Abstract
Poly(ADP-ribose)polymerase-1 (PARP-1) is a predominantly nuclear enzyme that exerts numerous functions in cellular physiology and pathology, from maintenance of DNA stability to transcriptional regulation. Through a proteomic analysis of PARP-1 co-immunoprecipitation complexes, we identified Mitofilin, a mitochondrial protein, as a new PARP-1 interactor. This result prompted us to further investigate the presence and the role of the enzyme in mitochondria. Using laser confocal microscopy and Western blot analysis of purified mitochondria, we demonstrated the mitochondrial localization of a fraction of PARP-1. Further, the effects of overexpressing or down-regulating Mitofilin showed that this protein promotes and is required for PARP-1 mitochondrial localization. We also report several lines of evidence suggesting that intramitochondrial PARP-1 plays a role in mitochondrial DNA (mtDNA) damage signaling and/or repair. First, we show that PARP-1 binds to different regions throughout the mtDNA. Moreover, we demonstrated that the depletion of either PARP-1 or Mitofilin, which abrogates the mitochondrial localization of the enzyme, leads to the accumulation of mtDNA damage. Finally, we show that DNA ligase III, known to be required for mtDNA repair, participates in a PARP-1-containing complex bound to mtDNA. This work highlights a new environment for PARP-1, opening the possibility that at least some of the nuclear functions of the enzyme can be also extended to mtDNA metabolism.
Introduction
Poly(ADP-ribose)polymerase-1 (PARP-1)5 is the best characterized member of a 17-protein family. PARP-1 has a highly conserved structural and functional organization including an N-terminal zinc finger DNA-binding domain, a nuclear localization signal, a central automodification domain, a Brca1 C-terminal domain involved in protein-protein interactions, and a C-terminal catalytic domain (1). PARP-1 catalyzes the polymerization of ADP-ribose units from donor NAD+ on acceptor proteins, leading to the formation of linear or branched polymers of ADP-ribose (PAR) (2). This post-translational modification can change the physico-chemical properties of PARP-1 targets, among which are PARP-1 itself and a variety of nuclear proteins involved in DNA metabolism and chromatin structure (2).
PARP-1 plays a key role in different cellular processes, such as DNA damage signaling (3), mitotic checkpoints (4), transcriptional regulation (5), cell death pathways (6), and cell proliferation (7, 8). The best studied role of PARP-1 is as a DNA damage sensor because its activity is strongly stimulated by its binding to damaged DNA (1). PARP-1 can recognize both single and double strand breaks (2), and the PAR synthesis that follows facilitates the initiation of DNA repair at DNA damage sites. This process involves both the recruitment of repair proteins, which bind to automodified PARP-1 (9, 10), and the modification of a number of DNA-associated proteins, such as histones, topoisomerases, and DNA polymerases (11).
It has long been questioned whether PARP-1, a predominantly nuclear enzyme, is also localized in other cell compartments, in particular in mitochondria (12). Mitochondria contain a significant proportion of the cellular NAD+, the substrate for PARP activity (13). Furthermore, increasing evidence indicates that mitochondrial DNA, which is constantly exposed to oxidative damage, is efficiently repaired through at least a subset of the mechanisms involved in nuclear DNA repair (14). A number of previous works reported the occurrence of intramitochondrial poly(ADP-ribosyl)ation (15–18). Moreover, it has been recently established that PARP-1 hyperactivation can cause the release of apoptosis-inducing factor (AIF) from mitochondria, triggering a caspase-independent cell death pathway (19). However, despite the clear involvement of PAR signaling in mitochondrial dysfunction, it is still debated whether these effects are mediated by a mitochondrially localized PARP-1. In fact, although some studies provided evidence for the presence of PARP-1 in mitochondria (18, 20, 21), others failed to detect the enzyme in this organelle, thus suggesting that the mitochondrial effects of PARP-1 activation are triggered by the nuclear enzyme (6, 22, 23).
In the course of a proteomic analysis of PARP-1 co-immunoprecipitating proteins, we identified Mitofilin, a transmembrane protein of the inner mitochondrial membrane (24, 25), as a new PARP-1 interactor. Although its function has not been extensively investigated, Mitofilin has been suggested to control mitochondrial cristae organization (26) and to participate in some mechanism of mitochondrial import related to maintenance of mitochondrial structure (27). In this work, we report that PARP-1 interacts with Mitofilin within mitochondria and that the mitochondrial localization of PARP-1 is regulated by the presence and abundance of Mitofilin. Moreover, we show that PARP-1 is associated with mitochondrial DNA, participating in a DNA ligase III-containing complex, and is involved in the maintenance of mitochondrial DNA integrity.
EXPERIMENTAL PROCEDURES
Cell Culture
Human diploid FB 1329 fibroblasts and HeLa cells were grown in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (Cambrex), 2 mm l-glutamine, 1% penicillin/streptomycin solution (v/v). When indicated, cells were treated with H2O2 400 μm for 1 h in Dulbecco's modified Eagle's medium without fetal bovine serum.
Western Blot
For Western blot analysis, total cellular extraction was performed in ice-cold RIPA with protease inhibitors, whereas mitochondrial extracts were obtained as described below. Proteins were resolved by electrophoresis in SDS-PAGE and transferred to nitrocellulose membranes by electroblotting. Membranes were blocked in 1% nonfat dry milk in phosphate-buffered saline containing 0.05% Tween 20 for 1 h at room temperature and incubated with the primary antibody overnight at 4 °C, and then membranes were washed three times and incubated in a 1:10,000 or 1:20,000 dilution peroxidase-conjugated anti-mouse or anti-rabbit antibodies (Bio-Rad). Proteins were detected using the ECL chemiluminescence system (Pierce).
The following primary antibodies used for Western blot were purchased from Santa Cruz Biotechnology: PARP-1 H-250 (sc-7150), Sp-1 (sc-59), and α-tubulin TU-02 (sc-8035). The following primary antibodies used for Western blot were purchased from Alexis Biochemicals: monoclonal anti-PARP-1 (C2-10) and monoclonal anti-mtHsp70 (JG1). The monoclonal anti-Mitofilin was from Mitoscience, the polyclonal anti-Mitofilin (PA1-16918) was from Affinity BioReagents, and the polyclonal anti-AIF (2267) was from ProSci Inc. The monoclonal anti DNA ligase III (6G9) was from Novus Biologicals.
