Abstract
Mitochondrial protein traffic requires precise recognition of the mitochondrial targeting signals by the import receptors on the mitochondrial surface including a general import receptor Tom20 and a receptor for presequence-less proteins, Tom70. Here we took a proteome-wide approach of mitochondrial protein import in vitro to find a set of presequence-containing precursor proteins for recognition by Tom70. The presequences of the Tom70-dependent precursor proteins were recognized by Tom20, whereas their mature parts exhibited Tom70-dependent import when attached to the presequence of Tom70-independent precursor proteins. The mature parts of the Tom70-dependent precursor proteins have the propensity to aggregate, and the presence of the receptor domain of Tom70 prevents their aggregate formation. Therefore Tom70 plays the role of a docking site for not only cytosolic chaperones but also aggregate-prone substrates to maintain their solubility for efficient transfer to downstream components of the mitochondrial import machineries.
Introduction
Eukaryotic cells consist of complex membrane-bounded compartments that are endowed with specific proteins. Among them, mitochondria are essential organelles and perform multiple functions ranging from energy generation to regulation of cell death. Mitochondria are estimated to consist of 1000–1500 different proteins, which represent 10–20% of proteins synthesized in eukaryotic cells. Because of the mitochondrial connection to several human diseases including neurodegenerative disorders and cancer, biogenesis and functional maintenance of mitochondria have gained widening interest, and efforts have been made to compile an extensive catalogue of the mitochondrial proteome (1).
The vast majority of mitochondrial proteins are encoded by the nuclear DNA, synthesized in the cytosol, and imported into the mitochondria, whereas only a handful of them are encoded by the mitochondrial DNA. Precise targeting of mitochondrial proteins requires recognition of mitochondrial targeting signals (2–4). The targeting signal for mitochondria is encoded within the protein itself. Most of the proteins that go across the outer and inner membranes to reach the matrix or inner membrane are synthesized as precursor proteins with an N-terminal cleavable presequence that contains a mitochondrial targeting signal. Many polytopic inner membrane proteins including those of the carrier protein family lack a presequence but instead have a mitochondrial targeting signal in the mature part as an internal signal.
Protein import into mitochondria is mediated by the translocators of the outer and inner mitochondrial membranes (5–8). Targeting information for mitochondria is recognized by the receptor subunits of the TOM40 complex including Tom20, Tom22, and Tom70 (4, 9, 10). Current understanding is that different types of targeting signals are recognized by different receptors. In general, targeting signals contained in the cleavable presequences are recognized by Tom20 and Tom22, and internal targeting signals of polytopic membrane proteins are recognized by Tom70 (9, 10). The structural analyses showed that the hydrophobic groove of Tom20 binds to the hydrophobic side of the eight-residue segment of the rat aldehyde dehydrogenase presequence, which appears to contain a five-residue motif as an element for Tom20 recognition, in the amphiphilic helical conformation (11–13). On the other hand, there are some cross-specificities between the two types of targeting signals and their cognate receptors. It has been reported that Tom70 recognizes, in addition to presequence-less polytopic membrane proteins, some presequence-containing precursor proteins including cytochrome c1, alcohol dehydrogenase III, and the α- and β-subunits of F1-ATPase (Atp1p and Atp2p); yet the presequences did not appear to be the sole determinant of those presequence-containing precursors to Tom70 (14–16). The purified presequence-containing adrenodoxin precursor loaded onto the cytosolic chaperone mitochondrial import-stimulating factor was shown to bind to Tom70 first and then to Tom20 during its import into mitochondria in vitro (17). However, in contrast to Tom20, little is known about the recognition of the mitochondrial targeting signal by Tom70, although a peptide scan analysis showed that Tom70 binds to multiple segments throughout the polypeptide chain of noncleavable phosphate carrier (18). Tom70 was also shown to function as a docking site for cytosolic chaperones such as Hsp70 (in yeast and mammals) and Hsp90 (mammals) to receive mitochondrial proteins (19).
In the present study, we took a proteome-wide approach of mitochondrial protein import in vitro to find a set of substrate proteins for recognition by Tom70. Translation products of total yeast RNA were imported into isolated wild type mitochondria and those without Tom70. Comparison of the imported proteins on two-dimensional electrophoresis gels between wild type and tom70Δ mitochondria allowed us to identify presequence-containing proteins affected by depletion of Tom70 systematically. Then we tried to elucidate the reason for the requirement of Tom70 for the import of the identified presequence-containing mitochondrial precursor proteins. The obtained results show that Tom70 is not required for targeting signal recognition but instead for maintenance of the solubility of the presequence-containing precursor proteins.
EXPERIMENTAL PROCEDURES
Plasmids and Yeast Strains
The yeast haploid strain W303-1A was used as a parental strain in this study. To construct the tom70Δ strain (MNA-mas70), the chromosomal TOM70 gene was disrupted with the URA3 marker. MNA-mas70 was transformed with the TRP1 plasmid pRS314/Tom70(WT), pRS314/Tom70(38TEV), pRS314/Tom70(98TEV), and pRS314/Tom70(246) to construct yeast strains Tom70W, Tom70T, Tom70T(98), and Tom70T(246), respectively. The plasmids used in this study are summarized in Table 1. Yeast cells were grown at 30 °C in lactate medium (0.3% (w/v) yeast extract, 0.1% (w/v) glucose, 0.05% (w/v) CaCl2·2H2O, 0.05% (w/v) NaCl, 0.06% (w/v) MgCl2·6H2O, 0.1% (w/v) KH2PO4, 0.1% (w/v) NH4Cl, 2% (w/v) lactic acid, pH 5.6) or SCLac medium (0.67% (w/v) yeast nitrogen base without amino acids, 0.5% (w/v) casamino acids, 2% (w/v) lactic acid, 0.05% (w/v) glucose, pH 5.6).
TABLE 1.
Plasmids used in this study
aa, amino acids.
