type Lhs1p efficiently reduced the thermal aggregation of luciferase, whereas bovine serum albumin was unable to prevent aggregation (Fig. 7A). To test whether efficient holdase activity requires nucleotide binding to Lhs1p, luciferase was incubated at 43 °C in the presence of limiting concentrations of untreated and GdnHCl-treated Lhs1p. GdnHCl-treated Lhs1p prevented luciferase aggregation with the same efficiency as untreated Lhs1p (Fig. 7B). In addition, the Lhs1p nucleotide binding mutants were also capable of preventing luciferase aggregation (Fig. 7C). These results demonstrate that Lhs1p can function as a nucleotide-independent holdase in vitro.
FIGURE 7.
Lhs1p functions as a nucleotide-independent holdase. A, firefly luciferase (0.6 μm) was incubated at 43 °C in the absence (●) or presence of 1.2 μm untreated Lhs1p (○) or 3 μm bovine serum albumin (Δ). Aggregation was followed by the increase in optical density at 320 nm. B, GdnHCl-treated Lhs1p and untreated Lhs1p prevent luciferase aggregation with similar efficiency. Maximal firefly luciferase (0.6 μm) aggregation upon incubation at 43 °C in the absence (●) or presence of increasing concentrations of untreated Lhs1p (○) or GdnHCl-treated Lhs1p (□) are indicated. C, the Lhs1p NBD mutations still allow holdase activity. Maximal firefly luciferase (0.6 μm) aggregation upon incubation at 43 °C in the absence (●) or presence of increasing concentrations of Lhs1p (○), Lhs1p(D26A) (□), or Lhs1p(G28A) (Δ). The mean ± S.D. (error bars) from three experiments is shown.
Lhs1p Nucleotide Binding Mutants Do Not Function in Vivo
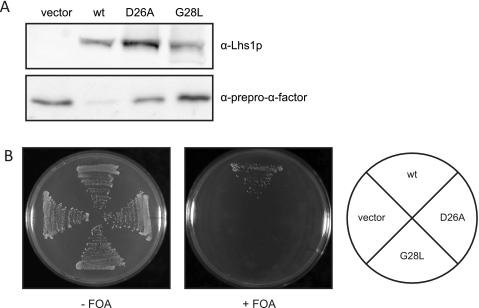
Although Lhs1p is not essential for viability, cells lacking LHS1 are defective in the translocation of a subset of proteins across the ER membrane (14). To determine whether nucleotide binding to Lhs1p is essential for its role in protein translocation, Δlhs1 strain JTY33 (13) was transformed with plasmids encoding either wild-type Lhs1p or the NBD mutants and the level of the post-translational translocation substrate prepro-α-factor in cell extracts was analyzed by immunoblotting. Both mutants showed a marked accumulation of the cytosolic precursor form of this substrate (Fig. 8A), demonstrating that the nucleotide binding mutations result in protein translocation defects. In addition to protein translocation defects, cells lacking LHS1 display a constitutive induction of the unfolded protein response (UPR) (14). The UPR results in transcriptional induction of a wide variety of genes, including ER chaperones and the other Kar2p nucleotide exchange factor Sil1p (13). To determine whether cells expressing the Lhs1p NBD mutants were able to grow in the absence of a functional UPR, we examined the ability of the mutants to support growth in the absence of IRE1, a gene required for activation of the UPR. Δlhs1Δire1 cells carrying the plasmid pRC43 (LHS1, URA3) (14) were transformed with plasmids carrying the NBD mutants and analyzed for growth after loss of pRC43 on 5′-fluoroorotic acid medium. After loss of pRC43, only a plasmid carrying wild-type LHS1 was able to rescue growth (Fig. 8B). Vector alone or plasmids expressing either the D26A or G28L mutant forms were unable to rescue the growth defect (Fig. 8B). These results demonstrate that the nucleotide binding mutations disrupt Lhs1p function in vivo.
FIGURE 8.