Immunoprecipitation Assay
Immunoprecipitation experiments were performed from 1 mg of cellular lysates in RIPA or from 200 μg of mitochondrial lysates using 1 μg of primary antibody. Samples were incubated with protein A/G-Sepharose (Amersham Biosciences) for 3 h at 4 °C on a rotating platform, and then the precleared samples were incubated with monoclonal (F1-23) or polyclonal (ALX-210-302 PARP-1 antibodies or polyclonal Mitofilin antibodies (all from Alexis Biochemicals) overnight with gentle rotation at 4 °C. Protein A/G-Sepharose was added, and samples were rocked again for 3–5 h prior to three washes with RIPA. Finally, the immunoprecipitated proteins were eluted with 1× Laemmli buffer and collected for Western blot analysis.
Mass Spectrometry (MS)
Large scale immunoprecipitated proteins were resolved on a 12% T-3.3% C SDS-PAGE separating gel (1 + 18 + 18 mm), revealed by SYPRO Ruby staining and visualized using a Typhoon 9200 laser scanner (GE Healthcare). Proteins were excised and digested with trypsin as described previously (28). Proteins were identified by matrix-assisted laser-desorption ionization (MALDI)-MS and MALDI-MS/MS (4700 Proteomics Analyzer; Applied Biosystems). Data were acquired in positive MS reflector mode. Five peptides (ABI4700 Calibration Mixture; Applied Biosystems) were used as calibration standards. Mass spectra were obtained from each sample by 30 subspectra accumulations (50 laser shots each) in a 750–4000 mass range. Five signal-to-noise best peaks of each spectrum were selected for MS/MS analysis. For MS/MS spectra, the collision energy was 1 keV, and the collision gas was air. The interpretations of both the MS and the MS/MS data were carried out by using the GPS Explorer software (Version 1.1, Applied Biosystems), which acts as an interface between the Oracle data base containing raw spectra and a local copy of the MASCOT search engine (Version 1.8). Peptide mass fingerprints obtained from MS analysis were used for protein identification in the Swiss-Prot non-redundant data base. All peptides mass values were considered monoisotopic, and mass tolerance was set at 30 ppm. One missed cleavage site was allowed, cysteines were considered carboamidomethylated, methionine was assumed to be partially oxidized, and serine, threonine, and tyrosine were assumed to be partially phosphorylated. Mascot (Matrix Science) scores greater than 65 were considered significant (p < 0.05). For MS/MS analysis, all peaks with a signal-to-noise ratio greater than 5 were searched against the NCBI data base using the same modifications as the MS data base, with a fragment tolerance less than 0.3 Da.
Confocal Analysis
For the immunofluorescence analysis, a Leica confocal microscope (laser-scanning TCS SP2) equipped with Ar/ArKr and HeNe lasers was utilized. After the treatments, cells were fixed in 30% methyl alcohol, 70% acetone and stained with PARP-1 (F1-23, Alexis Biochemicals), Mitofilin (Affinity BioReagents), or AIF (ProSci Inc.) as primary antibodies. Anti-mouse-Alexa Fluor 488 fluorescein dye and anti-rabbit Texas Red 594 rhodamine dye (both from Alexa Fluor, Invitrogen) were excited by laser light at 488-and 543-nm wavelength, respectively. The images were acquired utilizing the Leica confocal software. The images were scanned under a ×40 oil immersion objective. To avoid bleed-through effects, each dye was scanned independently. For acquisition of double immunofluorescence from transfected cells, optical spatial series, each composed of 10 optical sections with a step size of 1 μm, were performed. Images were electronically merged using the Leica confocal software and stored as TIFF files. Figures were assembled from the TIFF files using Adobe PhotoShop software.
The intensity correlation analysis of confocal images was performed using WCIF ImageJ software (National Institutes of Health, Bethesda, MD). At least five cytoplasmic areas for each experiment were analyzed, and the main values of Pearson's and Mander's coefficients were reported.
Gradient Purification
Mitochondria purification was performed as described before (29). Briefly, mitochondrial extracts were prepared from the cells (5 × 107) washed in a buffer containing 135 mm NaCl, 5 mm KCl, 25 mm Tris-Cl, pH 7.6 and allowed to swell for 10 min in ice-cold hypotonic calcium reticulocyte standard buffer (10 mm NaCl, 1.5 mm CaCl2, 10 mm Tris-HCl, pH 7.5, plus protease inhibitors). Cells were Dounce-homogenized with 70 strokes, with the addition of mannitol/sucrose (MS) buffer (210 mm mannitol, 70 mm sucrose, 5 mm Tris, pH 7.6) to stabilize mitochondria (2 ml of 2.5× for 3 ml of homogenate). Nuclear contaminants were removed by centrifugation at 600 × g for 15 min on ice. The supernatants were centrifuged at 6000 × g for 10 minutes. The pellet, corresponding to a mitochondrial enriched fraction, was suspended in RIPA and used for PARP-1-DNA ligase III complex-immunoprecipitation or suspended in 2 ml of MS buffer 1× and layered over a 1–1.5 m sucrose step gradient (10 mm Tris, 5 mm EDTA, pH 7.6, 2 mm dithiothreitol, plus protease inhibitors) for the purification of mitochondria. This gradient was centrifuged at 110,000 × g for 30 min at 4 °C. Mitochondria sediment at the interphase between 1 and 1.5 m sucrose. Fractions were collected from the gradient and subjected to dot blot analysis. The mitochondria-containing fractions were collected and analyzed for Western blot or used for immunoprecipitation of mitochondrial PARP-1.
siRNA and Overexpression Assay
siRNA transfection was performed with a mix containing 7 μl of Lipofectamine 2000 reagent (Invitrogen) plus 130 nm of siRNA oligonucleotides for Mitofilin and PARP-1. After 72 h, cells were fixed for immunofluorescence analysis and collected for Western blot or for direct PCR (DPCR). The siRNA oligonucleotide against human PARP-1 (30) was: 5′-AAG ATA GAG CGT GAA GGC GAA-3′ (nucleotides 2671–2691). The siRNA oligonucleotide against human Mitofilin (26) was: 5′-AAT TGC TGG AGC TGG CCT TTT-3′.
For Mitofilin overexpression, HeLa cells were seeded in a 35-mm dish and then transfected with a mix containing 10 μl of Lipofectamine 2000 reagent and 4 μg of pCMV6-XL5/Mitofilin purchased from OriGene. After 48 h, cells were fixed for immunofluorescence analysis and collected for Western blot.