| Plasmid | Vector | Relevant characteristics | Source or reference |
|---|---|---|---|
| pSu9-DHFR | pGEM-4Z | Presequence of Su9 (1–69 aa) and DHFR (187 aa) | Ref. 30 |
| pHsp60 | pGEM-4Z | Hsp60 precursor (1–572 aa) | This study |
| pCox4p | pGEM-4Z | Cox4 precursor (1–155 aa) | Ref. 28 |
| pAco1p | pGEM-4Z | Aco1 precursor (1–778 aa) | This study |
| pAtp1p | pGEM-4Z | Atp1 precursor (1–545 aa) | This study |
| pCit1p | pGEM-4Z | Cit1 precursor (1–479 aa) | This study |
| pDld2p | pGEM-4Z | Dld2 precursor (1–530 aa) | This study |
| pIdh1p | pGEM-4Z | Idh1 precursor (1–360 aa) | This study |
| pAtp2p | pGEM-4Z | Atp2 precursor (1–511 aa) | This study |
| pAco1p-DHFR | pGEM-4Z | Aco1 precursor (1–778 aa) and DHFR (187 aa) | This study |
| pAtp1p-DHFR | pGEM-4Z | Atp1 precursor (1–545 aa) and DHFR (187 aa) | This study |
| pCit1p-DHFR | pGEM-4Z | Cit1 precursor (1–479 aa) and DHFR (187 aa) | This study |
| pDld2p-DHFR | pGEM-4Z | Dld2 precursor (1–530 aa) and DHFR (187 aa) | This study |
| pIdh1p-DHFR | pGEM-4Z | Idh1 precursor (1–360 aa) and DHFR (187 aa) | This study |
| pAtp2p-DHFR | pGEM-4Z | Atp2 precursor (1–511 aa) and DHFR (187 aa) | This study |
| pAco1(5)-DHFR | pGEM-4Z | Presequence of Aco1 (1–28 aa) and DHFR (187 aa) | This study |
| pAtp1(5)-DHFR | pGEM-4Z | Presequence of Atp1 (1–47 aa) and DHFR (187 aa) | This study |
| pCit1(5)-DHFR | pGEM-4Z | Presequence of Cit1 (1–42 aa) and DHFR (187 aa) | This study |
| pDld2(5)-DHFR | pGEM-4Z | Presequence of Dld2 (1–49 aa) and DHFR (187 aa) | This study |
| pIdh1(5)-DHFR | pGEM-4Z | Presequence of Idh1 (1–34 aa) and DHFR (187 aa) | This study |
| pAtp2(5)-DHFR | pGEM-4Z | Presequence of Atp2 (1–37 aa) and DHFR (187 aa) | This study |
| pAco1(5)-Hsp60 | pGEM-4Z | Presequence of Aco1 (1–28 aa) and mature part of Hsp60 (22–572 aa) | This study |
| pAtp1(5)-Hsp60 | pGEM-4Z | Presequence of Atp1 (1–47 aa) and mature part of Hsp60 (22–572 aa) | This study |
| pCit1(5)-Hsp60 | pGEM-4Z | Presequence of Cit1 (1–42 aa) and mature part of Hsp60 (22–572 aa) | This study |
| pDld2(5)-Hsp60 | pGEM-4Z | Presequence of Dld2 (1–49 aa) and mature part of Hsp60 (22–572 aa) | This study |
| pIdh1(5)-Hsp60 | pGEM-4Z | Presequence of Idh1 (1–34 aa) and mature part of Hsp60 (22–572 aa) | This study |
| pAtp2(5)-Hsp60 | pGEM-4Z | Presequence of Atp2 (1–37 aa) and mature part of Hsp60 (22–572 aa) | This study |
| pAco1(5)-Cox4p | pGEM-4Z | Presequence of Aco1 (1–28 aa) and mature part of Cox4 (26–155 aa) | This study |
| pAtp1(5)-Cox4p | pGEM-4Z | Presequence of Atp1 (1–47 aa) and mature part of Cox4 (26–155 aa) | This study |
| pCit1(5)-Cox4p | pGEM-4Z | Presequence of Cit1 (1–42 aa) and mature part of Cox4 (26–155 aa) | This study |
| pDld2(5)-Cox4p | pGEM-4Z | Presequence of Dld2 (1–49 aa) and mature part of Cox4 (26–155 aa) | This study |
| pIdh1(5)-Cox4p | pGEM-4Z | Presequence of Idh1 (1–34 aa) and mature part of Cox4 (26–155 aa) | This study |
| pAtp2(5)-Cox4p | pGEM-4Z | Presequence of Atp2 (1–37 aa) and mature part of Cox4 (26–155 aa) | This study |
| pCox4-Aco1p | pGEM-4Z | Presequence of Cox4 (1–25 aa) and mature part of Aco1 (24–778 aa) | This study |
| pCox4-Atp1p | pGEM-4Z | Presequence of Cox4 (1–25 aa) and mature part of Atp1 (43–545 aa) | This study |
| pCox4-Cit1p | pGEM-4Z | Presequence of Cox4 (1–25 aa) and mature part of Cit1 (38–479 aa) | This study |
| pCox4-Dld2p | pGEM-4Z | Presequence of Cox4 (1–25 aa) and mature part of Dld2 (45–530 aa) | This study |
| pCox4-Idh1p | pGEM-4Z | Presequence of Cox4 (1–25 aa) and mature part of Idh1 (30–360 aa) | This study |
| pCox4-Atp2p | pGEM-4Z | Presequence of Cox4 (1–25 aa) and mature part of Atp2 (33–511 aa) | This study |
| pSu9-Aco1p | pGEM-4Z | Presequence of Su9 (1–69 aa) and mature part of Aco1 (24–778 aa) | This study |
| pSu9-Atp1p | pGEM-4Z | Presequence of Su9 (1–69 aa) and mature part of Atp1 (43–545 aa) | This study |
| pSu9-Cit1p | pGEM-4Z | Presequence of Su9 (1–69 aa) and mature part of Cit1 (38–479 aa) | This study |
| pSu9-Dld2p | pGEM-4Z | Presequence of Su9 (1–69 aa) and mature part of Dld2 (45–530 aa) | This study |
| pSu9-Idh1p | pGEM-4Z | Presequence of Su9 (1–69 aa) and mature part of Idh1 (30–360 aa) | This study |
| pSu9-Atp2p | pGEM-4Z | Presequence of Su9 (1–69 aa) and mature part of Atp2 (33–511 aa) | This study |
| pRS314/Tom70(WT) | pRS314 | Expression of wild type Tom70 under the control of own promoter | This study |
| pRS314/Tom70(38TEV) | pRS314 | Expression of Tom70(38TEV); TEV protease recognition sequence was inserted between residues 38 and 39 | This study |
| pRS314/Tom70(98TEV) | pRS314 | Expression of Tom70(98TEV); TEV protease recognition sequence was inserted between residues 98 and 99 | This study |
| pRS314/Tom70(246TEV) | pRS314 | Expression of Tom70(246TEV); TEV protease recognition sequence was inserted between residues 246 and 247 | This study |
| pRS314/Tom20(WT) | pRS314 | Expression of wild type Tom20 under the control of own promoter | Ref. 10 |
| pRS314/Tom20(73TEV) | pRS314 | Expression of Tom20(73TEV); TEV protease recognition sequence was inserted between residues 73 and 74 | Ref. 10 |
Two-dimensional Gel Electrophoresis
Isoelectric focusing (IEF)5 for the first dimension was performed using Immobiline DryStrip (GE Healthcare Biosciences) and the IPGphor IEF system (GE Healthcare Biosciences). After import reaction, mitochondria were reisolated and solubilized with rehydration buffer (0.5% IPG buffer (GE Healthcare Biosciences), 2% CHAPS, 7 m urea, 2 m thiourea, 20 mm dithiothreitol, 1% SB3–10), and the solubilized proteins were applied to Immobiline DryStrip (18 cm) rehydrated with 350 μl of rehydration buffer. IEF was conducted on the IPGphor with a current limit of 50 μA/strip at 20 °C with the following focusing program: a gradient to 500 V for 5 min, a gradient to 4000 V for 85 min, and a gradient to 8000 V for 5 h (pI 4–7) or 10 h (pI 6–9). The strips were then treated with equilibration buffer (50 mm Tris-HCl, pH 8.8, 6 m urea, 30% glycerol, 2% SDS) for 15 min and subjected to SDS-PAGE for the second dimension.
