Abstract
Invasion of hepatocytes by Plasmodium sporozoites is a prerequisite for establishment of a malaria natural infection. The molecular mechanisms underlying sporozoite invasion are largely unknown. We have previously reported that CD81 is required on hepatocytes for infection by Plasmodium falciparum and Plasmodium yoelii sporozoites. CD81 belongs to the tetraspanin superfamily of transmembrane proteins. By interacting with each other and with other transmembrane proteins, tetraspanins may play a role in the lateral organization of membrane proteins. In this study, we investigated the role of the two major molecular partners of CD81 in hepatocytic cells, CD9P-1/EWI-F and EWI-2, two transmembrane proteins belonging to a novel subfamily of immunoglobulin proteins. We show that CD9P-1 silencing increases the host cell susceptibility to P. yoelii sporozoite infection, whereas EWI-2 knock-down has no effect. Conversely, overexpression of CD9P-1 but not EWI-2 partially inhibits infection. Using CD81 and CD9P-1 chimeric molecules, we demonstrate the role of transmembrane regions in CD81-CD9P-1 interactions. Importantly, a CD9P-1 chimera that no longer associates with CD81 does not affect infection. Based on these data, we conclude that CD9P-1 acts as a negative regulator of P. yoelii infection by interacting with CD81 and regulating its function.
Introduction
Malaria remains the most important parasitic human disease, responsible for nearly one million deaths each year (1). Plasmodium natural infection is initiated by the inoculation of sporozoites in the host by a female Anopheles mosquito. Within the first hours of infection the motile sporozoites reach the liver and invade hepatocytes, where they further differentiate into a replicative exo-erythrocytic form (EEF)5 that will ultimately give rise to thousands of merozoites that initiate the pathogenic erythrocytic cycle. Plasmodium sporozoites invade hepatocytes actively by forming a membrane-bound compartment called the parasitophorous vacuole (PV), where the parasite resides for further intracellular development (2, 3). The nature of the molecular interactions mediating sporozoite invasion still remains elusive. Two well-characterized sporozoite surface proteins, the circumsporozoite protein and the thrombospondin-related adhesive protein, are known to interact with the liver heparan sulfate proteoglycans (HSPGs) (2, 3). HSPGs play a role in the initial sequestration of the sporozoites in the liver sinusoids (4, 5), and activate sporozoites for productive invasion, leading to proteolytic processing of the circumsporozoite protein (6). Two sporozoite proteins, P36 and P36p/P52, were shown to play a major role during infection of hepatocytes (7–9). Both belong to a Plasmodium-specific family characterized by the presence of domains with six conserved cysteines, but their precise function during sporozoite invasion and/or maintenance of the PV early after invasion remains unknown.
To date, the only host molecule clearly identified as being essential for sporozoite invasion is CD81. Indeed, we have previously reported that CD81 is required for invasion of hepatocytes by sporozoites of human Plasmodium falciparum and rodent Plasmodium yoelii. This was evidenced by the lack of infection of CD81-deficient mouse hepatocytes by P. yoelii sporozoites (10) and by the inhibition of P. yoelii and P. falciparum sporozoite invasion by antibodies or siRNA targeting mouse and human CD81, respectively (10, 11). Another host cell surface protein, the scavenger receptor class B type I (SR-BI), also plays a role during sporozoite invasion (12, 13). SRBI regulates CD81 expression level as well as membrane cholesterol (12), which is known to modulate CD81 ability to support Plasmodium infection (11). This suggests that SRBI acts indirectly during sporozoite infection through regulation of CD81 expression and function.
CD81 belongs to a family of largely distributed integral membrane proteins called tetraspanins. These proteins display four transmembrane domains delimiting two extracellular domains of unequal size and three short intracellular regions. Tetraspanins have been implicated in various biological processes such as cell adhesion, migration, cell fusion, co-stimulation, signal transduction, and differentiation (reviewed in Refs. 14–16). They also play a role in infection by different pathogenic agents and notably several viruses including HIV. It is interesting to note that CD81 is essential for the infection of hepatocytic cells by the hepatitis C virus (HCV) (17, 18) although the exact mechanism by which CD81 mediates HCV infection is still unclear.
In all these processes, the precise function of tetraspanins remains unknown. A current model is that through their mutual interaction and the association with other surface molecules tetraspanins organize a network of interaction referred to as the “tetraspanin web” (14–16). Within this network of interactions, tetraspanins form primary complexes with a limited number of proteins called tetraspanin partners. These tetraspanin-partner interactions are direct and highly specific. For example, CD9P-1 (EWI-F, FPRP) and EWI-2 (PGRL), two related proteins of the immunoglobulin superfamily, associate directly only with CD81 and CD9 (19–22). These primary interactions resist detergents such as digitonin (and in some cases Triton X-100) and typically occur at a high stoichiometry (14). From the recent analysis of the dynamics of the tetraspanin web (23, 24), we suggested that the transient association of various primary complexes would facilitate the assembly of more complex proteolipidic structures upon proper stimulation (14). In this study, we have examined whether the two major CD81 partners, CD9P-1 and EWI-2, play a role in the infection of hepatocytic cells by Plasmodium sporozoites.
EXPERIMENTAL PROCEDURES
Antibodies
The anti-human CD81 (TS81) CD82 (TS82b), and CD9P-1 (1F11) were previously described (19). The anti-mouse CD9 (4.1F12) and CD81 mAbs (MT81 and MT81w) have been described elsewhere (11). A goat polyclonal anti-mEWI-2 was purchased from R&D systems and an anti-mouse α5 integrin mAb MFR5 from BD Pharmingen (Le Pont de Claix, France). For generation of 8G1 (anti-mouse CD9P-1 mAb), a rat was injected intraperitoneally twice with 107 L-CD9 cells, and a final boost was performed 3 weeks later with CD9-containing complexes collected from a Brij 97 lysate of ∼109 L-CD9 cells. Spleen cells were fused with P3 × 63AG8 mouse myeloma cells to generate hybridomas according to standard techniques.