DPCR
For DPCR HeLa cells were collected by trypsin treatment and centrifuged at 2000 × g for 5 min. The pellet was washed twice and suspended in phosphate-buffered saline to count cells. Amplification products were obtained by PCR of 103 and 5 × 102 whole cells using an Expand high fidelity PCR kit (Roche Applied Science) following the previously published method (31). 300 nm of the L2 (5′-GCC CGT ATT TAC CCT ATA GC-3′) and H3 (5′-GTC TAG GGC TGT TAG AAG TC-3′) primers (32) was used to amplify the target 5595-bp product of the human mitochondrial genome. Template, primers, and 5 μl of 10× buffer and distilled H2O were mixed and heated for 7 min at 98 °C. The mixture was immediately placed on ice, at which time the remaining reagents were added. The PCR cycling profile was a pre-PCR incubation at 94 °C for 2 min and then 10 cycles of 94 °C for 15 s, 50 °C for 30 s, and 68 °C for 4 min, which was followed by 15 cycles of 94 °C for 15 s, 50 °C for 30 s, and 68 °C for 4 min, with a 20-s extension per cycle. PCRs were performed in triplicate for each sample and for three independent experiments. As a control, a 157-bp fragment located in the COXII gene (nucleotides 8465–8621) was amplified with the following primers designed by Primer3 (33): COX, forward, 5′-CAC CTA CCT CCC TCA CCA AA-3′, and COX, reverse, 5′-GGG ATC AAT AGA GGG GGA AA-3′. The PCR cycling profile was: a treatment of 7 minutes at 98 °C, pre-PCR of 2 minutes at 94 °C, then 25 cycles of 94 °C for 15 s, 54 °C for 30 s, and 72 °C for 30 s.
Chromatin Immunoprecipitation (ChIP) and Sequential Chromatin Immunoprecipitation
Cells were treated with formaldehyde (1% final concentration), added directly to the culture dishes, to cross-link protein complexes to the DNA. The reaction was stopped by adding glycine to a final concentration of 0.125 m for 5 min at room temperature. Cells were washed with cold phosphate-buffered saline, scraped, lysed in 5 mm EDTA, 50 mm Tris-HCl (pH 8.0), SDS 1% supplemented with protease and phosphatase inhibitors, and rocked on ice for 10 min. Total lysates were sonicated to obtain chromatin fragments of an average length of 200–800 bp and centrifuged at 10,000 rpm for 10 min at 4 °C. The sonicated supernatant fractions were diluted 10-fold with dilution buffer (5 mm EDTA, 50 mm Tris-HCl (pH 8.0), 0.5% Nonidet P-40, 200 mm NaCl supplemented with protease and phosphatase inhibitors). After determining the DNA concentrations, 150 μg of chromatin was incubated with protein A/G-Sepharose for 3 h at 4 °C on a rotating platform. Saturation with salmon sperm was avoided due to its high content of mitochondrial DNA. The precleared chromatin samples were centrifuged at 14,000 rpm for 5 min and incubated with PARP-1 or DNA ligase III antibodies or without antibody overnight with gentle rotation at 4 °C. Before washing, an aliquot of the supernatant of the no antibody control was taken as input sample. After washing with a low salt wash buffer (0.1% SDS, 1% Triton, 2 mm EDTA, 20 mm Tris-HCl, pH 8.0, 150 mm NaCl), high salt wash buffer (0.1% SDS, 1% Triton, 2 mm EDTA, 20 mm Tris-HCl, pH 8.0, 500 mm NaCl), LiCl wash buffer (1% Nonidet P-40, 1 mm EDTA, 10 mm Tris-HCl, pH 8.0, 250 mm LiCl, 1% sodium deoxycholate), and Tris-EDTA 1× buffer (10 mm Tris-Cl, pH 7.4, and 1 mm EDTA), pellets were dissolved in 300 μl of elution buffer (1% SDS, 0.1 m NaHCO3). In the sequential chromatin immunoprecipitation experiments, pellets were eluted with 50 μl of 10 mm dithiothreitol, 20-fold diluted in ReChIP buffer (1% Triton, 2 mm EDTA, 20 mm Tris-HCl (pH 8.0), 150 mm NaCl), and immunoprecipitated with the second antibody. The samples treated with RNase A for 10 min at room temperature were incubated at 67 °C overnight to reverse the protein-DNA cross-linking. Then NaHCO3 was neutralized with 6 μl of Tris-HCl 1 m (pH 6–7.5). After treatment with proteinase K, the DNA was extracted with phenol-chloroform, precipitated with isopropyl alcohol, and suspended in distilled water. The following primers were used for amplification a 284-bp (nucleotides 16,248–16,544) fragment of the D-loop region (34) (d-loop forward, 5′-CCC CTC ACC CAC TAG GAT AC-3′, and d-loop, reverse, 5′-ACG TGT GGG CTA TTT AGG C-3′); a 157-bp fragment located in the COXII gene and a 225-bp fragment in the ND2 gene (nucleotides 4563–4777); and ND2, forward, 5′-GCC CTA GAA ATA AAC ATG CTA-3′, and ND2, reverse, 5′-GGG CTA TTC CTA GTT TTA TT-3′ (35).
RESULTS
PARP-1 Interacts with the Mitochondrial Protein Mitofilin
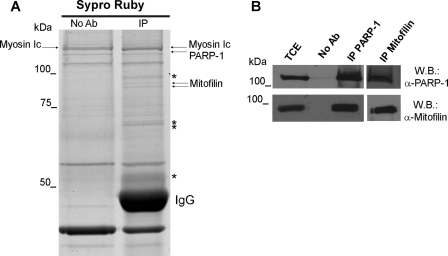
Searching for new PARP-1-interacting proteins, we carried out the immunoprecipitation of endogenous PARP-1 from whole cell extracts of human diploid fibroblasts. PARP-1 was specifically immunoprecipitated utilizing different antibodies (see “Experimental Procedures”). Unexpectedly, only a few bands were detectable after immunoprecipitation with the monoclonal antibody F1-23. Instead, a complex protein profile, consistent with the ability of PARP-1 to interact with a variety of partners (5), was revealed by the polyclonal antibody (data not shown). This finding, likely explained by the ability of the monoclonal antibody to select for a few specific interactions, gave us the chance to undertake the identification of selected PARP-1 interactors. The immunoprecipitated complex and the relative negative control were resolved by SDS-PAGE, visualized by Sypro-Ruby staining, and analyzed by MALDI-TOF/TOF (tandem time-of-flight) mass spectrometry. In particular, a combined approach using peptide mass fingerprint and tandem mass spectrometry was performed to identify the interacting proteins (Fig. 1A). Through this analysis, a doublet of near 85 kDa was recognized as the two previously described isoforms of the mitochondrial inner membrane protein Mitofilin (25) (supplemental Table 1). The ability of PARP-1 and Mitofilin to interact with each other was validated by immunoprecipitation/Western blot analysis. The use of a Mitofilin-specific antibody confirmed the presence of the protein in PARP-1 immunoprecipitates with both monoclonal antibodies (Fig. 1B) and polyclonal antibodies (data not shown). Moreover, as shown in the same figure, the reverse immunoprecipitation revealed the presence of PARP-1 in Mitofilin immunoprecipitates, thus proving the in vivo association of the two proteins.