The D26A and G28L mutations affect Lhs1p function in vivo. A, JTY33 (Δlhs1) transformed with either empty vector or HIS3-based plasmids containing LHS1, lhs1(D26A), or lhs1(G28L) was grown to midlog phase in yeast nitrogen base (YNB) with appropriate supplements. Whole cells extracts were analyzed by immunoblotting with antibodies against Lhs1p and prepro-α-factor. B, the ability of the Lhs1p mutant to complement the lethal defect associated with the Δire1Δlhs1 double mutation was examined by transforming JTY44 (Δire1Δlhs1) containing pRC43 (LHS1, URA3) with either empty vector or HIS3-based plasmids containing LHS1, lhs1(D26A) or lhs1(G28L), followed by counterselection of pRC43 on 5′-fluoroorotic acid (FOA) medium.
DISCUSSION
The Lhs1p nucleotide binding domain contains many residues that are similar throughout the Hsp70 superfamily. Especially the residues important for ATP binding and hydrolysis in canonical Hsp70s are highly conserved. Although this strict sequence conservation suggests the NBD is important for Lhs1p function, its exact role has not been clearly identified. Here we show that nucleotide binding to Lhs1p is essential for its function as a Kar2p nucleotide exchange factor. We found that Lhs1p was purified as a relatively stable ATP-bound complex. Removal of bound ATP was made possible by reversible denaturation whereupon the resulting apoLhs1p showed a much reduced interaction with Kar2p. This interaction was restored following incubation of apoLhs1p with nucleotide. This finding was confirmed with two different mutations in the nucleotide binding domain of Lhs1p that disrupted nucleotide binding. Both D26A and G28L mutant forms of Lhs1p showed reduced interaction with Kar2p and were unable to provide any nucleotide exchange activity in vitro. Nucleotide binding therefore appears to be essential for the NEF activity of Lhs1p.
The stability of the ATP-bound form of Lhs1p was particularly striking. Bound ATP could not be removed even after prolonged dialysis, neither did we find evidence for significant spontaneous exchange with competitor nucleotide even when the latter was present in substantial excess. However, by developing a reliable method for generating apoLhs1p we have been able to demonstrate that it is the nucleotide-bound form of Lhs1p that interacts with Kar2p. However, the interaction between Lhs1p-ATP and Kar2p was shown to be inhibited by the addition of ATP. Using an ATP-binding mutant form of Kar2p (G246D) we were then able to demonstrate that this inhibitory effect of ATP was now lost. Taken together, our data demonstrate that nucleotide-bound Lhs1p binds Kar2p only when the latter is in its nucleotide-free or ADP-bound states. This would be consistent with the function of Lhs1p as a nucleotide exchange factor for Kar2p.
Like Lhs1p, members of the cytosolic Hsp110 subfamily also function as nucleotide exchange factors for canonical Hsp70s (20, 21). Nucleotide binding mutations in the Hsp110 Sse1p have similarly been reported to affect the interaction with the Hsp70s and to reduce nucleotide exchange activity (21, 42). Recent structural data demonstrate that Sse1p interacts with Hsp70s via its NBD and the helical bundle domain of the substrate binding domain (39, 40) and this interaction can only occur when the Sse1-NBD is in a closed, nucleotide bound, conformation (32, 39, 40). In a pulldown assay, both ADP and ATP binding to Lhs1p appear to stimulate the interaction between Lhs1p and Kar2p to a similar extent. In contrast, ATP binding to Sse1p was reported to support the interaction between Sse1p and its Hsp70 partner Ssa1p more efficiently than ADP binding (32). It is possible that Lhs1p and Sse1p respond slightly different to either ADP or ATP binding as protease protection of Lhs1p was found to be similar in the presence of ATP and ADP, whereas ADP-bound Sse1p is more susceptible to proteolysis than ATP-bound Sse1p (22). These data appear to indicate some differences in the properties of the ER-lumenal Lhs1p from its cytosolic counterpart.