Construction of Chimeric Molecules
Plasmids encoding CD9, CD82, hCD9P-1, and hEWI-2 were previously described (19, 20). The mCD9P-1 plasmid was generated in our laboratory after RT-PCR on RNA purified from LM cells. All chimeras were engineered by classical PCR protocols using cDNA coding the human molecules as a template and entirely sequenced. The chimeras CD82 × 81 and CD82LEL81 were inserted into the pCNC retroviral vector (25). CD9P-1/MHC class I chimeras were inserted into pcDNA3 (Invitrogen, Cergy Pontoise, France). The junctions are as follows: CD82 × 81: CD82-NMGK·IAKD-CD81; CD82LEL81: same junction and CD81-GKLY·IILG-CD82; CD9P-1/HLA: CD9P-1-AFKYP·IVGIV-MHC; CD9P-1/cytHLA: CD9P-1-SSHW·CRRK-MHC.
Parasites and Cells
P. yoelii (265BY strain) and Plasmodium berghei (ANKA strain) sporozoites were obtained from dissection of infected Anopheles stephensi mosquito salivary glands. The human hepatocarcinoma cells stably expressing CD81 (HepG2/CD81) has been previously described (26). This cell line as well as the mouse hepatoma cells Hepa1-6 (ATCC CRL-1830) were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal calf serum (Biowest, Nuaillé, France), 2 mm glutamine, and antibiotics (Invitrogen).
Small-interfering RNA (siRNA) and Plasmid Transfections, Flow Cytometry
We used small double-stranded RNA oligonucleotides targeting mouse and human CD9P-1 (5′-CAU UUG AGA UGA CUU GCA A dTdT-3′), and mouse and human EWI-2 (5′-GUU CUC CUA UGC UGU CUU U dTdT-3′). A non-functional siRNA targeting human CD82 (5′-ACC TCC TCC AGC TCG CTT AdTdT-3′) was used as a control siRNA throughout the study. Cells (5–10 × 106 cells in 400 μl of RPMI) were transfected with 200 pmol of siRNA by electroporation (300V, 500 μF) using the Gene Pulser apparatus (Bio-Rad, Ivry, France). Following siRNA transfection, cells were cultured for 48 h before flow cytometry analysis or sporozoite infection. Hepa1-6 cells were transfected with plasmids using Lipofectamine 2000 reagent according to the manufacturer's recommendations, and all experiments were performed 24–48 h later. Immunolabeling and flow cytometry analysis were performed as described previously (11). Hepa1-6 transfectants stably expressing hCD9P-1 were selected in the presence of G418. Two different populations of hCD9P-1-positive cells were sorted using a FACS-VANTAGE-DIVA (Becton-Dickinson-BDIS) after immunolabeling with the anti-hCD9P-1 mAb.
Sporozoite Infection Assay
Hepa1-6 or HepG2 cells (1.5 × 105 per well) were seeded in 8-chamber plastic Lab-Tek slides (Nalge Nunc International, Cergy Pontoise, France), 24–48 h prior to inoculation with P. yoelii (2–5 × 104 sporozoites per well). After 3 h at 37 °C, the cells were washed and further incubated in fresh medium for 24–36 h before immunolabeling and counting of EEFs as described (10). All experiments were done in triplicates.
Immunoprecipitation
Surface labeling of cells with EZ-link-Sulfo-NHS-LC-biotin (Pierce) was performed as described previously (19, 20). Cells, biotin-labeled or not, were lysed at 4 °C in lysis buffer containing 30 mm Tris, pH 7.4, 150 mm NaCl, 0.02% NaN3, protease inhibitors, and either 1% Brij97 (Sigma) or 1% digitonin (high purity; Calbiochem, San Diego, CA). Immunoprecipitations were then performed using protein G-Sepharose beads (Amersham Biosciences) as described (19, 20). The immunoprecipitates were separated by SDS-PAGE (5–15% gel) under non-reducing conditions and transferred to a nitrocellulose membrane (GE Healthcare).
Immunoprecipitates were analyzed by Western blotting using biotinylated mAbs and Alexa Fluor 680-labeled streptavidin or specific mAb labeled with Alexa 680 (Invitrogen), and the data were acquired using the Odyssey Infrared Imaging System (LI-COR Biosciences). Biotin-labeled surface proteins were revealed using Alexa Fluor 800-labeled streptavidin (Invitrogen).