FIGURE 1.
Interaction of PARP-1 with Mitofilin. A, PARP-1 immunoprecipitation was performed from human fibroblasts FB1329 using the monoclonal antibody F1-23 and SYPRO Ruby staining for proteomic analysis of co-immunoprecipitated proteins. Arrows indicate the protein identified by mass spectrometry. Bands marked with asterisks were identified as degradation products of IgG. No Ab, total cellular extracts incubated with protein A/G-Sepharose without antibody; IP, immunoprecipitation with the corresponding antibodies. B, PARP-1 and Mitofilin Western blots (W.B.) on PARP-1 and Mitofilin immunoprecipitates. TCE, total cellular extract.
PARP-1 and Mitofilin Interact within Mitochondria
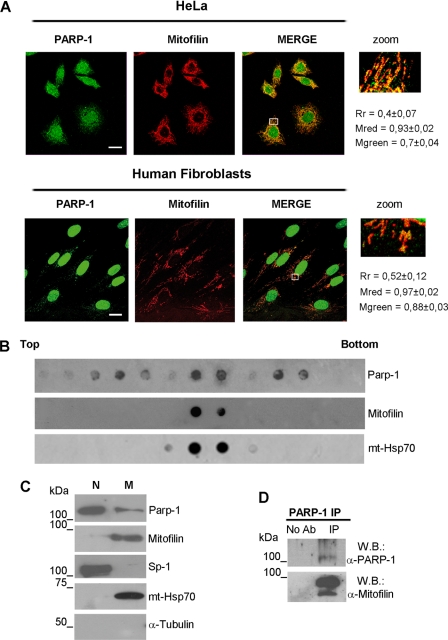
The identification of a mitochondrial protein as a PARP-1 interactor prompted us to investigate whether PARP-1 and Mitofilin co-localize into mitochondria. For this purpose, we performed double immunofluorescence staining and confocal laser analysis of the two proteins both in HeLa cells and in human diploid fibroblasts. Representative images are reported in Fig. 2A. PARP-1 (visualized in green) shows, as expected, the maximum of fluorescence in the nuclei but also a less intense extranuclear staining. In both cell types, the Mitofilin fluorescence (stained in red) appeared with the rod-shaped feature that is typical of mitochondrial morphology. The merged images show that the majority of extranuclear PARP-1 co-localizes with Mitofilin (orange-yellow fluorescence). Additional co-immunostaining analysis (supplemental Fig. 1) revealed that the enzyme also co-localizes with the mitochondrial transcription factor A (36). These results strongly support the mitochondrial localization of a fraction of PARP-1. To better investigate this issue, we analyzed the presence of PARP-1 in purified mitochondria. Human fibroblasts were lysed by hypotonic buffer, and the cytoplasmic fraction was separated from nuclei by differential centrifugation. Subsequently, mitochondria were purified by density sedimentation with a two-step sucrose gradient. The gradient was fractionated and analyzed by dot blot with antibodies against Mitofilin or PARP-1 to identify the mitochondria-containing fractions and the enzyme distribution. mtHsp70 was used as a further mitochondrial marker. As shown in Fig. 2B, Mitofilin was strictly located in the two mitochondrial fractions, whereas PARP-1 showed a distribution defining three peaks, one of which coincided with the two Mitofilin-containing fractions. The two PARP-1-positive/Mitofilin-negative peaks likely reflect the association of PARP-1 with other cytoplasmic structures. This hypothesis is consistent with the observation that a small proportion of the cytoplasmic enzyme shows an extramitochondrial localization, as also observed in Fig. 2A. The two PARP-1-positive fractions that were also positive to Mitofilin were pooled together and analyzed by Western blot, in parallel with the nuclear samples obtained from the cellular lysis. As shown in Fig. 2C, this assay confirmed the presence of PARP-1 in mitochondria. The analysis of subcellular markers indicated that the mitochondrial fraction, positive for mtHsp70 and Mitofilin, was negative for Sp1 and α-tubulin, used as nuclear and cytoplasmic markers, respectively, thus allowing us to exclude the contamination with other cellular compartments. Finally, we performed PARP-1 immunoprecipitation from purified mitochondria, and the Western blot analysis for the presence of Mitofilin revealed a strong signal, further confirming that just the mitochondrial PARP-1 is interacting with Mitofilin (Fig. 2D).
FIGURE 2.
PARP-1 interacts with Mitofilin within mitochondria. A, confocal laser scanning microscopy of double immunofluorescence staining performed on HeLa (upper panel) or FB 1329 (lower panel) cells. Fixed cells were labeled with α-PARP-1 (green) and α-Mitofilin (red). Bars indicate 20 μm. Rectangle-marked areas of each merge were enlarged in the zoom panels. Cytoplasmic co-localization was measured as described under “Experimental Procedures.” Rr, Pearson's correlation coefficient; Mred, Mander's co-localization coefficient for red; Mgreen, Mander's co-localization coefficient for green. B, mitochondria were purified from FB 1329 cells by a two-step sucrose gradient. The distribution of PARP-1, Mitofilin, and mtHsp70 in the gradient fractions was analyzed with the corresponding antibodies by dot blot. C, the nuclear fraction (N), obtained from differential centrifugation, and the mitochondrial fraction (M), obtained after sucrose gradient, were analyzed by Western blot with the listed antibodies. D, Mitofilin and PARP-1 Western blot (W.B.) on PARP-1 immunoprecipitates obtained from purified mitochondria. No Ab, mitochondrial extracts incubated with protein G-Sepharose without antibody; IP, immunoprecipitation with α-PARP-1 antibody.