Import Assays
Mitochondria were isolated from W303-1A (WT) and MNA-mas70 (tom70Δ) strains grown in lactate medium at 30 °C. Radiolabeled precursor proteins were synthesized with rabbit reticulocyte lysate by coupled transcription/translation in the presence of [35S]methionine. WT or tom70Δ mitochondria (0.1 mg protein/ml) were incubated with radiolabeled precursor proteins in import buffer (250 mm sucrose, 10 mm MOPS-KOH, pH 7.2, 80 mm KCl, 2 mm potassium Pi, 2 mm methionine, 5 mm dithiothreitol, 5 mm MgCl2, 2 mm ATP, 2 mm NADH, 1% BSA) at 25 °C. The import reaction was stopped by the addition of 20 μg/ml valinomycin. Protease treatment was performed by incubating the mitochondria with 100 μg/ml proteinase K for 20 min on ice, which was inactivated by subsequent addition of 1 mm phenylmethylsulfonyl fluoride. The mitochondria were reisolated by centrifugation and washed once with SEM buffer (250 mm sucrose, 1 mm EDTA, 10 mm MOPS-KOH, pH 7.2), and the proteins were analyzed by SDS-PAGE and radioimaging with a Storm 860 image analyzer (Amersham Biosciences). Most import experiments were performed at least twice for different preparation of mitochondria to confirm reproducibility and independence of different mitochondrial preparation.
Total RNA Preparation
Yeast wild type cells (W303-1A) were grown in lactate medium to logarithmic phase, washed with diethyl pyrocarbonate-treated water, and resuspended in TES buffer (10 mm Tris-HCl, pH 7.5, 10 mm EDTA, 0.5% SDS). An equal volume of water-saturated phenol solution was added to the suspension, and the mixture was incubated at 65 °C for 45 min with occasional vortexing. After centrifugation, the aqueous phase was recovered and extracted twice with chloroform, and then total RNAs were precipitated by ethanol precipitation.
Chaperone Assays for Tom70d
The cytosolic domain (residues 39–617) of Tom70 with the C-terminal His10 tag (Tom70d) was overexpressed in Escherichia coli strain BL21 (DE3) using the vector pET-16b (Novagen), and purified by nickel-nitrilotriacetic acid and Mono-Q 5/50 columns. Radiolabeled proteins were synthesized with PURESYSTEM by coupled transcription/translation in the presence of [35S]methionine at 37 °C for 1 h. The reaction mixture contained no additional protein, 5 μm Tom70d, or 5 μm BSA. The translation products were centrifuged at 20,000 × g for 20 min at 4 °C.
RESULTS
Proteome-wide Import Analyses Reveal Substrates for Tom70
To perform a global search for substrate proteins for Tom70 recognition in mitochondrial protein import, we isolated total RNA from wild type yeast cells, which was used for in vitro translation in the presence of [35S]Met with reticulocyte lysate. The mixture of radiolabeled proteins was then subjected to in vitro import analyses with mitochondria from WT cells and those from tom70-deletion mutant cells (tom70Δ). To analyze the roles of receptors by mitochondrial import assays, experimental conditions should be optimized to make the receptor-mediated step rate-limiting in the import process (10, 20). We thus lowered the concentrations of mitochondria to the range that rendered the import rate of ADP/ATP carrier (AAC), a presequence-less substrate for the Tom70 recognition, dependent on the concentrations of mitochondria. Under this condition, import of AAC into tom70Δ mitochondria was retarded as compared with that into WT mitochondria (data not shown). We adopted this condition of mitochondrial concentrations (≤0.25 mg of protein/ml) for the import of the translation products of total yeast RNA into WT and tom70Δ mitochondria at 25 °C for 5 min. After the import reaction, the mitochondria were treated with protease to remove proteins outside mitochondria, reisolated by centrifugation, and solubilized with 2% CHAPS, 7 m urea, and 2 m thiourea. The protease-protected, presumably imported proteins were separated by two-dimensional gel electrophoresis with denaturing IEF in the first dimension and SDS-PAGE in the second dimension and were detected by radioimaging (Fig. 1A). When valinomycin, which dissipates the membrane potential (ΔΨ) across the inner membrane, was added before incubation of the translation products with mitochondria, most of the spots disappeared, suggesting that they represent proteins that require ΔΨ for import (data not shown).
FIGURE 1.
In vitro import of total RNA translation products into WT and tom70Δ mitochondria. A, total RNA was isolated from W303-1A cells and translated with reticulocyte lysate in the presence of [35S]Met. The radiolabeled translation products were incubated with wild type (WT) or tom70Δ (Δ70) mitochondria (0.25 mg of protein/ml) at 25 °C for 5 min. The mitochondria were treated with 100 μg/ml proteinase K for 20 min on ice and reisolated. The protease-protected imported proteins were subjected to two-dimensional gel electrophoresis (IEF (pI 4–7) for the first dimension and SDS-PAGE with 7–14% gel for the second dimension) followed by radioimaging. The insets are the same regions of the two-dimensional gel images around spots 3, 9, 13, and 22 (for the numbers of the spots, see supplemental Fig. S1) for WT and tom70Δ mitochondria, and the intensities of the spots highlighted with open circles were quantified and compared between WT (blue bar) and tom70Δ (pink bar) mitochondria. The intensities for WT were set to 1.0. The error bars represent S.D. values from three independent experiments. B, radiolabeled translation products of yeast total RNA were imported into WT and tom70Δ mitochondria and subjected to two-dimensional gel electrophoresis using a pI 4.7 strip or pI 6–9 strip for IEF and a 7% gel or 9% gel for SDS-PAGE followed by radioimaging as in A. Intensities of the 114 spots for imported protease-protected proteins resolved on two-dimensional gel analyses were quantified and plotted for wild type (WT, blue bar) and tom70Δ (pink bar) mitochondria. For the numbers of the spots, see supplemental Fig. S1. The intensities for WT were set to 1.0. The error bars represent S.D. values from three independent experiments.
In total, as many as 114 spots of the imported proteins were resolved by two-dimensional gel electrophoresis under four different conditions (supplemental Fig. S1). When we compare the two-dimensional gel electrophoresis patterns of imported proteins between WT mitochondria and tom70Δ mitochondria, we find several spots exhibiting lower intensities with tom70Δ mitochondria than with WT mitochondria. Four typical examples, spots 3, 9, 13, and 22, are shown in Fig. 1A. The resolved 114 spots were thus quantified and compared between WT mitochondria and tom70Δ mitochondria (Fig. 1B). Intensities of spots 3, 6, 9, 13, 22, 30, 41, 67, 71, 72, 73, 82, 83, 90, 91, 102, and 109 for tom70Δ mitochondria were <75% of those for WT mitochondria. These spots represent proteins whose import into mitochondria was strongly affected by deletion of the TOM70 gene. Interestingly, some proteins corresponding to spots 21, 74, and 86 were more efficiently (> 2-fold) imported into tom70Δ mitochondria than into WT mitochondria. We confirmed that the levels of Tom20, Tom40, or Tom71, a minor isoform of Tom70, did not change in tom70Δ mitochondria (supplemental Fig. S2). Perhaps an unknown adaptation may have been caused by deletion of the TOM70 gene as previously observed for deletion of Tom20 (21). This point will be discussed later.