In addition to its NEF activity Lhs1p can also function as a holdase (this study). Unlike the Kar2p interaction, nucleotide binding is not required for Lhs1p substrate binding as both GdnHCl-treated Lhs1p and the Lhs1p nucleotide-binding mutants are efficient holdases. This observation is consistent with studies on deletion mutants of Hsp110 (29) and Grp170 (19), which indicate that the substrate binding domain alone is sufficient for holdase activity. Importantly, the holdase activity of the nucleotide-binding mutants is not sufficient for function in vivo. Δlhs1 cells expressing Lhs1p(D26A) or Lhs1p(G28L) show a dramatic accumulation of prepro-α-factor, suggesting that the translocation defects of Δlhs1 cells are primarily caused by the lack of Lhs1p NEF activity. This is consistent with the observation that overexpression of Sil1p can partially compensate the translocation defect of a Δlhs1 strain (13). However, Sil1p overexpression does not suppress the Δlhs1 translocation defect completely (13) and it cannot be excluded that substrate binding to Lhs1p plays a role during protein translocation as well. In such a scenario, both Lhs1p and Kar2p could associate with the translocation peptide, either sequentially or simultaneously as has been proposed for the Hsp110·Hsp70 complex (39). The observation that without UPR induction of Sil1p, both NBD mutations are lethal clearly indicates that the Lhs1p nucleotide exchange activity is indispensible for Lhs1p function in vivo.
Acknowledgments
We thank Neil Bullied (University of Manchester, UK) for the use of the Cary 300 Biospectrophotometer and Christopher Sellick, Michael Spiller, Gabriella Forte, Barrie Wilkinson, and Martin Pool (University of Manchester, UK) for helpful discussions and technical assistance.
This work was supported by The Wellcome Trust.
- ER
- endoplasmic reticulum
- NBD
- nucleotide binding domain
- NEF
- nucleotide exchange factor
- UPR
- unfolded protein response
- GST
- glutathione S-transferase
- AMP-PNP
- 5′-adenylyl-β,γ-imidodiphosphate
- GdnHCl
- guanidine HCl.
REFERENCES
- 1.Simons J. F., Ferro-Novick S., Rose M. D., Helenius A. (1995) J. Cell Biol. 130, 41–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vogel J. P., Misra L. M., Rose M. D. (1990) J. Cell Biol. 110, 1885–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kabani M., Kelley S. S., Morrow M. W., Montgomery D. L., Sivendran R., Rose M. D., Gierasch L. M., Brodsky J. L. (2003) Mol. Biol. Cell 14, 3437–3448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nishikawa S. I., Fewell S. W., Kato Y., Brodsky J. L., Endo T. (2001) J. Cell Biol. 153, 1061–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Denic V., Quan E. M., Weissman J. S. (2006) Cell 126, 349–359 [DOI] [PubMed] [Google Scholar]
- 6.Schmid D., Baici A., Gehring H., Christen P. (1994) Science 263, 971–973 [DOI] [PubMed] [Google Scholar]
- 7.Szabo A., Langer T., Schröder H., Flanagan J., Bukau B., Hartl F. U. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 10345–10349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liberek K., Marszalek J., Ang D., Georgopoulos C., Zylicz M. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 2874–2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kabani M., Beckerich J. M., Gaillardin C. (2000) Mol. Cell. Biol. 20, 6923–6934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steel G. J., Fullerton D. M., Tyson J. R., Stirling C. J. (2004) Science 303, 98–101 [DOI] [PubMed] [Google Scholar]
- 11.Tyson A. R., Jr., Stirling C. J. (1997) Trends Cell Biol. 7, 277–282 [DOI] [PubMed] [Google Scholar]
- 12.Easton D. P., Kaneko Y., Subjeck J. R. (2000) Cell Stress Chaperones 5, 276–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tyson J. R., Stirling C. J. (2000) EMBO J. 19, 6440–6452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Craven R. A., Egerton M., Stirling C. J. (1996) EMBO J. 15, 2640–2650 [PMC free article] [PubMed] [Google Scholar]