Mitofilin Regulates the Mitochondrial Localization of PARP- 1
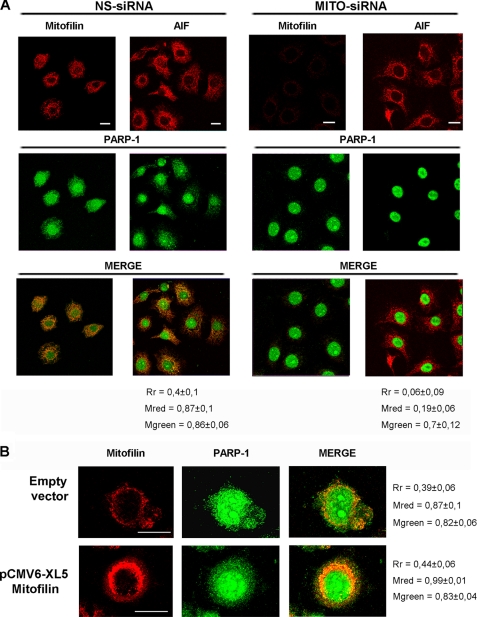
It has been previously shown that Mitofilin associates with outer membrane proteins implicated in mitochondrial import (27). In light of this finding, we asked whether the interaction of PARP-1 with Mitofilin could be relevant in regulating its mitochondrial localization. To investigate this hypothesis, we first analyzed the effects of Mitofilin knockdown on PARP-1 localization. HeLa cells were transfected with siRNA specific for Mitofilin or with nonspecific siRNA as a control. Western blot on total cellular lysates, collected 72 h after transfection, confirmed a good efficiency of Mitofilin depletion (more than 10-fold decrease; supplemental Fig. 2). To investigate the PARP-1 localization, we analyzed siRNA-transfected cells by double fluorescence staining and confocal laser microscopy (Fig. 3A). We used AIF as a mitochondrial marker in the absence of Mitofilin. Remarkably, cells in which Mitofilin was silenced presented a clear difference in the PARP-1 localization. Indeed, the extranuclear PARP-1 was significantly reduced, and its co-localization with AIF was suppressed, indicating a loss of mitochondrial localization for PARP-1. It is important to notice that the mitochondrial localization of AIF is preserved, thus allowing us to exclude a general alteration in mitochondrial trafficking.
FIGURE 3.
Mitofilin is required for mitochondrial localization of PARP-1. A, HeLa cells were transfected with Mitofilin-specific (MITO-siRNA) or with nonspecific (NS-siRNA) siRNAs. After 72 h, cells were fixed for α-PARP-1/α-Mitofilin or α-PARP-1/α-AIF double immunofluorescence and confocal laser scanning analysis. AIF staining was shown as a mitochondrial marker in the absence of Mitofilin. B, HeLa cells were transfected with the pCMV6-XL5 vector containing the full-length human Mitofilin cDNA (OriGene) or with the empty vector. After 48 h, cells were fixed and labeled with α-PARP-1 and α-Mitofilin for confocal laser scanning microscopy. Bars indicate 20 μm. Cytoplasmic co-localization was measured as described under “Experimental Procedures.” Rr, Pearson's correlation coefficient; Mred, Mander's ccoefficient for red; Mgreen, Mander's co-localization coefficient for green.
To obtain further evidence of the role of Mitofilin in regulating PARP-1 mitochondrial localization, we analyzed the effect of Mitofilin overexpression. HeLa cells were transfected with a pCMV6-XL5 vector containing the human full-length Mitofilin cDNA. Also, in this case, the protein levels were determined by Western blot on total cellular lysates (supplemental Fig. 3), confirming a significant increase of Mitofilin protein levels (about 2.5-fold) when compared with control cells. As reported in Fig. 3B, the analysis of double immunofluorescence-stained cells by confocal laser microscopy clearly showed that Mitofilin overexpression causes an increase of PARP-1 in the extranuclear compartment. Taken together, these results demonstrated that the localization of PARP-1 in mitochondria is dependent on the presence and abundance of Mitofilin.
Mitochondrial PARP-1 Is Necessary for mtDNA Integrity
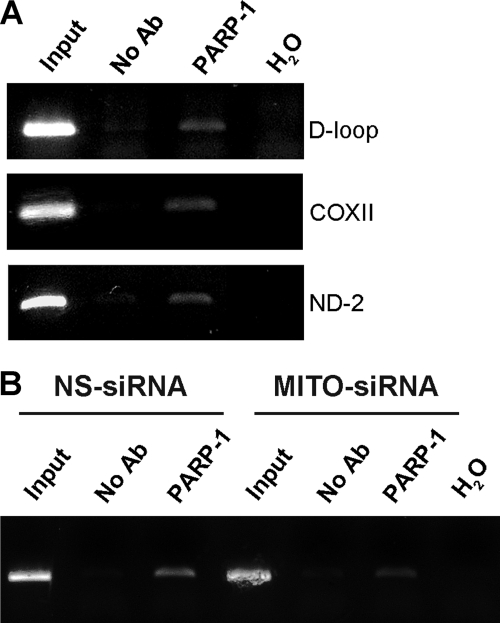
On the basis of the well established nuclear functions of the enzyme, we hypothesized a possible role for mtPARP-1 in mitochondrial DNA (mtDNA) metabolism. At first, we investigated whether PARP-1 was associated with mtDNA. To this end, we employed a modified ChIP assay (see “Experimental Procedures”). DNA, extracted from PARP-1 immunoprecipitates and control samples, was analyzed by PCR with primer sets specific for three different fragments of mtDNA: one comprised in the regulatory D-loop region, another in the ND2 gene, and another one in COXII gene. As shown in Fig. 4A, the amplification of all the fragments in the PARP-1 cross-linked sample indicated that the enzyme contacts mtDNA at least in three different regions. As expected, this binding is significantly reduced after Mitofilin knockdown, which prevents the mitochondrial localization of PARP-1 (Fig. 4B). This finding, denoting that PARP-1 binds to both regulatory and coding sequences throughout the mitochondrial genome, suggests that the enzyme may exert a global role in mtDNA organization and/or stability.
FIGURE 4.
Mitochondrial PARP-1 is associated to mitochondrial DNA. A, PCR analysis after chromatin immunoprecipitation with an antibody against PARP-1 showed the amplification of the desired product size in a different region of the mtDNA. DNA sample lane descriptions are as follows. Input, samples isolated from total lysates prior to antibody pulldown as an internal positive control; No Ab, samples isolated after pull down with no antibody; PARP-1, samples isolated after PARP-1 pull down; H2O, PCR negative control. B, HeLa cells were transfected with Mitofilin siRNA or control siRNA and analyzed as in A. The densitometric analysis of band intensity (ImageJ software) indicated that immunoprecipitation of Mitofilin-specific siRNA (MITO-siRNA)/nonspecific (NS-siRNA) siRNA = 0.5.