For identification of the spots in the two-dimensional gel electrophoresis radioimaging, we compared them with those in the two-dimensional gel electrophoresis maps for proteins associated with isolated yeast mitochondria,6 where ∼90 different protein spots were assigned to specific yeast proteins by in-gel trypsin digestion followed by mass spectrometry measurements. By referring to even this limited information on the two-dimensional gel electrophoresis maps, we could find possible candidates for the proteins whose import appeared to depend on Tom70. We thus tentatively assigned spots 3, 6, 9, 13, 67, 73, 82, and 102 to specific mitochondrial proteins, Atp2p (F1-ATPase β-subunit), Tom40, Ssc1p (mitochondrial Hsp70), Dld2p (d-lactase dehydrogenase), Cit1p (citrate synthase), Atp1p (F1-ATPase α-subunit), Aco1 (mitochondrial aconitase), and Idh1 (isocitrate dehydrogenase), respectively. Among these eight proteins, only Atp1p and Atp2p were previously reported to depend on Tom70 for their import (14). Except for Tom40, these proteins are synthesized as precursors with a cleavable N-terminal presequence. It is to be noted under the present conditions of two-dimensional gel electrophoresis that most of the known presequence-less substrates for Tom70 such as AAC and other inner membrane carrier proteins are too hydrophobic to be resolved in the two-dimensional gel (data not shown).
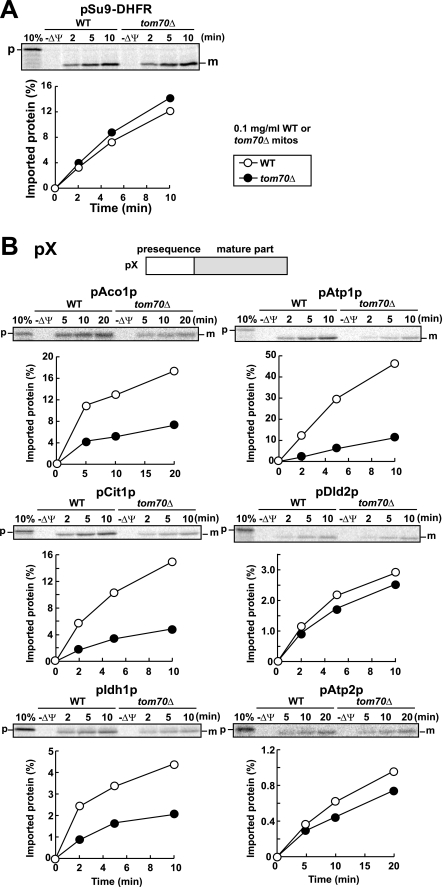
To further confirm the above assignments, we performed in vitro import of the assigned mitochondrial proteins into WT mitochondria and tom70Δ mitochondria individually. The assigned proteins were synthesized in vitro with reticulocyte lysate in the presence of [35S]Met, and radiolabeled proteins were imported into WT mitochondria and tom70Δ mitochondria. As a control, pSu9-DHFR, a fusion protein between the presequence of subunit 9 of Neurospora crassa Fo-ATPase (pSu9) and mouse DHFR, which does not require Tom70 for import, was imported with similar or slightly higher efficiency into tom70Δ mitochondria than WT mitochondria (Fig. 2A). On the other hand, precursors of Aco1p (pAco1p), Atp1p (pAtp1p), Cit1p (pCit1p), and Idh1p (pIdh1p) were imported into tom70Δ mitochondria with lower efficiency than into WT mitochondria, whereas import rates of precursors to Atp2p (pAtp2p) and Dld2p (pDld2p) into tom70Δ mitochondria were similar to those imported into WT mitochondria (Fig. 2B). The different Tom70 dependence between pAtp1p and pAtp2p may be due to the difference in the presequence lengths or properties (e.g. isoeletric points) of the mature domains. Therefore we judge that at least four proteins, pAco1p, pAtp1p, pCit1p, and pIdh1p, are presequence-containing mitochondrial precursor proteins that require Tom70 for their import into mitochondria.
FIGURE 2.
In vitro import of pSu9-DHFR and pX into WT and tom70Δ mitochondria. Radiolabeled pSu9-DHFR (A) or radiolabeled precursor proteins pAco1p, pAtp1p, pCit1p, pDld2p, pIdh1p, and pAtp2p (B) were incubated with wild type (WT, open circles) or tom70Δ (filled circles) mitochondria (0.1 mg of protein/ml) at 25 °C for indicated times. The mitochondria were treated with proteinase K and imported, protease-protected proteins were analyzed by SDS-PAGE and radioimaging. The amounts of radiolabeled proteins added to each reaction are set to 100%. p, precursor forms; m, mature forms; 10%, 10% of the input; −ΔΨ, valinomycin was added.
Tom70-dependent Precursors Contain Targeting Signals in the Presequences
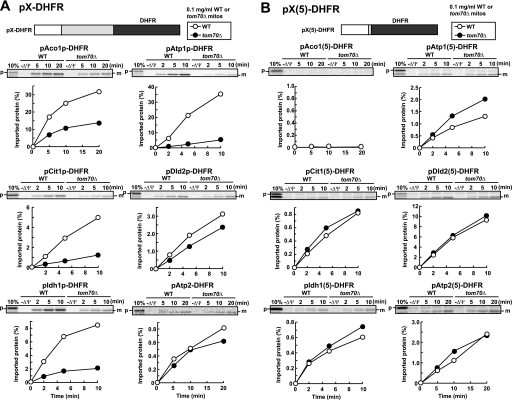
Does Tom70 recognize the presequence or mature part of the identified Tom70-dependent presequence-containing precursor proteins (pX, where pX is the full-length precursor to protein X)? To address this question, we tested the Tom70 dependence in the import of the fusion proteins (pX(5)-DHFR) between the presequence of pX (plus five residues in the mature domain carrying the presequence cleavage site) and a nonmitochondrial protein DHFR as a passenger domain. As controls, the pX-DHFR fusion proteins consisting of the full-length precursor proteins (pAco1p, pAtp1p, pCit1p, pDld2p, pIdh1p, and pAtp2p) and DHFR were imported efficiently into mitochondria (Fig. 3A). Import of pAco1p-DHFR, pAtp1p-DHFR, pCit1p-DHFR, and pIdh1p-DHFR (like pAco1p, pAtp1p, pCit1p, and pIdh1p) depended on Tom70, whereas that of pDld2p-DHFR or pAtp2p-DHFR (like pDld2p or pAtp2p) did not depend on Tom70. Now the DHFR fusion proteins with a presequence of the Tom70-dependent precursors, pAco1p, pAtp1p, pCit1p, and pIdh1p (pAco1(5)-DHFR, pAtp1(5)-DHFR, pCit1(5)-DHFR, and pIdh1(5)-DHFR, respectively) were hardly or inefficiently imported into mitochondria (note that the scales for y axes differ for different charts). On the other hand, pDld2(5)-DHFR and pAtp2(5)-DHFR were imported into mitochondria more efficiently than corresponding full-length fusion proteins, pDld2p-DHFR and pAtp2p-DHFR (Fig. 3B). The impaired import of pAco1(5)-DHFR, pAtp1(5)-DHFR, pCit1(5)-DHFR, and pIdh1(5)-DHFR was not due to aggregate formation or sequestering of the presequence in the DHFR part (supplemental Fig. S3, A and B). The mature parts of pAco1p, pAtp1p, pCit1p, or pIdh1p lacking the presequence were not imported into WT or tom70Δ mitochondria at all (data not shown).
FIGURE 3.