RESULTS
CD9P-1/FPRP and EWI-2/PGRL Are the Major Partners of CD81 in Human and Mouse Hepatocytic Cells
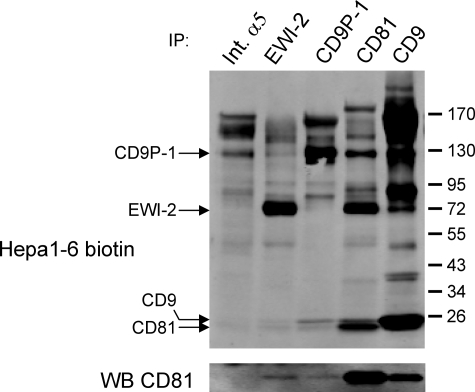
We have previously shown that the two major surface proteins co-immunoprecipitated with CD81 in primary human hepatocytes correspond to the two main partners of CD81, CD9P-1 and EWI-2, two members of a novel subfamily of immunoglobulin proteins (20). Here, we analyzed the molecular partners of CD81 in Hepa1-6 cells, a model cell line supporting P. yoelii sporozoite infection (27). We performed co-immunoprecipitation experiments using digitonin lysates from biotin-labeled cells. Under these conditions, tetraspanins to tetraspanins interactions are not conserved (or strongly reduced) and only partners directly associated in primary complexes coprecipitate with tetraspanins (28). In Hepa1-6 cells, like in human hepatocytes, CD81 co-immunoprecipitated two major proteins with molecular masses consistent with that of CD9P-1 (125 kDa) and EWI-2 (65 kDa) (Fig. 1). The high molecular mass protein indeed co-migrates with the major protein immunoprecipitated by a new mAb 8G1 specific for mouse CD9P-1 (see “Experimental Procedures”), and the 65 kDa protein co-migrates with the major protein immunoprecipitated by an antibody specific for EWI-2. The interaction of CD9P-1 with CD81 is further demonstrated by the ability of 8G1 to co-immunoprecipitate a fraction of CD81 (Fig. 1, lower panel). As a control, an anti-integrin α5 antibody did not co-immunoprecipitate CD81.
FIGURE 1.
CD9P-1 and EWI-2 are CD81 major partners in mouse hepatocytic cells. After cell surface biotinylation, Hepa1-6 cells were lysed with digitonin, and immunoprecipitations (IP) were performed as indicated on the top of each lane. The antibodies used for immunoprecipitations were MFR5 (Int. α5), a polyclonal antibody to EWI-2, 8G1 (CD9P-1), MT81 (CD81), and 4.1. F12 (CD9). The proteins in the immunoprecipitates were revealed using IRdye800-conjugated streptavidin. Bands corresponding to CD9P-1, EWI-2, CD81, and CD9 are indicated on the left. In the lower panel, the presence of CD81 in the different IPs was analyzed by Western blot using Alexa 680-conjugated CD81 antibody (MT81). CD81 is present in both CD9P-1 and EWI-2 IP but not in the integrin α5 (Int. α5) IP.
The Expression Level of CD9P-1 Modulates Hepatocyte Susceptibility to P. yoelii Sporozoites
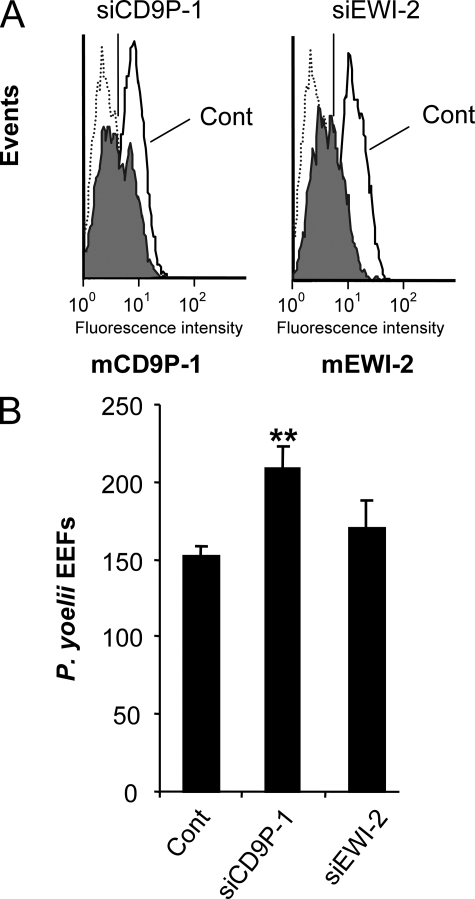
To test whether CD9P-1 and EWI-2 play a role during infection by Plasmodium sporozoites, we first used siRNA to specifically knock-down the expression of these molecules in Hepa1-6 cells. We could achieve a ∼65–70% inhibition of mCD9P-1 and EWI-2 expression as shown by FACS analysis (Fig. 2A). Interestingly, whereas mEWI-2 silencing had no significant effect on P. yoelii infection, the knock-down of mCD9P-1 significantly increased the number of P. yoelii-infected cells, up to 35% (Fig. 2B).
FIGURE 2.
The knock-down of CD9P-1 expression increases the infection of Hepa1-6 cells by P. yoelii sporozoites. Hepa1-6 cells were transfected with a siRNA oligonucleotide targeting CD9P-1 (siCD9P-1) or EWI-2 (siEWI-2) or with a control siRNA (cont). A, the efficiency of silencing was determined 48 h later by indirect immunofluorescence and flow cytometry analysis using anti-CD9P-1 mAb (8G1, left panel) or anti-EWI-2 polyclonal antibody (right panel). For both, silencing yields ∼65–70% inhibition of the expression level of their target. B, 48 h after transfection, the cells were incubated with P. yoelii sporozoites, and the level of infection was determined by quantification of the number of EEFs 36 h later (mean of triplicate wells ± S.D.). **, p < 0.01 as compared with control.