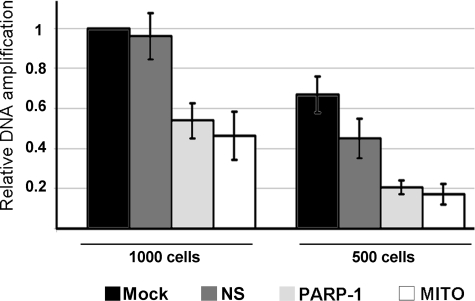
mtDNA is prone to accumulate damage due to the lack of protective histones and to the presence of high levels of reactive oxygen species (37). To explore the hypothesis that PARP-1 was required for the maintenance of mtDNA integrity, we examined the effects of depleting the enzyme or preventing its mitochondrial localization through Mitofilin knockdown. The mtDNA state was analyzed by DPCR from whole cell lysates. This technique, which avoids the damaging manipulation of DNA during isolation (31), allows us to detect DNA damage by examining the efficiency of Taq/Tgo DNA polymerase mix in amplifying long fragments of mtDNA. Decreased amplification of long fragments indicates accumulation of DNA damage that causes polymerase stalling (31, 32). In particular, we amplified two mtDNA fragments corresponding, respectively, to a 5-kbp region extending from the ATPase 8 gene to the ND5 gene or to a 157-bp region in the COXII gene (to normalize the mtDNA). We performed DPCR on cells transfected with PARP-1 or Mitofilin siRNAs. As shown in Fig. 5, PARP-1 knockdown results in a significantly decreased amplification of the 5-kbp fragment, indicative of DNA damage accumulation, within 3 days from transfection. A similar effect is evident when Mitofilin is depleted, and consequently, the mitochondrial localization of PARP-1 is specifically abrogated. This result indicates that the presence of PARP-1 within mitochondria is necessary for protecting mtDNA from damage or for promoting its repair.
FIGURE 5.
Mitochondrial PARP-1 is required for maintenance of DNA stability. HeLa cells were mock-transfected (Mock) or transfected with Mitofilin (MITO), PARP-1 (PARP-1), or nonspecific (NS) siRNAs and collected after 72 h for DPCR from whole cells. Two different amounts of cells (1000 and 500) were employed to ensure the reliability of the assay. Two pairs of primers were used to amplify two fragments of mtDNA corresponding, respectively, to a 157-bp region (used to normalize the mtDNA content) or to a 5-kbp region. The relative amplification efficiencies, calculated by densitometric analysis (ImageJ software) are shown as percentages of the control (Mock). The graph reports the mean values from three independent experiments.
It has been recently recognized that mtDNA can be efficiently repaired through several mechanisms, among which base excision repair is the best characterized. Moreover, a subset of the proteins involved in nuclear DNA repair has been isolated from mitochondria (14).
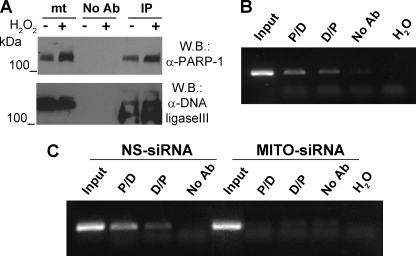
The activity of DNA ligase III, necessary for joining the free DNA ends, is the final step in the base excision repair pathway (14). It has been previously reported that DNA ligase III is localized to mitochondria and that its depletion through antisense strategy affects mtDNA maintenance (38). Using the modified ChIP protocol described above, we have found that DNA ligase III is bound to mtDNA (supplemental Fig. 4), thus supporting its role in mtDNA repair. It is worthy of note that the recruitment of DNA ligase III on DNA strand breaks during nuclear DNA repair involves its interaction with PARP-1 (10). On the basis of these observations, we investigated whether PARP-1 and DNA ligase III interact within mitochondria. To this end, we performed immunoprecipitation/Western blot analysis from mitochondrial extracts. As shown in Fig. 6A, DNA ligase III was efficiently co-immunoprecipitated with PARP-1 not only in the presence of oxidative stress, known to induce high levels of mtDNA damage (39), but also in basal conditions.
FIGURE 6.
Mitochondrial PARP-1 is associated with DNA Ligase III on mtDNA. A, PARP-1 was specifically immunoprecipitated from a mitochondria-enriched fraction of HeLa cells treated or not with 400 μm H2O2. Western blot (W.B.) analysis was performed with α-PARP-1 and α-DNA ligase III antibodies. mt, mitochondrial extract; No Ab, mitochondrial extracts incubated with protein G-Sepharose without antibody; IP, immunoprecipitation with α-PARP-1 antibody. B, sequential ChIP analysis of mtDNA from HeLa cells. Soluble chromatin was prepared from HeLa cells and divided into two chromatin aliquots, which were immunoprecipitated (first round of ChIP) with antibodies to PARP-1 and DNA ligase III, respectively. Immunocomplexes from PARP-1 and DNA ligase III were reimmunoprecipitated (second round of ChIP) with reciprocal antibodies (P/D and D/P). A fragment of the D-loop region was amplified by PCR. Input, samples isolated from total lysates prior to antibody pulldown as an internal positive control; No Ab, samples isolated after pulldown with no antibody; H2O, PCR negative control. C, sequential ChIP analysis of mtDNA from HeLa cell transfected with Mitofilin siRNA or control siRNA and analyzed as in B.
Next we determined whether the two proteins participate in the same protein complex associated with mitochondrial DNA by performing sequential ChIP experiments. The PARP-1-bound chromatin was immunoprecipitated with a specific antibody against DNA ligase III. The result shown in Fig. 6B indicates that PARP-1 and DNA ligase III co-occupy the D-loop region in the mtDNA. Moreover, reversal of the order of antibodies in sequential ChIP yielded equivalent results. Again, Mitofilin depletion prevented the association of the PARP-1-DNA ligase III complex to mtDNA (Fig. 6C). Interestingly, single-step ChIP assays showed that the absence of Mitofilin, and thus of mitochondrial PARP-1, impaired the binding of DNA ligase III to mtDNA (supplemental Fig. 4). These observations support the conclusion that mitochondrial PARP-1 functionally interacts with DNA ligase III, most probably regulating its association with mtDNA and promoting mtDNA repair.
DISCUSSION
Using mass spectrometry analysis of PARP-1 co-immunoprecipitating proteins, we identified Mitofilin as a novel PARP-1 interactor. To our knowledge, this is the first time that PARP-1 is found to associate to a mitochondrial protein. This finding prompted us to reinvestigate the subcellular localization of PARP-1 because, despite several evidences, there is a general “skeptical attitude” toward the mitochondrial localization of the enzyme (12). The results of both confocal microscopy and cell fractionation experiments clearly indicated that PARP-1 and Mitofilin interact within mitochondria.