In vitro import of pX-DHFR and pX(5)-DHFR into wild type and tom70Δ mitochondria. A, radiolabeled fusion proteins pAco1p-DHFR, pAtp1p-DHFR, pCit1p-DHFR, pDld2p-DHFR, pIdh1p-DHFR, and pAtp2p-DHFR were imported into wild type (WT) or tom70Δ mitochondria (0.1 mg of protein/ml) and analyzed as in Fig. 2. B, radiolabeled fusion proteins pAco1(5)-DHFR, pAtp1(5)-DHFR, pCit1(5)-DHFR, pDld2(5)-DHFR, pIdh1(5)-DHFR, and pAtp2(5)-DHFR were imported into wild type (WT) or tom70Δ mitochondria (0.1 mg of protein/ml) and analyzed as in Fig. 2. The amounts of radiolabeled proteins added to each reaction are set to 100%. p, precursor forms; m, mature forms; 10%, 10% of the input; −ΔΨ, valinomycin was added.
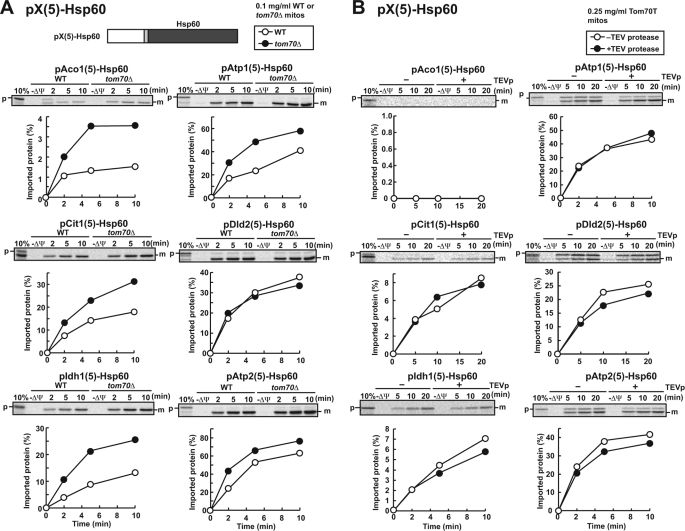
Because tight folding of the DHFR domain may well hamper efficient import into mitochondria, we tested the import of fusion proteins containing the mature domains of authentic mitochondrial precursor proteins of Cox4p (pCox4p, cytochrome oxidase subunit IV) and Hsp60 (pHsp60), which are more susceptible to protease digestion than DHFR (supplemental Fig. S4). Although pX(5)-Cox4p fusion proteins with the presequence of pAco1p, pAtp1p, pCit1p, or pIdh1p were still hardly or not imported into mitochondria (supplemental Fig. S5), the fusion proteins pX(5)-Hsp60 with any of the six presequences (pAco1(5)-Hsp60, pAtp1(5)-Hsp60, pCit1(5)-Hsp60, pDld2(5)-Hsp60, pIdh1(5)-Hsp60, and pAtp2(5)-Hsp60) were imported into mitochondria (Fig. 4A), suggesting that the presequences of pAco1p, pAtp1p, pCit1p, pDld2p, pIdh1p, and pAtp2p have sufficient information to target passenger proteins to mitochondria. The low ability of those presequences to direct tightly folded DHFR domain to mitochondria is likely due to their short lengths (23, 42, 37, 44, 29, and 32 residues long for pAco1p, pAtp1p, pCit1p, pDld2p, pIdh1p, and pAtp2, respectively), because presequences shorter than ∼70 residues cannot engage with mitochondrial Hsp70 in the matrix without prior unfolding of the mature domains (22, 23). On the other hand, because the N-terminal 69 residues of the mature part of pHsp60 appear protease-susceptible and -labile (supplemental Fig. S4), even the short presequences (23–44 residues) can reach the matrix without unfolding of the core part of the mature domain of pHsp60, so that import efficiency of pX(5)-Hsp60 became high. The different import efficiency between the pX(5)-Cox4p fusion proteins and pX(5)-Hsp60 fusion proteins may reflect a difference in the folding status between Cox4p and Hsp60p, as indicated by the moderate protease resistance of pCox4p as compared with highly protease-sensitive pHsp60 (supplemental Fig. S4).
FIGURE 4.
In vitro import of pX(5)-Hsp60 into WT and tom70Δ mitochondria or into mitochondria without the receptor domain of Tom70. A, radiolabeled fusion proteins pAco1(5)-Hsp60, pAtp1(5)-Hsp60, pCit1(5)-Hsp60, pDld2(5)-Hsp60, pIdh1(5)-Hsp60, and pAtp2(5)-Hsp60 were imported into wild type (WT) or tom70Δ mitochondria (0.1 mg of protein/ml) and analyzed as in Fig. 2. B, radiolabeled fusion proteins pAco1(5)-Hsp60, pAtp1(5)-Hsp60, pCit1(5)-Hsp60, pDld2(5)-Hsp60, pIdh1(5)-Hsp60, and pAtp2(5)-Hsp60 were incubated with TEV protease (TEVp)-untreated (open circles) or TEVp-treated (filled circles) Tom70T mitochondria (0.25 mg of protein/ml) at 25 °C for the indicated times. The mitochondria were treated with proteinase K, and the imported proteins were analyzed by SDS-PAGE and radioimaging. The amounts of radiolabeled proteins added to each reaction are set to 100%. p, full-length proteins; m, Hsp60; 10%, 10% of the input; −ΔΨ, valinomycin was added.
It is to be noted that the pX(5)-Hsp60 fusion proteins tend to be imported into tom70Δ mitochondria more efficiently than WT mitochondria (Fig. 4A). This unexpected observation could reflect secondary effects or adaptation caused by deletion of the TOM70 gene because some mitochondrial proteins were imported into tom70Δ mitochondria more efficiently than into WT mitochondria in our initial screening as well (Fig. 1B). To test this interpretation, we took the approach with TEV protease (TEVp) to remove the receptor domain after isolation of mitochondria (10). Briefly, we introduced a cleavage site specific for TEVp between the N-terminal transmembrane segment and the following C-terminal receptor domain of Tom70 (Tom70T mitochondria), so that we could delete the cytosolic receptor domain of Tom70 in vitro after isolating mitochondria by TEVp treatment. Pretreatment of Tom70T mitochondria with 76 μg/ml TEVp at 30 °C for 20 min resulted in the removal of >90% of the receptor domain of Tom70 (supplemental Fig. S6). Although deletion of the receptor domain of Tom70 after isolation of mitochondria did not affect the import of pSu9-DHFR, import of full-length pAco1p, pAtp1p, pCit1p, and pIdh1p were retarded after deletion of the Tom70 receptor domain as compared with the case without TEVp treatment (supplemental Fig. S7). Now pAtp1(5)-Hsp60, pCit1(5)-Hsp60, and pIdh1(5)-Hsp60 (and pDld2(5)-Hsp60 and pAtp2(5)-Hsp60, as well) were imported into Tom70T mitochondria after TEV protease treatment with an efficiency similar or smaller to that without TEV protease treatment (Fig. 4B). Therefore enhanced import rates of some precursor proteins into tom70Δ mitochondria are most likely due to adaptation of the cells to compensate for the effects of depletion of Tom70.