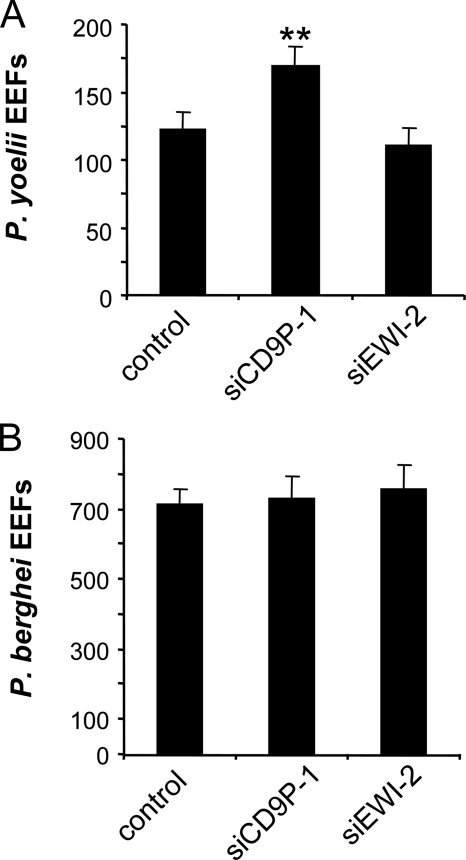
To ensure that this effect of CD9P-1 silencing on P. yoelii infection is not cell type-dependent, similar experiments were performed in human hepatocarcinoma HepG2 cells transduced to stably express CD81, which also support P. yoelii sporozoite infection (29). As seen with Hepa1-6 cells, the knock-down of human CD9P-1 in HepG2/CD81 cells increased the susceptibility to P. yoelii infection, whereas EWI-2 siRNA had no effect (Fig. 3A). Importantly, the knock-down of CD9P-1 in HepG2/CD81 cells had no consequence on infection by P. berghei sporozoites, which is CD81-independent (30) (Fig. 3B).
FIGURE 3.
Silencing of CD9P-1 only affects CD81-dependent sporozoite infection. HepG2/CD81 were transfected with siRNA targeting CD9P-1 (siCD9P-1), EWI-2 (siEWI-2), or with a control siRNA (control) 48 h before infection assay. A, quantification of P. yoelii EEFs (mean of triplicate wells ± S.D.). B, quantification of P. berghei EEFs. **, p < 0.01 as compared with control.
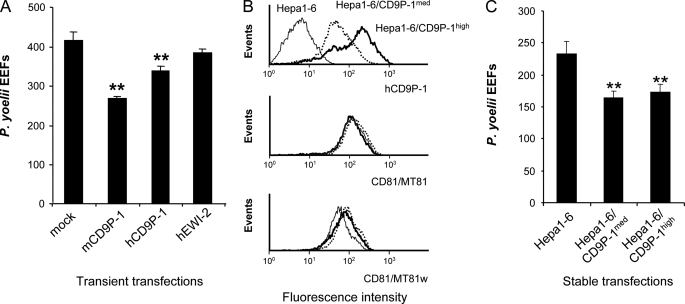
The above data suggest that CD9P-1 is a negative regulator of P. yoelii infection. To further confirm this hypothesis, we then tested the effects of overexpressing CD9P-1 and EWI-2 by transient plasmid transfection in Hepa1-6 cells. As shown in Fig. 4A, overexpression of both mouse and human CD9P-1 significantly reduced the susceptibility of Hepa1-6 cells to P. yoelii sporozoite infection. In contrast, overexpression of human EWI-2 did not modify the number of infected cells. Finally, to further investigate the role of CD9P-1 on infection by sporozoites, we performed stable transfections to generate two Hepa1-6 cell lines expressing different levels of hCD9P-1, Hepa1-6/CD9P-1med and Hepa1-6/CD9P-1high. As shown in Fig. 4C, both cell populations were less susceptible to P. yoelii infection than parental Hepa1-6 cells, confirming the inhibitory effect of CD9P-1 overexpression observed under transient transfection conditions. Interestingly, despite expressing ∼4 times more CD9P-1 (Fig. 4B), Hepa1-6/CD9P-1high cells were not less susceptible to infection than Hepa1-6/CD9P-1med cells (Fig. 4C). Importantly, the expression level of CD9 (not shown) and CD81 was not altered by overexpression of CD9P-1 (Fig. 4B). In addition, the binding of MT81w, a mAb that specifically recognizes a fraction of CD81 associated with the tetraspanin web (11), was also unchanged (Fig. 4B). Altogether these results indicate that CD9P-1 plays a negative role on P. yoelii sporozoite infection without interfering with CD81 expression.
FIGURE 4.
Transfection of CD9P-1 inhibits the infection of Hepa1-6 cells by P. yoelii sporozoites. A, Hepa1-6 cells were transiently transfected with plasmids coding for mouse CD9P-1 (mCD9P-1), human CD9P-1 (hCD9P-1), human EWI-2 (hEWI-2), or with the empty vector (mock), 24 h before infection with P. yoelii sporozoites. The number of EEF (mean ± S.D.) was determined as described under “Experimental Procedures.” B, flow cytometry analysis of hCD9P-1 expression in two sorted populations stably expressing medium (Hepa1-6/CD9P-1med) or high (Hepa1-6/CD9P-1high) levels of CD9P-1. The expression level of CD81 is controlled using two different CD81 mAb (MT81 and MT81w). C, Hepa1-6 cells as well as these two populations of CD9P-1-expressing cells were infected with P. yoelii sporozoites, and the number of EEFs was determined (mean of triplicate wells ± S.D.). **, p < 0.01 as compared with control cells.