Mitofilin is anchored to the mitochondrial inner membrane, with a small N-terminal domain protruding in the mitochondrial matrix and a long coiled-coil domain in the intermembrane space (25). The finding that Mitofilin is complexed with some proteins of the outer membrane, known to be implicated in mitochondrial import, suggested its possible involvement in protein translocation across the two mitochondrial membranes (27). In support of this hypothesis, we have observed (data not shown) that Mitofilin co-immunoprecipitates with the mitochondrial matrix ATPase mtHsp70, a key regulator of protein import (40). Interestingly, preliminary results also indicated that PARP-1 interacts with mtHsp70, leading to the suggestion that the enzyme could be translocated into the mitochondrial matrix through a mechanism involving its interaction with Mitofilin. Consistently with this assumption, we found that the localization of PARP-1 in mitochondria of human cells is dependent on the presence and abundance of Mitofilin. Further investigation is necessary to clarify the mechanism by which PARP-1, which has no canonical mitochondria-targeting sequence, is imported in these organelles. In this regard, it has to be noted that more than 50% of the mitochondrial proteins do not use the classical import pathway that requires the recognition of a specific sequence (41). Furthermore, many nuclear proteins and transcription factors, such us NF-κB and AP-1, have been detected in mitochondria despite lacking canonical mitochondria-targeting sequences, indicating the existence of still unknown mechanisms of intracellular trafficking (42).
We report several lines of evidence suggesting that intramitochondrial PARP-1 plays a role in mtDNA damage signaling and/or repair. First, we have demonstrated that PARP-1 binds to different regions throughout the mtDNA. Most importantly, we have found that DNA ligase III, involved in the sealing of strand breaks, participates in the PARP-1-containing complex bound to mtDNA. It has been previously shown that DNA ligase III preferentially associates with active PARP-1 (10). Consistently, it has been reported that mitochondria contain a basal PARP enzymatic activity higher than that found in nuclei (18). This finding, together with our results, would be in agreement with a role of mitochondrial PARP-1 as a sensor of persistent DNA damage, a natural condition suffered by mtDNA even in the absence of genotoxic stimuli. Our observation that the depletion of mitochondrial PARP-1 causes the accumulation of mtDNA damage strongly supports that the enzyme plays a direct role in the maintenance of mtDNA integrity. This finding is in line with previous works indicating an involvement of poly(ADP-ribosyl)ation in protecting mtDNA against damage after exposure to genotoxic stimuli (43, 44). However, because genotoxic treatments also lead to extensive nuclear DNA damage, which in turn causes mitochondrial dysfunction and mtDNA damage, these works did not exclude an indirect role of nuclear PARP-1 in maintaining the integrity of mtDNA. Instead, we found that just mtPARP-1, as demonstrated through its specific depletion, is responsible for mtDNA stability.
In light of the numerous PARP-1 nuclear functions, we do not exclude that the enzyme can be involved in multiple aspects of mtDNA metabolism. In particular, considering its co-localization with transcription factor A, a multifunction component of mitochondrial nucleoids, and its association with the D-loop region, it is tempting to hypothesize additional roles of PARP-1 in regulating transcription and/or replication of mtDNA.
This work highlights the existence of a functional link between Mitofilin, DNA ligase III, PARP-1, and mtDNA. A growing amount of evidence suggests that Mitofilin can play an essential role in neurodegenerative disorders because decreased protein levels have been found in fetal Down syndrome brains (45) and in a series of models of human disease (46). In line with this evidence, we observed that Mitofilin depletion, through the abrogation of mitochondrial localization of PARP-1, causes an accumulation of mtDNA damage that is a critical event in neurodegeneration. Our present finding that Mitofilin regulates PARP-1 mitochondrial localization may open a new way to look for functional determinants responsible for the accumulation of mtDNA damage, suggesting new fields of future investigation and therapeutic approaches in many human disorders.
Supplementary Material
Acknowledgments
We thank S. De Grossi, C. Ramina, and Dr. M. Senni for technical assistance at the Confocal Microscopy facility of the Histology and Medical Embryology Department, Sapienza University. Dr. T. Alonzi (National Institute for Infectious Diseases “L. Spallanzani,” IRCCS) is gratefully acknowledged for helpful suggestions and discussion of the results. We thank Dr. A. Busanello (Pasteur Institute-Fondazione Cenci Bolognetti, Department of Cellular Biotechnologies and Haematology, University of Rome “La Sapienza”) for useful insight on ChIP and sequential ChIP.
This work was supported by Grant 4179 from the Associazione Italiana per la Ricerca sul Cancro (AIRC) and the Agenzia Spaziale Italiana from Molecules to Man (ASI MOMA) project.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4 and Table 1.
- PARP-1
- poly(ADP-ribose)polymerase-1
- PAR
- poly(ADP-ribose)
- AIF
- apoptosis-inducing factor
- mt
- mitochondrial
- siRNA
- small interfering RNA
- DPCR
- direct PCR
- MS
- mass spectrometry
- MS/MS
- tandem MS
- MALDI
- matrix-assisted laser desorption/ionization
- RIPA
- radioimmune precipitation buffer
- ChIP
- chromatin immunoprecipitation.