Although the Presequence Is Recognized by Tom20, the Mature Part Is Responsible for Tom70 Dependence in Mitochondrial Import
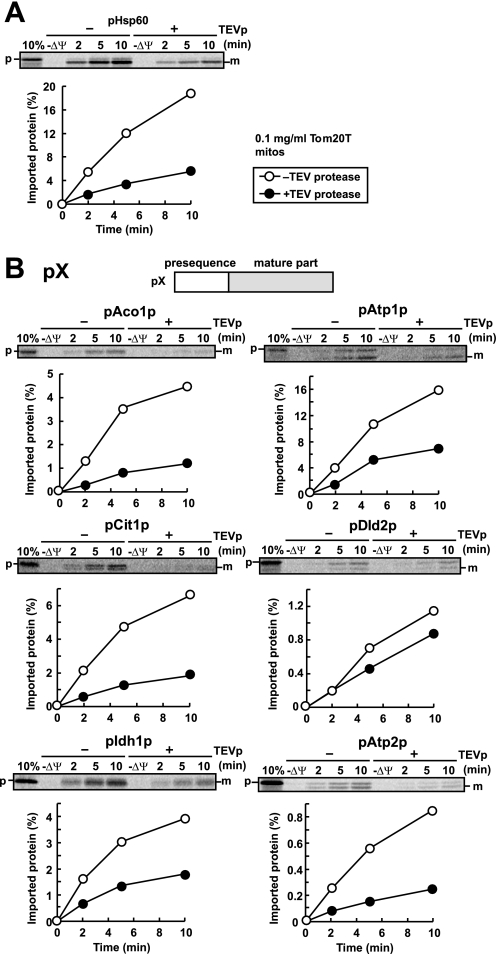
If Tom70 is dispensable for the import of pX(5)-Hsp60, whose presequences pX(5) are derived from the Tom70-dependent precursor proteins, does Tom20 recognize those presequences? To address this question, we utilized Tom20 with a TEVp cleavage site between the N-terminal transmembrane segment and the C-terminal receptor domain of Tom20 (Tom20T), which allows us to analyze the effects of deletion of the Tom20 receptor domain as in the case of Tom70 (10). Import of pHsp60 into Tom20T mitochondria after TEVp treatment was retarded compared with Tom20T mitochondria without TEVp treatment (Fig. 5A). The import of pAco1p, pAtp1p, pCit1p, pIdh1p, and pAtp2p was also retarded by TEV protease treatment of Tom20T mitochondria (Fig. 5B), indicating that the targeting signals in the presequences of those precursor proteins are recognized by Tom20. The import of pDld2p was not markedly dependent on the TEVp treatment, perhaps because of its significantly low import efficiency.
FIGURE 5.
In vitro import of pSu9-DHFR and pX into mitochondria without the receptor domain of Tom20. Radiolabeled pSu9-DHFR (A) or radiolabeled precursor proteins pAco1p, pAtp1p, pCit1p, pDld2p, pIdh1p, and pAtp2p (B) were incubated with TEVp-untreated (open circles) or TEVp-treated (filled circles) Tom20T mitochondria (0.25 mg of protein/ml) at 25 °C for the indicated times. The mitochondria were treated with proteinase K, and the imported proteins were analyzed by SDS-PAGE and radioimaging. The amounts of radiolabeled proteins added to each reaction are set to 100%. p, precursor forms; m, mature forms; 10%, 10% of the input; −ΔΨ, valinomycin was added.
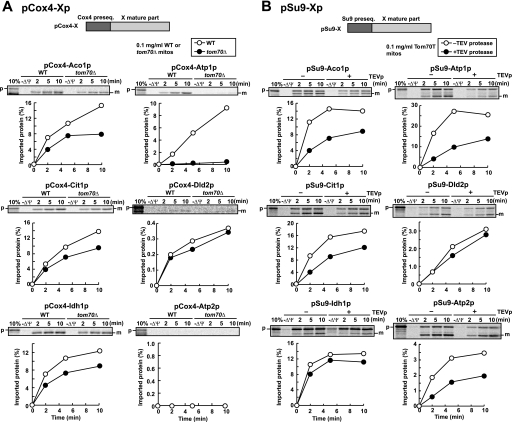
Evidently, the presequences of the Tom70-dependent precursor proteins contain mitochondrial targeting signals that are recognized by Tom20. Then which is responsible for the Tom70 dependence in mitochondrial import, the presequence or mature part? To answer this question, we tested the import of the fusion proteins consisting of the presequence of Tom70-independent precursor proteins followed by the mature domain of the Tom70-dependent precursors, pAco1p, pAtp1p, pCit1p, and pIdh1p into WT and tom70Δ mitochondria. We chose the short (25-residue) presequence of a Tom70-independent precursor protein, pCox4p. pCox4-DHFR, a fusion protein between the Cox4 presequence and DHFR, and the pCox4 precursor (pCox4p) were imported into tom70Δ mitochondria with similar efficiency or slightly higher efficiency than WT mitochondria (data not shown). The fusion proteins (pCox4-X) with the pCox4 presequence followed by the mature parts of the Tom70-dependent precursors, pAco1p, pAtp1p, pCit1p, and pIdh1p (pCox4-Aco1p, pCox4-Atp1p, pCox4-Cit1p, and pCox4-Idh1p, respectively) were imported into WT mitochondria more efficiently than into tom70Δ mitochondria despite the lack of the original presequence (Fig. 6A). Import efficiency of pCox4-Dld2p and pCox4-Atp2p was too low to compare import rates between WT and tom70Δ mitochondria, probably because of tight folding of the mature parts or aggregate formation. To increase import efficiency by efficient engagement with mtHsp70 (22, 23), we made fusion proteins consisting of the long (69 residues) pSu9 presequence followed by the mature parts of pAco1p, pAtp1p, pCit1p, pDld2p, pIdh1p, and pAtp2p (pSu9-Aco1p, pSu9-Atp1p, pSu9-Cit1p, pSu9-Dld2p, pSu9-Idh1p, and pSu9-Atp2p, respectively), and tested their import. All of these fusion proteins were imported into WT and tom70Δ mitochondria with higher efficiency than the corresponding pCox4-Xp fusion proteins (supplemental Fig. S8). However, the import rates into WT mitochondria were similar to or even lower than tom70Δ mitochondria, suggesting that adaptation caused by the TOM70 deletion masked a possible decrease in import efficiency arising from the depletion of Tom70. Therefore we tested the import of those pSu9-X fusion proteins into Tom70T mitochondria after TEV protease treatment to remove the receptor domain of Tom70. Now pSu9-Aco1p, pSu9-Atp1p, pSu9-Cit1p, and pSu9-Atp2p were clearly imported into mitochondria without TEV protease treatment more efficiently than those with TEV protease treatment (Fig. 6B). It is interesting to note that pSu9-Atp2p was imported into mitochondria in a Tom70-dependent manner, whereas pAtp2p on its own did not significantly depend on Tom70 for its import. This may suggest that the import ability of the pAtp2p presequence, which is lower than that of the pSu9 presequence, may mask the Tom70 dependence under the present in vitro import conditions. Taken together, we conclude that although different presequences affect the import efficiency of different mature domains differently, the mature parts of pAco1p, pAtp1p, pCit1p, and pIdh1p (and pAtp2p) are at least partly responsible for the Tom70 dependence in their import into mitochondria.
FIGURE 6.