The Interaction of CD81 with CD9P-1 Requires Transmembrane Regions
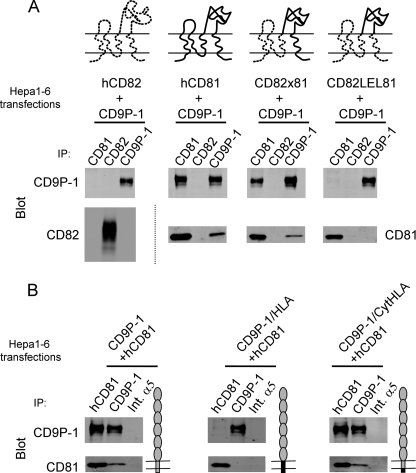
As an initial step to determine whether the effect of CD9P-1 on infection is related to its association with CD81, we first characterized the structural determinants of CD81-CD9P-1 interaction. We took advantage of the absence of interaction between the tetraspanin CD82 and CD9P-1 after lysis in digitonin, and tested the ability of chimeric human CD81/CD82 molecules to associate with CD9P-1 in transiently transfected Hepa1-6 cells. The chimeras were similarly expressed on the cell surface as determined by flow cytometric analysis of transfected cells (data not shown). The chimeric molecule consisting of the first half of CD82 joined to the large extracellular loop (LEL), fourth TM region, and C-terminal cytoplasmic end of CD81 (CD82 × 81) still interacted with CD9P-1 in the same proportion as CD81. In contrast, no interaction was observed with a chimeric molecule in which only the LEL of CD82 was replaced by that of CD81 (CD82LEL81) (Fig. 5A), pointing out the importance of the fourth TM domain and/or C-terminal cytoplasmic end of CD81 to confer its interaction with CD9P-1.
FIGURE 5.
The fourth TM domain of CD81 and the TM domain of CD9P-1 are required for CD81-CD9P-1 interaction. Hepa1-6 cells were transiently transfected with plasmids encoding different constructs as indicated. All constructs encode the human molecules. 48 h after transfection, the cells were lysed with digitonin, and immunoprecipitations were performed as indicated at the top of each lane. Immunoprecipitates were analyzed by Western blot using biotin-labeled mAbs (CD9P-1 and CD81 or CD82) and Alexa 680-labeled streptavidin. A, analysis of the CD81 regions contributing to the interaction with CD9P-1 using CD82/CD81 chimeric molecules. A schematic representation of CD82/CD81 molecules is shown above each panel. B, analysis of the human CD9P-1 regions contributing to the interaction with CD81 using CD9P-1/MHC Class I chimeric molecules (CD9P-1/HLA or CD9P-1/CytHLA). A schematic representation of CD9P-1/MHC Class I molecules is shown on the right of each panel.
We then generated a chimeric molecule in which the TM domain and the cytoplasmic region of hCD9P-1 were substituted by the equivalent moiety of human MHC class I molecule (CD9P-1/HLA). CD9P-1/HLA is totally defective for its association with CD81 (Fig. 5B). In contrast, a chimera in which only the cytoplasmic portion of CD9P-1 was replaced by the cytoplasmic part of MHC class I molecule (CD9P-1/CytHLA) interacted with hCD81 at the same level as WT CD9P-1 (Fig. 5B), indicating that the cytoplasmic domain of CD9P-1 is not required for interaction with CD81. Altogether these data strongly suggest that the interaction between CD9P-1 and CD81 occurs via the fourth TM domain of CD81 and the TM region of CD9P-1.
CD9P-1 Exerts a Negative Effect on Sporozoite Infection by Interacting with CD81
The CD9P-1/HLA chimera, which is expressed on the cell surface but does not associate with CD81, constitutes a unique tool that allowed us to investigate whether the inhibitory effect of CD9P-1 on sporozoite infection is linked to its interaction with CD81.
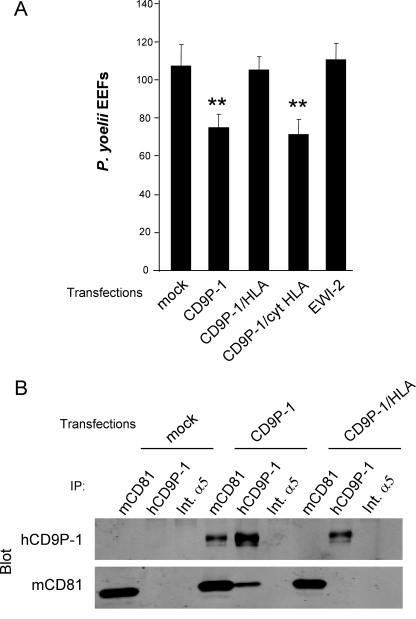
Whereas transient transfection of CD9P-1 in Hepa1-6 cells reduced infection, transfection of CD9P-1/HLA had no significant effect on the number of infected cells (Fig. 6A). Importantly, we checked that hCD9P-1 transfected in Hepa1-6 cells was able to interact with the endogenous mCD81, whereas, under the same conditions, CD9P-1/HLA was not (Fig. 6B). Finally, the chimeric molecule CD9P-1/cytHLA, which has retained a normal ability to interact with CD81, had the same inhibitory effect as CD9P-1 (Fig. 6A). This result indicates that the cytoplasmic region of CD9P-1 is not involved and confirms that the negative effect of CD9P-1 on P. yoelii infection operates via interaction of CD9P-1 with CD81.
FIGURE 6.
A CD9P-1 mutant defective for the association with CD81 has no effect on sporozoite infection of hepatocytic cells. A, Hepa1-6 cells were transiently transfected with plasmids coding for CD9P-1, CD9P-1/HLA, CD9P-1/CytHLA, EWI-2, or empty vector (mock), 24 h before infection with P. yoelii sporozoites. The number of EEF-infected cells (mean ± S.D.) was determined as described under “Experimental Procedures.” **, p < 0.01 as compared with control cells. B, analysis of interaction between human CD9P-1 and endogenous mCD81. Hepa1-6 cells were transiently transfected with CD9P-1, CD9P-1/HLA, or empty vector (mock). After 48 h, the cells were lysed in 1% Brij97 before immunoprecipitation with mAbs against mCD81 (MT81), hCD9P-1 (1F11) or mouse integrin α5 (Int.α5) as a control, as indicated at the top of each lane. Immunoprecipitates were analyzed by Western blotting using biotin-labeled mAbs (hCD9P-1, mCD81) and Alexa 680-labeled streptavidin.