REFERENCES
- 1.Kim M. Y., Zhang T., Kraus W. L. (2005) Genes Dev. 19, 1951–1967 [DOI] [PubMed] [Google Scholar]
- 2.D'Amours D., Desnoyers S., D'Silva I., Poirier G. G. (1999) Biochem. J. 342, 249–268 [PMC free article] [PubMed] [Google Scholar]
- 3.Malanga M., Althaus F. R. (2005) Biochem. Cell Biol. 83, 354–364 [DOI] [PubMed] [Google Scholar]
- 4.Saxena A., Saffery R., Wong L. H., Kalitsis P., Choo K. H. (2002) J. Biol. Chem. 277, 26921–26926 [DOI] [PubMed] [Google Scholar]
- 5.Kraus W. L. (2008) Curr. Opin. Cell Biol. 20, 294–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu S. W., Wang H., Poitras M. F., Coombs C., Bowers W. J., Federoff H. J., Poirier G. G., Dawson T. M., Dawson V. L. (2002) Science 297, 259–263 [DOI] [PubMed] [Google Scholar]
- 7.Simbulan-Rosenthal C. M., Rosenthal D. S., Luo R., Samara R., Espinoza L. A., Hassa P. O., Hottiger M. O., Smulson M. E. (2003) Oncogene 22, 8460–8471 [DOI] [PubMed] [Google Scholar]
- 8.Carbone M., Rossi M. N., Cavaldesi M., Notari A., Amati P., Maione R. (2008) Oncogene 27, 6083–6092 [DOI] [PubMed] [Google Scholar]
- 9.Masson M., Niedergang C., Schreiber V., Muller S., Menissier-de Murcia J., de Murcia G. (1998) Mol. Cell. Biol. 18, 3563–3571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leppard J. B., Dong Z., Mackey Z. B., Tomkinson A. E. (2003) Mol. Cell. Biol. 23, 5919–5927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dantzer F., Schreiber V., Niedergang C., Trucco C., Flatter E., De La Rubia G., Oliver J., Rolli V., Ménissier-de Murcia J., de Murcia G. (1999) Biochimie 81, 69–75 [DOI] [PubMed] [Google Scholar]
- 12.Scovassi A. I. (2004) FASEB J. 18, 1487–1488 [DOI] [PubMed] [Google Scholar]
- 13.Di Lisa F., Ziegler M. (2001) FEBS Lett. 492, 4–8 [DOI] [PubMed] [Google Scholar]
- 14.Larsen N. B., Rasmussen M., Rasmussen L. J. (2005) Mitochondrion 5, 89–108 [DOI] [PubMed] [Google Scholar]
- 15.Burzio L. O., Sáez L., Cornejo R. (1981) Biochem. Biophys. Res. Commun. 103, 369–375 [DOI] [PubMed] [Google Scholar]
- 16.Masmoudi A., Islam F., Mandel P. (1988) J. Neurochem. 51, 188–193 [DOI] [PubMed] [Google Scholar]
- 17.Masmoudi A., Mandel P. (1987) Biochemistry 26, 1965–1969 [DOI] [PubMed] [Google Scholar]
- 18.Du L., Zhang X., Han Y. Y., Burke N. A., Kochanek P. M., Watkins S. C., Graham S. H., Carcillo J. A., Szabó C., Clark R. S. (2003) J. Biol. Chem. 278, 18426–18433 [DOI] [PubMed] [Google Scholar]
- 19.Wang Y., Dawson V. L., Dawson T. M. (2009) Exp. Neurol. 218, 193–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mosgoeller W., Steiner M., Hozák P., Penner E., Wesierska-Gadek J. (1996) J. Cell Sci. 109, 409–418 [DOI] [PubMed] [Google Scholar]
- 21.Lai Y., Chen Y., Watkins S. C., Nathaniel P. D., Guo F., Kochanek P. M., Jenkins L. W., Szabó C., Clark R. S. (2008) J. Neurochem. 104, 1700–1711 [DOI] [PubMed] [Google Scholar]
- 22.Cipriani G., Rapizzi E., Vannacci A., Rizzuto R., Moroni F., Chiarugi A. (2005) J. Biol. Chem. 280, 17227–17234 [DOI] [PubMed] [Google Scholar]
- 23.Poitras M. F., Koh D. W., Yu S. W., Andrabi S. A., Mandir A. S., Poirier G. G., Dawson V. L., Dawson T. M. (2007) Neuroscience 148, 198–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Odgren P. R., Toukatly G., Bangs P. L., Gilmore R., Fey E. G. (1996) J. Cell Sci. 109, 2253–2264 [DOI] [PubMed] [Google Scholar]
- 25.Gieffers C., Korioth F., Heimann P., Ungermann C., Frey J. (1997) Exp. Cell Res. 232, 395–399 [DOI] [PubMed] [Google Scholar]
- 26.John G. B., Shang Y., Li L., Renken C., Mannella C. A., Selker J. M., Rangell L., Bennett M. J., Zha J. (2005) Mol. Biol. Cell 16, 1543–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie J., Marusich M. F., Souda P., Whitelegge J., Capaldi R. A. (2007) FEBS Lett. 581, 3545–3549 [DOI] [PubMed] [Google Scholar]
- 28.Mancone C., Amicone L., Fimia G. M., Bravo E., Piacentini M., Tripodi M., Alonzi T. (2007) Proteomics 7, 143–154 [DOI] [PubMed] [Google Scholar]
- 29.Dohi T., Beltrami E., Wall N. R., Plescia J., Altieri D. C. (2004) J. Clin. Invest. 114, 1117–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kameoka M., Nukuzuma S., Itaya A., Tanaka Y., Ota K., Ikuta K., Yoshihara K. (2004) J. Virol. 78, 8931–8934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Milano J., Day B. J. (2000) Nucleic Acids Res. 28, 968–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Velsor L. W., Kovacevic M., Goldstein M., Leitner H. M., Lewis W., Day B. J. (2004) Toxicol. Appl. Pharmacol. 199, 10–19 [DOI] [PubMed] [Google Scholar]
- 33.Rozen S., Skaletsky J. H. (2000) Bioinformatics Methods and Protocols: Methods in Molecular Biology, pp. 365–386, Humana Press, Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- 34.Dement G. A., Maloney S. C., Reeves R. (2007) Exp. Cell Res. 313, 77–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Atilano S. R., Coskun P., Chwa M., Jordan N., Reddy V., Le K., Wallace D. C., Kenney M. C. (2005) Invest Ophthalmol. Vis. Sci. 46, 1256–1263 [DOI] [PubMed] [Google Scholar]
- 36.Kang D., Kim S. H., Hamasaki N. (2007) Mitochondrion 7, 39–44 [DOI] [PubMed] [Google Scholar]
- 37.Salazar J. J., Van Houten B. (1997) Mutat. Res. 385, 139–149 [DOI] [PubMed] [Google Scholar]
- 38.Lakshmipathy U., Campbell C. (2001) Nucleic Acids Res. 29, 668–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yakes F. M., Van Houten B. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 514–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strub A., Lim J. H., Pfanner N., Voos W. (2000) Biol. Chem. 381, 943–949 [DOI] [PubMed] [Google Scholar]
- 41.Bolender N., Sickmann A., Wagner R., Meisinger C., Pfanner N. (2008) EMBO Rep. 9, 42–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Psarra A. M., Sekeris C. E. (2008) Biochim. Biophys. Acta 1783, 1–11 [DOI] [PubMed] [Google Scholar]
- 43.Jarrett S. G., Boulton M. E. (2007) Ophthalmic. Res. 39, 213–223 [DOI] [PubMed] [Google Scholar]
- 44.Druzhyna N., Smulson M. E., LeDoux S. P., Wilson G. L. (2000) Diabetes 49, 1849–1855 [DOI] [PubMed] [Google Scholar]
- 45.Myung J. K., Gulesserian T., Fountoulakis M., Lubec G. (2003) Cell. Mol. Biol. 49, 739–746 [PubMed] [Google Scholar]
- 46.Van Laar V. S., Dukes A. A., Cascio M., Hastings T. G. (2008) Neurobiol. Dis. 29, 477–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.