In vitro import of mature parts of pX fused to a Tom70-independent presequence depends on Tom70. A, radiolabeled fusion proteins pCox4-Aco1p, pCox4-Atp1p, pCox4-Cit1p, pCox4-Dld2p, pCox4-Idh1p, and pCox4-Atp2p were imported into wild type (WT) or tom70Δ mitochondria (0.1 mg of protein/ml) and analyzed as in Fig. 2. B, radiolabeled fusion proteins pSu9-Aco1p, pSu9-Atp1p, pSu9-Cit1p, pSu9-Dld2p, and pSu9-Atp2p were imported into Tom70T mitochondria (0.1 mg of protein/ml) with or without TEVp treatment (right panels) and analyzed as described for Fig. 2. The amounts of radiolabeled proteins added to each reaction are set to 100%. p, precursor forms; m, mature forms. 10%, 10% of the input; −ΔΨ, valinomycin was added.
Tom70 Increases Solubility of Aggregate-prone Mature Domains of Tom70-dependent Precursors
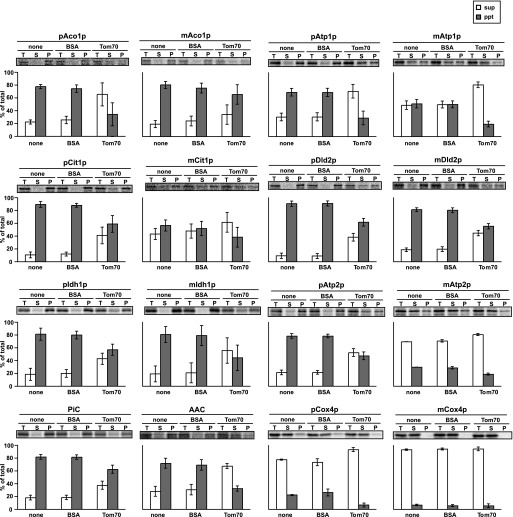
Why do the mature parts of the presequence-containing pAco1p, pAtp1p, pCit1p, and pIdh1p cause Tom70 dependence in their import? Because Tom70 functions as a docking site for the cytosolic chaperone Hsp70 in yeast (19), we first asked whether those Tom70-dependent precursors tend to aggregate. To address this question, we took advantage of the use of PURESYSTEM, a cell-free bacterial translation system reconstituted from purified recombinant components and ribosomes but free of any chaperones (24). Briefly we synthesized the Tom70-dependent precursor proteins in vitro with PURESYSTEM and analyzed the solubility of the translated proteins by centrifugation. Although pCox4, which does not depend on Tom70 in its import, is soluble in the absence of chaperones, the Tom70-dependent precursor proteins, pAco1p, pAtp1p, pCit1p, and pIdh1p (and pDld2p and pAtp2p, as well) form aggregates in the absence of chaperones (Fig. 7, none). Although the tendency of the Tom70-dependent precursor proteins to form aggregates decreased with the removal of the presequence, the corresponding mature domains of pAco1p, pCit1p, pDld2p, and pIdh1p still show a propensity to form aggregates (Fig. 7). Therefore those precursor proteins with aggregate-prone mature parts likely rely on the cytosolic chaperone Hsp70 to remain soluble or import-competent and, as a consequence, require Tom70 as a docking site for the chaperone-precursor complexes on the mitochondrial surface as well.
FIGURE 7.
Tom70 prevents aggregation of newly synthesized Tom70-dependent mitochondrial proteins. Mitochondrial precursor proteins pAco1p, pAtp1p, pCit1p, pDld2p, pIdh1p, pAtp2p, and pCox4p; their mature parts, mAco1p, mAtp1p, mCit1p, mDld2p, mIdh1p, mAtp2p, and mCox4p; and presequence-less Tom70-dependent carrier proteins, AAC and phosphate carrier, were synthesized in vitro at 37 °C for 1 h by coupled transcription/translation reactions with PURESYSTEM and [35S]Met in the absence (none) or presence of 5 μm purified Tom70d (Tom70) or 5 μm BSA. The translation products were subjected to centrifugation at 20,000 × g for 20 min at 4 °C, and pellet (gray bars, ppt) and supernatant (white bar, sup) fractions were separated. The proteins were analyzed by SDS-PAGE and radioimaging. T, total; S, supernatant; P, pellet. The error bars represent S.D. values from three independent experiments.
We then asked whether the role of Tom70 in the import of the Tom70-dependent precursors is more than a docking site for the cytosolic chaperones. We thus tested the solubility of those precursor proteins synthesized with PURESYSTEM in the presence or absence of the recombinant cytosolic domain of Tom70 (Tom70d) or BSA. The presence of the cytosolic domain of Tom70, but not BSA, increased the amounts of the soluble forms (Fig. 7, BSA and Tom70) in a dose (Tom70d)-dependent manner (supplemental Fig. S9). Even the mature domains, mAco1p, mmAtp1p, mDld2p, mIdh1p, and mAtp2p, became more soluble in the presence of Tom70d than BSA (Fig. 7). As a control, AAC, a highly aggregate-prone presequence-less, but Tom70-dependent membrane protein, exhibited increased solubility in the presence of Tom70d as well, whereas aggregate formation of phosphate carrier, another presequence-less aggregate-prone carrier protein, was not relieved by Tom70d (Fig. 7). Therefore, the cytosolic domain of Tom70 has a chaperone-like function to maintain solubility of many, if not all, aggregate-prone substrate proteins.
DISCUSSION
The mitochondrial outer membrane translocator, the TOM40 complex, contains at least three import receptors, Tom20 and Tom22 for presequence-containing precursor proteins and Tom70 for presequence-less proteins. Nevertheless, previous observations that some presequence-containing precursor proteins also depend on Tom70 for their import had been left out of further investigations. In the present study, we took a proteome-wide approach of mitochondrial protein import in vitro and identified several new presequence-containing proteins (pAco1p, pAtp1p, pCit1p, pDld2p, pIdh1p, and pAtp2p) that require Tom70 for their import into mitochondria. By testing their in vitro import into WT and tom70Δ mitochondria, we found that pAco1p, pAtp1p, pCit1p, and pIdh1p indeed depended on Tom70 for their import. Tom70 dependence in import was found for only the pSu9 fusion protein for the mature part of pAtp2p, but not pDld2p. Therefore, identification of the presequence-containing Tom70 substrates by in vitro import of total RNA translation products followed by two-dimensional gel analyses was confirmed by individual in vitro import for pAco1p, pAtp1p, pCit1p, and pIdh1p (and pAtp2p), but not for pDld2p. The spot for Dld2p in two-dimensional gel analyses (spot 3 in Fig. 1A) may be overlapped by other proteins. We then performed systematic analyses of in vitro import of fusion proteins containing various combinations of the presequences or mature parts of Tom70-independent precursor proteins or nonmitochondrial DHFR and the presequences, mature parts, or full-length forms of the identified Tom70-dependent precursor proteins. We thus found that the presequences of the Tom70-dependent precursor proteins require the general presequence receptor Tom20 for their import, whereas the mature parts are responsible for the Tom70 dependence for their import. When synthesized in the reconstituted, cell-free translation system lacking cytosolic chaperones, both full-length forms and mature parts of the Tom70-dependent precursor proteins exhibited formation of insoluble aggregates, which were suppressed by the presence of the receptor domain of Tom70, suggesting that Tom70 has a chaperone-like function. Although a chaperone-like function of Tom70 was previously speculated, direct experimental evidence was lacking (15, 25).