- 15.Saris N., Holkeri H., Craven R. A., Stirling C. J., Makarow M. (1997) J. Cell Biol. 137, 813–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baxter B. K., James P., Evans T., Craig E. A. (1996) Mol. Cell. Biol. 16, 6444–6456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamilton T. G., Flynn G. C. (1996) J. Biol. Chem. 271, 30610–30613 [DOI] [PubMed] [Google Scholar]
- 18.Weitzmann A., Volkmer J., Zimmermann R. (2006) FEBS Lett. 580, 5237–5240 [DOI] [PubMed] [Google Scholar]
- 19.Park J., Easton D. P., Chen X., MacDonald I. J., Wang X. Y., Subjeck J. R. (2003) Biochemistry 42, 14893–14902 [DOI] [PubMed] [Google Scholar]
- 20.Raviol H., Sadlish H., Rodriguez F., Mayer M. P., Bukau B. (2006) EMBO J. 25, 2510–2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dragovic Z., Broadley S. A., Shomura Y., Bracher A., Hartl F. U. (2006) EMBO J. 25, 2519–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raviol H., Bukau B., Mayer M. P. (2006) FEBS Lett. 580, 168–174 [DOI] [PubMed] [Google Scholar]
- 23.Huang P., Gautschi M., Walter W., Rospert S., Craig E. A. (2005) Nat. Struct. Mol. Biol. 12, 497–504 [DOI] [PubMed] [Google Scholar]
- 24.Conz C., Otto H., Peisker K., Gautschi M., Wölfle T., Mayer M. P., Rospert S. (2007) J. Biol. Chem. 282, 33977–33984 [DOI] [PubMed] [Google Scholar]
- 25.Sikorski R. S., Hieter P. (1989) Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McClellan A. J., Endres J. B., Vogel J. P., Palazzi D., Rose M. D., Brodsky J. L. (1998) Mol. Biol. Cell 9, 3533–3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fullerton D. M. (2004) Analysis of the Interactions of Hsp70 ER Lumenal Chaperones. Ph.D. thesis, University of Manchester, UK [Google Scholar]
- 28.Corsi A. K., Schekman R. (1997) J. Cell Biol. 137, 1483–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oh H. J., Easton D., Murawski M., Kaneko Y., Subjeck J. R. (1999) J. Biol. Chem. 274, 15712–15718 [DOI] [PubMed] [Google Scholar]
- 30.Wilkinson B. M., Tyson J. R., Reid P. J., Stirling C. J. (2000) J. Biol. Chem. 275, 521–529 [DOI] [PubMed] [Google Scholar]
- 31.Gao B., Greene L., Eisenberg E. (1994) Biochemistry 33, 2048–2054 [DOI] [PubMed] [Google Scholar]
- 32.Andréasson C., Fiaux J., Rampelt H., Mayer M. P., Bukau B. (2008) J. Biol. Chem. 283, 8877–8884 [DOI] [PubMed] [Google Scholar]
- 33.Theyssen H., Schuster H. P., Packschies L., Bukau B., Reinstein J. (1996) J. Mol. Biol. 263, 657–670 [DOI] [PubMed] [Google Scholar]
- 34.Wei J., Gaut J. R., Hendershot L. M. (1995) J. Biol. Chem. 270, 26677–26682 [DOI] [PubMed] [Google Scholar]
- 35.Kassenbrock C. K., Kelly R. B. (1989) EMBO J. 8, 1461–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chappell T. G., Konforti B. B., Schmid S. L., Rothman J. E. (1987) J. Biol. Chem. 262, 746–751 [PubMed] [Google Scholar]
- 37.Flaherty K. M., DeLuca-Flaherty C., McKay D. B. (1990) Nature 346, 623–628 [DOI] [PubMed] [Google Scholar]
- 38.Liu Q., Hendrickson W. A. (2007) Cell 131, 106–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Polier S., Dragovic Z., Hartl F. U., Bracher A. (2008) Cell 133, 1068–1079 [DOI] [PubMed] [Google Scholar]
- 40.Schuermann J. P., Jiang J., Cuellar J., Llorca O., Wang L., Gimenez L. E., Jin S., Taylor A. B., Demeler B., Morano K. A., Hart P. J., Valpuesta J. M., Lafer E. M., Sousa R. (2008) Mol. Cell 31, 232–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flaherty K. M., Wilbanks S. M., DeLuca-Flaherty C., McKay D. B. (1994) J. Biol. Chem. 269, 12899–12907 [PubMed] [Google Scholar]
- 42.Shaner L., Wegele H., Buchner J., Morano K. A. (2005) J. Biol. Chem. 280, 41262–41269 [DOI] [PubMed] [Google Scholar]