DISCUSSION
Infection of hepatocytes by sporozoites is a crucial step in the Plasmodium life cycle within the mammalian host. We have demonstrated before that the tetraspanin CD81 expressed on the surface of host hepatocytes plays an essential role during Plasmodium sporozoite invasion. Like for other tetraspanins, the function of CD81 is thought to be linked to its ability to associate with other transmembrane molecules. In this study, we show that CD9P-1, a direct partner of CD81, negatively regulates the infection of hepatocytic cells by P. yoelii sporozoites, whereas the closely related molecule EWI-2, another CD81 partner, does not influence infection.
How does CD9P-1 inhibit Plasmodium infection? In one hypothesis, CD9P-1 would interfere with some signaling pathways required for infection. However, this hypothesis is not supported by the finding that the cytoplasmic domain of CD9P-1 is not necessary for its inhibitory effect. To date, the only known cytoplasmic proteins interacting with CD9P-1 are the ERM (Ezrin-Radixin-Moesin) proteins (31), which function as membrane-microfilament linkers (32). Host actin cytoskeleton was recently shown to be involved during Plasmodium sporozoite invasion (33), but the role of ERM during infection was not investigated. Our data showing that replacement of the cytoplasmic end of CD9P-1, which interacts with ERM, has no effect on the inhibitory effect of CD9P-1, clearly indicate that the association of CD9P-1 with ERM proteins does not contribute to its ability to inhibit infection.
An important finding is the demonstration that a chimeric CD9P-1 molecule not associating with CD81 lacks the ability to inhibit P. yoelii infection. This strongly suggests that CD9P-1 modulates CD81 function during P. yoelii infection. This is consistent with the finding that CD9P-1 expression does not regulate the CD81-independent infection of HepG2 cells by P. berghei. The key role of CD81 was recently strengthened by the demonstration that the effect of SR-BI depletion on P. yoelii infection was, at least in part, a consequence of a decreased CD81 expression (12). This is not the case for CD9P-1 because the effect of CD9P-1 silencing (data not shown) or overexpression (Fig. 4B, middle panel) on infection was observed without any change in CD81 surface expression level. We also considered the hypothesis that CD9P-1 could modify the CD81 membrane environment. Indeed, using a particular CD9 mAb, C9BB that is distinct from other anti-CD9 antibodies in terms of binding affinity and preference for clustered CD9, Yang et al. (34) have shown that the ectopic expression of CD9P-1 and EWI-2 can modify the cell surface CD9 organization. Similarly to this CD9 mAb, MT81w is a mAb that specifically recognizes a fraction of CD81 associated with other tetraspanins and thus can be used as a tool to probe the changes in the organization of CD81 at the cell membrane (11). For example, using MT81w as a marker for tetraspanin-tetraspanin interactions, we provided clear evidence that the membrane cholesterol regulates the organization of cell surface tetraspanins complexes, and we showed a correlation between MT81w binding and infection level (11). We did not observe any changes in the binding of MT81w to Hepa1-6 cells either after CD9P-1 silencing (data not shown) or after its stable overexpression (Fig. 4B, bottom panel). This strongly suggests that the modulation of CD9P-1 expression level does not modify CD81 organization, although we cannot exclude the possibility of changes not detected by MT81w.
Given the ability of tetraspanins to interact with many other integral proteins to functionally modulate these proteins (14–16) and in the absence of evidence that CD81 could be a receptor for sporozoites, one major hypothesis is that CD81 modulates the activity of a partner molecule that may function as a sporozoite receptor. In this context, could CD9P-1 compete with such a putative receptor for association with CD81? This seems unlikely for several reasons. (i) Silencing of CD9P-1 increases infection despite the fact that CD9P-1 is expressed at a much lower level than CD81 and thus associates with only a small fraction of CD81 (see Fig. 1). (ii) We have tested the susceptibility to infection of two sorted populations of CD9P-1-expressing cells differing in their level of CD9P-1 expression by a factor of ∼4. The reduction of infection observed in both populations was partial and very similar (about 30%), arguing against a simple competition phenomenon. (iii) We have demonstrated that the interaction of CD81 with CD9P-1 required the 4th transmembrane region of CD81. However, according to the modeling of Seigneuret (35), this region lies opposite the extracellular region previously determined to play a key role in infection (36). This evidence is rather compatible with the hypothesis that CD9P-1 could form a complex with both CD81 and another still unidentified partner, the function of which would be altered in the presence of CD9P-1. However, as yet, we have failed to collect evidence for the existence of such a putative partner, which could be because of technical limitations.
Relatively few studies have analyzed the regions of tetraspanins involved in the interaction with their molecular partners. The best characterized tetraspanin/partner pair is CD151/integrin α3β1, and several studies have revealed the importance of the CD151 large extracellular domain for this interaction (37, 38). This is consistent with the LEL of tetraspanins being the most variable region of these molecules (14–16). Our results clearly reveal that other regions of tetraspanins contribute to interactions with their partners. Indeed, we have demonstrated that the interaction of CD81 with CD9P-1 relies on the 4th transmembrane domain of CD81 and the transmembrane domain of CD9P-1. Interestingly, CD9P-1, like EWI-2, has a GXXX(G/A/S) motif involved in transmembrane helix-helix association (39). CD81, in contrast to other tetraspanins, does not have such a motif in TM4, but it has an AXXXA motif that mediates α-helix interaction in soluble proteins (40). Further work is required to determine whether these motifs mediate CD81/CD9P-1 interaction. Other tetraspanins may also interact with their partner proteins through transmembrane helix interactions. Indeed, the interaction of CD9 with EWI-2 was shown to be mediated by two independent domains. One is located in the LEL, and the second one is located in the TM2/TM3 region, which is mainly composed of transmembrane regions (20). In addition, the analysis of urothelial plaques, hexagonally packed 16-nm uroplakin particles that cover the urothelial apical surface, by cryo-electron microscopy at 6 Å resolution revealed that the non-tetraspanin uroplakins were in contact with both the LEL and the transmembrane domains (most likely TM3 and/or TM4) of their tetraspanin partners (41).