Many mitochondrial precursor proteins tend to form aggregates because of the properties of their presequences, mature parts, or both (26). Therefore aggregate-prone mitochondrial precursor proteins require cytosolic chaperones including yeast Hsp70 (17, 27, 28) to maintain their solubility or import competence. Tom70 is known to function as a docking site for those chaperones to recruit the chaperone-substrate complexes to the TOM40 complex (19). Now the present study indicated that although Tom70 can recognize substrates with internal targeting signals as their cognate receptor, it also receives some substrates as a chaperone even if they do not have internal signals. This is in good accordance with the previous bioinformatics-based proposal that hydrophobic mitochondrial proteins, irrespective of the presence of presequences, could be substrates for Tom70 (17). Perhaps a cytosolic chaperone and its substrate may still form a complex on Tom70 for a while. Then after dissociation of the chaperone-substrate complex, Tom70 likely maintains solubility of the bound substrate protein for efficient transfer to downstream components such as Tom20, Tom22, and Tom40 of the TOM40 complex. Indeed the previous study of peptide scanning analyses showed that some of the Tom70-binding peptides of its substrate, phosphate carrier, are rather hydrophobic (18).
The crystal structure of the cytosolic domain of yeast Tom70 contains a putative binding pocket for substrate proteins, which consists of many hydrophobic residues (25, 29). It is likely that this pocket would provide a binding site for the solvent-exposed hydrophobic region of the unfolded or loosely folded substrate proteins, thereby preventing them from aggregate formation. Systematic analyses to measure the effects of amino acid replacement of the binding pocket of Tom70 on aggregate formation of newly synthesized substrate proteins in the absence of cytosolic chaperones will reveal a more detailed structural basis for the chaperone function of Tom70.
Supplementary Material
Acknowledgments
We thank members of the Endo laboratory for discussions and comments.
This work was supported by grants-in aid for scientific research from Ministry of Education, Culture, Sports, Science, and Technology, a Core Research for Evolutional Science and Technology grant from Japan Science and Technology Agency, and a grant from the Uehara Memorial Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S9.
K. Terao, H. Yamamoto, M. Uchida, S. Kitamura, S. Nishikawa, and T. Endo, unpublished results.
- IEF
- isoelectric focusing
- AAC
- ADP/ATP carrier
- TEVp
- TEV protease
- BSA
- bovine serum albumin
- WT
- wild type
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- MOPS
- 4-morpholinepropanesulfonic acid
- DHFR
- dihydrofolate reductase.
REFERENCES
- 1.Meisinger C., Sickmann A., Pfanner N. (2008) Cell 134, 22–24 [DOI] [PubMed] [Google Scholar]
- 2.Schatz G., Dobberstein B. (1996) Science 271, 1519–1526 [DOI] [PubMed] [Google Scholar]
- 3.Pfanner N., Geissler A. (2001) Nat. Rev. Mol. Cell Biol. 2, 339–349 [DOI] [PubMed] [Google Scholar]
- 4.Endo T., Kohda D. (2002) Biochim. Biophys. Acta 1592, 3–14 [DOI] [PubMed] [Google Scholar]
- 5.Koehler C. M. (2004) Annu. Rev. Cell Dev. Biol. 20, 309–335 [DOI] [PubMed] [Google Scholar]
- 6.Bohnert M., Pfanner N., van der Laan M. (2007) FEBS Lett. 581, 2802–2810 [DOI] [PubMed] [Google Scholar]
- 7.Neupert W., Herrmann J. M. (2007) Ann. Rev. Biochem. 76, 723–749 [DOI] [PubMed] [Google Scholar]
- 8.Endo T., Yamano K. (2009) Biol. Chem. 390, 723–730 [DOI] [PubMed] [Google Scholar]
- 9.Lithgow T., Glick B. S., Schatz G. (1995) Trends Biochem. Sci. 20, 98–101 [DOI] [PubMed] [Google Scholar]
- 10.Yamano K., Yatsukawa Y., Esaki M., Hobbs A. E., Jensen R. E., Endo T. (2008) J. Biol. Chem. 283, 3799–3807 [DOI] [PubMed] [Google Scholar]
- 11.Abe Y., Shodai T., Muto T., Mihara K., Torii H., Nishikawa S., Endo T., Kohda D. (2000) Cell 100, 551–560 [DOI] [PubMed] [Google Scholar]
- 12.Muto T., Obita T., Abe Y., Shodai T., Endo T., Kohda D. (2001) J. Mol. Biol. 306, 137–143 [DOI] [PubMed] [Google Scholar]
- 13.Saitoh T., Igura M., Obita T., Ose T., Kojima R., Maenaka K., Endo T., Kohda D. (2007) EMBO J. 26, 4777–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hines V., Brandt A., Griffiths G., Horstmann H., Brütsch H., Schatz G. (1990) EMBO J. 9, 3191–3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hines V., Schatz G. (1993) J. Biol. Chem. 268, 449–454 [PubMed] [Google Scholar]
- 16.Hachiya N., Mihara K., Suda K., Horst M., Schatz G., Lithgow T. (1995) Nature 376, 705–709 [DOI] [PubMed] [Google Scholar]
- 17.Chan N. C., Likić V. A., Waller R. F., Mulhern T. D., Lithgow T. (2006) J. Mol. Biol. 358, 1010–1022 [DOI] [PubMed] [Google Scholar]
- 18.Brix J., Rüdiger S., Bukau B., Schneider-Mergener J., Pfanner N. (1999) J. Biol. Chem. 274, 16522–16530 [DOI] [PubMed] [Google Scholar]
- 19.Young J. C., Hoogenraad N. J., Hartl F. U. (2003) Cell 112, 41–50 [DOI] [PubMed] [Google Scholar]
- 20.Lithgow T., Schatz G. (1995) J. Biol. Chem. 270, 14267–14269 [DOI] [PubMed] [Google Scholar]
- 21.Lithgow T., Junne T., Wachter C., Schatz G. (1994) J. Biol. Chem. 269, 15325–15330 [PubMed] [Google Scholar]
- 22.Matouschek A., Azem A., Ratliff K., Glick B. S., Schmid K., Schatz G. (1997) EMBO J. 16, 6727–6736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sato T., Esaki M., Fernandez J. M., Endo T. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 17999–18004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shimizu Y., Inoue A., Tomari Y., Suzuki T., Yokogawa T., Nishikawa K., Ueda T. (2001) Nat. Biotechnol. 19, 751–755 [DOI] [PubMed] [Google Scholar]
- 25.Wu Y., Sha B. (2006) Nat. Struct. Mol. Biol. 13, 589–593 [DOI] [PubMed] [Google Scholar]
- 26.Endo T., Mitsui S., Roise D. (1995) FEBS Lett. 359, 93–96 [DOI] [PubMed] [Google Scholar]
- 27.Deshaies R. J., Koch B. D., Werner-Washburne M., Craig E. A., Schekman R. (1988) Nature 332, 800–805 [DOI] [PubMed] [Google Scholar]
- 28.Asai T., Takahashi T., Esaki M., Nishikawa S., Ohtsuka K., Nakai M., Endo T. (2004) J. Biol. Chem. 279, 19464–19470 [DOI] [PubMed] [Google Scholar]
- 29.Li J., Qian X., Hu J., Sha B. (2009) J. Biol. Chem. 284, 23852–23859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanamori T., Nishikawa S., Nakai M., Shin I., Schultz P. G., Endo T. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3634–3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.