CD81 is essential for the infection of hepatocytic cells not only by several Plasmodium species but also by the hepatitis C virus. However, the expression of this molecule in non-hepatic cell lines does not lead to HCV entry, suggesting that additional molecule(s) regulate HCV cellular tropism. Very recently, Rocha-Perugini et al. (42) have identified a cleavage product of EWI-2, referred to as EWI-2wint, which associates with CD81 and inhibits its interaction with the HCV envelope glycoproteins. Ectopic expression of this molecule in a hepatoma cell line susceptible to HCV infection blocked viral entry. By contrast, expression of CD9P-1 has no effect on HCV infection.6 This further suggests that CD81 supports Plasmodium and HCV infections through different mechanisms.
Acknowledgments
We thank J. Chaker, E. Giboyau, K. Farhati, and T. Houpert for technical assistance.
This work was supported in part by grants from the Association Nouvelle Recherche Biomédicale-Vaincre le Cancer, the Association pour la Recherche sur le Cancer, the Agence Nationale de Recherche sur le Sida et les Hépatites virales (ANRS), the Agence Nationale de la Recherche (ANR), and by European Union FP5 Contract Number QLK2-CT-2002-00774.
L. Cocquerel, unpublished data.
- EEF
- exo-erythrocytic form
- PV
- parasitophorous vacuole
- LEL
- large extracellular loop
- TM
- transmembrane
- siRNA
- small interfering RNA
- HCV
- hepatitis C virus
- mAb
- monoclonal antibody
- WT
- wild type
- FACS
- fluorescent-activated cell sorting.
REFERENCES
- 1.Guinovart C., Navia M. M., Tanner M., Alonso P. L. (2006) Curr. Mol. Med. 6, 137–140 [DOI] [PubMed] [Google Scholar]
- 2.Prudêncio M., Rodriguez A., Mota M. M. (2006) Nat. Rev. Microbiol. 4, 849–856 [DOI] [PubMed] [Google Scholar]
- 3.Silvie O., Mota M. M., Matuschewski K., Prudêncio M. (2008) Curr. Opin. Microbiol. 11, 352–359 [DOI] [PubMed] [Google Scholar]
- 4.Frevert U. (2004) Trends Parasitol. 20, 417–424 [DOI] [PubMed] [Google Scholar]
- 5.Pradel G., Garapaty S., Frevert U. (2002) Mol. Microbiol. 45, 637–651 [DOI] [PubMed] [Google Scholar]
- 6.Coppi A., Tewari R., Bishop J. R., Bennett B. L., Lawrence R., Esko J. D., Billker O., Sinnis P. (2007) Cell Host. Microbe. 2, 316–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishino T., Chinzei Y., Yuda M. (2005) Mol. Microbiol. 58, 1264–1275 [DOI] [PubMed] [Google Scholar]
- 8.Labaied M., Harupa A., Dumpit R. F., Coppens I., Mikolajczak S. A., Kappe S. H. (2007) Infect. Immun. 75, 3758–3768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Schaijk B. C., Janse C. J., van Gemert G. J., van Dijk M. R., Gego A., Franetich J. F., van, de, Vegte-Bolmer M., Yalaoui S., Silvie O., Hoffman S. L., Waters A. P., Mazier D., Sauerwein R. W., Khan S. M. (2008) PLoS. ONE. 3, e3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silvie O., Rubinstein E., Franetich J. F., Prenant M., Belnoue E., Rénia L., Hannoun L., Eling W., Levy S., Boucheix C., Mazier D. (2003) Nat. Med. 9, 93–96 [DOI] [PubMed] [Google Scholar]
- 11.Silvie O., Charrin S., Billard M., Franetich J. F., Clark K. L., van Gemert G. J., Sauerwein R. W., Dautry F., Boucheix C., Mazier D., Rubinstein E. (2006) J. Cell Sci. 119, 1992–2002 [DOI] [PubMed] [Google Scholar]
- 12.Yalaoui S., Huby T., Franetich J. F., Gego A., Rametti A., Moreau M., Collet X., Siau A., van Gemert G. J., Sauerwein R. W., Luty A. J., Vaillant J. C., Hannoun L., Chapman J., Mazier D., Froissard P. (2008) Cell Host Microbe 4, 283–292 [DOI] [PubMed] [Google Scholar]
- 13.Rodrigues C. D., Hannus M., Prudêncio M., Martin C., Gonçalves L. A., Portugal S., Epiphanio S., Akinc A., Hadwiger P., Jahn-Hofmann K., Röhl I., van, Gemert G. J., Franetich J. F., Luty A. J., Sauerwein R., Mazier D., Koteliansky V., Vornlocher H. P., Echeverri C. J., Mota M. M. (2008) Cell Host Microbe 4, 271–282 [DOI] [PubMed] [Google Scholar]
- 14.Charrin S., Le Naour F., Silvie O., Milhiet P. E., Boucheix C., Rubinstein E. (2009) Biochem. J. 420, 133–154 [DOI] [PubMed] [Google Scholar]
- 15.Hemler M. E. (2005) Nat. Rev. Mol. Cell. Biol. 6, 801–811 [DOI] [PubMed] [Google Scholar]
- 16.Levy S., Shoham T. (2005) Nat. Rev. Immunol. 5, 136–148 [DOI] [PubMed] [Google Scholar]
- 17.Cocquerel L., Voisset C., Dubuisson J. (2006) J. Gen. Virol. 87, 1075–1084 [DOI] [PubMed] [Google Scholar]
- 18.Bartosch B., Cosset F. L. (2006) Virology 348, 1–12 [DOI] [PubMed] [Google Scholar]
- 19.Charrin S., Le Naour F., Oualid M., Billard M., Faure G., Hanash S. M., Boucheix C., Rubinstein E. (2001) J. Biol. Chem. 276, 14329–14337 [DOI] [PubMed] [Google Scholar]
- 20.Charrin S., Le Naour F., Labas V., Billard M., Le Caer J. P., Emile J. F., Petit M. A., Boucheix C., Rubinstein E. (2003) Biochem. J. 373, 409–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stipp C. S., Kolesnikova T. V., Hemler M. E. (2001) J. Biol. Chem. 276, 40545–40554 [DOI] [PubMed] [Google Scholar]
- 22.Stipp C. S., Orlicky D., Hemler M. E. (2001) J. Biol. Chem. 276, 4853–4862 [DOI] [PubMed] [Google Scholar]
- 23.Espenel C., Margeat E., Dosset P., Arduise C., Le Grimellec C., Royer C. A., Boucheix C., Rubinstein E., Milhiet P. E. (2008) J. Cell Biol. 182, 765–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barreiro O., Zamai M., Yáñez-Mó M., Tejera E., López-Romero P., Monk P. N., Gratton E., Caiolfa V. R., Sánchez-Madrid F. (2008) J. Cell Biol. 183, 527–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soneoka Y., Cannon P. M., Ramsdale E. E., Griffiths J. C., Romano G., Kingsman S. M., Kingsman A. J. (1995) Nucleic Acids Res. 23, 628–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bartosch B., Vitelli A., Granier C., Goujon C., Dubuisson J., Pascale S., Scarselli E., Cortese R., Nicosia A., Cosset F. L. (2003) J. Biol. Chem. 278, 41624–41630 [DOI] [PubMed] [Google Scholar]
- 27.Mota M. M., Rodriguez A. (2000) Exp. Parasitol. 96, 257–259 [DOI] [PubMed] [Google Scholar]
- 28.Serru V., Le Naour F., Billard M., Azorsa D. O., Lanza F., Boucheix C., Rubinstein E. (1999) Biochem. J. 340, 103–111 [PMC free article] [PubMed] [Google Scholar]
- 29.Silvie O., Greco C., Franetich J. F., Dubart-Kupperschmitt A., Hannoun L., van Gemert G. J., Sauerwein R. W., Levy S., Boucheix C., Rubinstein E., Mazier D. (2006) Cell Microbiol. 8, 1134–1146 [DOI] [PubMed] [Google Scholar]
- 30.Silvie O., Franetich J. F., Boucheix C., Rubinstein E., Mazier D. (2007) Int. J. Parasitol. 37, 173–182 [DOI] [PubMed] [Google Scholar]
- 31.Sala-Valdés M., Ursa A., Charrin S., Rubinstein E., Hemler M. E., Sánchez-Madrid F., Yáñez-Mó M. (2006) J. Biol. Chem. 281, 19665–19675 [DOI] [PubMed] [Google Scholar]
- 32.Charrin S., Alcover A. (2006) Front. Biosci. 11, 1987–1997 [DOI] [PubMed] [Google Scholar]
- 33.Gonzalez V., Combe A., David V., Malmquist N. A., Delorme V., Leroy C., Blazquez S., Ménard R., Tardieux I. (2009) Cell Host Microbe 5, 259–272 [DOI] [PubMed] [Google Scholar]
- 34.Yang X. H., Kovalenko O. V., Kolesnikova T. V., Andzelm M. M., Rubinstein E., Strominger J. L., Hemler M. E. (2006) J. Biol. Chem. 281, 12976–12985 [DOI] [PubMed] [Google Scholar]
- 35.Seigneuret M. (2006) Biophys. J. 90, 212–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yalaoui S., Zougbédé S., Charrin S., Silvie O., Arduise C., Farhati K., Boucheix C., Mazier D., Rubinstein E., Froissard P. (2008) PLoS. Pathog. 4, e1000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kazarov A. R., Yang X., Stipp C. S., Sehgal B., Hemler M. E. (2002) J. Cell Biol. 158, 1299–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berditchevski F., Gilbert E., Griffiths M. R., Fitter S., Ashman L., Jenner S. J. (2001) J. Biol. Chem. 276, 41165–41174 [DOI] [PubMed] [Google Scholar]
- 39.Curran A. R., Engelman D. M. (2003) Curr. Opin. Struct. Biol. 13, 412–417 [DOI] [PubMed] [Google Scholar]
- 40.Kleiger G., Grothe R., Mallick P., Eisenberg D. (2002) Biochemistry 41, 5990–5997 [DOI] [PubMed] [Google Scholar]
- 41.Min G., Wang H., Sun T. T., Kong X. P. (2006) J. Cell Biol. 173, 975–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rocha-Perugini V., Montpellier C., Delgrange D., Wychowski C., Helle F., Pillez A., Drobecq H., Le Naour F., Charrin S., Levy S., Rubinstein E., Dubuisson J., Cocquerel L. (2008) PLoS ONE 3, e1866. [DOI] [PMC free article] [PubMed] [Google Scholar]