Abstract
Structure-function analysis has revealed the mechanism of yeast RNA polymerase II transcription at 8-oxoguanine (8-oxoG), the major DNA lesion resulting from oxidative stress. When polymerase II encounters 8-oxoG in the DNA template strand, it can misincorporate adenine, which forms a Hoogsteen bp with 8-oxoG at the active center. This requires rotation of the 8-oxoG base from the standard anti- to an uncommon syn-conformation, which likely occurs during 8-oxoG loading into the active site. The misincorporated adenine escapes intrinsic proofreading, resulting in transcriptional mutagenesis that is observed directly by mass spectrometric RNA analysis.
Introduction
Reactive oxygen species damage DNA by converting guanine to 8-oxoguanine (8-oxoG).3 8-oxoG is the major mutagenic lesion (1) and can form a Watson-Crick bp with cytosine, but also a Hoogsteen bp with adenine (see Fig. 1a) (2). Mammalian polymerase (pol) II incorporates cytosine or adenine opposite 8-oxoG and can bypass the lesion (3–5). Bypass is enhanced by the 3′-RNA cleavage factor TFIIS, although this apparently does not lead to a substantial removal of misincorporated adenine (6, 7).
FIGURE 1.
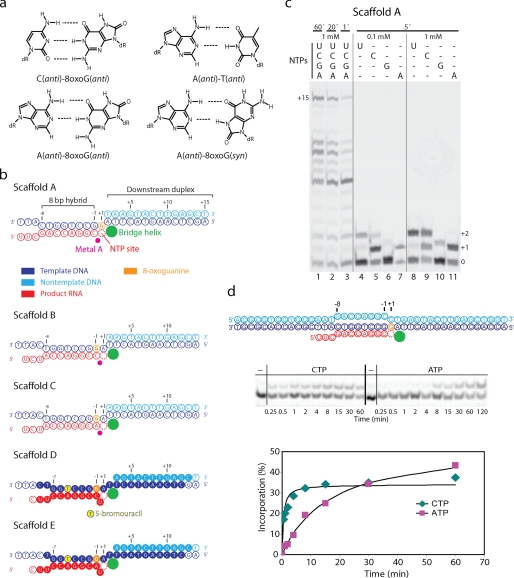
8-oxoG directs slow adenine misincorporation. a, base pairing properties of 8-oxoG. b, nucleic acid scaffolds (color coding used throughout). RNA was 5′-end-labeled with 6-carboxyfluorescein and a UGCAU linker. Filled circles denote nucleotides present in the structures (see Fig. 3). c, RNA extension with EC A. d, time courses of incorporation with CTP or misincorporation with ATP. The indicated complementary scaffold was used. The DNA non-template was 5′-end-labeled with biotin using a TTTTT linker. The RNA was 5′-end-labeled with 6-carboxyfluorescein and a UGCAU linker.
Here, we investigated the molecular basis of 8-oxoG transcription by pol II. We show that pol II from the yeast Saccharomyces cerevisiae can misincorporate adenine at 8-oxoG and that the misincorporated adenine escapes intrinsic polymerase proofreading and remains in the transcript upon elongation, resulting in transcriptional mutagenesis. The mutant transcript was observed directly by mass spectrometry. We then used x-ray analysis to show that the misincorporated adenine forms a Hoogsteen bp with 8-oxoG at the pol II active center. In contrast, 8-oxoG forms a standard Watson-Crick bp with a correctly incorporated cytosine. Finally, we suggest a mechanism for mutagenic transcription and its suppression by TFIIS.
EXPERIMENTAL PROCEDURES
Sample Preparation
S. cerevisiae pol II containing a hexahistidine-tagged Rpb3 subunit (strain kindly provided by the M. Kashlev laboratory) was purified as described (8). Briefly, cells were lysed by bead beating. The lysate was cleared by centrifugation and ultracentrifugation. The cleared lysate was precipitated by the addition of saturated ammonium sulfate. The pellet was subjected to nickel-nitrilotriacetic acid affinity chromatography. The eluted protein was subjected to anion exchange chromatography (Mono Q, GE Healthcare). The last elution peak was collected and concentrated. The concentrated pol II was precipitated by the addition of ammonium sulfate, and the pellets were stored at −80 °C. Recombinant TFIIS was prepared as described (9). DNA strands containing 8-oxoG were synthesized by BioSpring.
Elongation Complex Assembly and RNA Extension and Cleavage Assays
For RNA extension assays, synthetic oligonucleotides were annealed for scaffold assembly as described (10). Elongation complexes (ECs) were obtained by incubating pol II with 2 m eq of nucleic acid scaffold in transcription buffer (20 mm HEPES (pH 7.6), 60 mm (NH4)2SO4, 8 mm MgSO4, 10 μm ZnCl2, 10% (v/v) glycerol, and 10 mm dithiothreitol) at 20 °C for 30 min. For RNA extension assays, different amounts of NTPs were added, and the mixture was incubated at 28 °C as described (10). ECs containing complete complementary scaffolds were assembled essentially as described (11). Briefly, the DNA non-template was 5′-end-labeled with biotin using a TTTTT linker. The RNA was 5′-end-labeled with 6-carboxyfluorescein and a UGCAU linker. For EC assembly, pol II was incubated with a hybrid of the DNA template strand annealed to the RNA (2-fold excess) for 15 min at 20 °C, subsequently with the biotinylated DNA non-template strand (4-fold excess) for 10 min at 25 °C, and finally with recombinant Rpb4/7 (5-fold excess) for 10 min at 25 °C. Bead-based RNA extension assays were carried out as described (12) with minor changes. In summary, beads (Dynabeads MyOneTM streptavidin T1, Invitrogen) were added and incubated for 30 min at 25 °C. Beads were subsequently washed with transcription buffer containing 0.1% Triton-X, transcription buffer containing 0.2 m (NH4)2SO4, and transcription buffer. Beads were resuspended in transcription buffer and incubated with NTPs at 28 °C. The reaction was stopped by the addition of 100 mm EDTA. The beads were transferred into urea loading buffer, and samples were loaded onto a 20% polyacrylamide gel containing 7 m urea. The RNA products 5′-end-labeled with 6-carboxyfluorescein were visualized with a Typhoon 9400 scanner (GE Healthcare). Gel bands were quantified using ImageQuant (GE Healthcare). The data points were fitted assuming Michaelis-Menten kinetics using SigmaPlot. For MALDI time-of-flight analysis, reactions were incubated with the indicated NTPs, stopped, and analyzed as described (10). Because the amounts of product required for MALDI analysis could not be obtained with the use of complementary scaffolds, we used minimal scaffolds.
Crystal Structure Analysis
The 8-oxoG-containing scaffolds were co-crystallized, and the structures were determined essentially as described (13) with minor changes. The crystallization solution lacked magnesium ions (200 mm ammonium acetate, 300 mm sodium acetate, 50 mm HEPES (pH 7.0), 4–6% (w/v) polyethylene glycol 6000, and 5 mm tris(2-carboxyethyl)phosphine). Diffraction data were collected at a wavelength of 0.919 Å at the protein crystallography beamline X06SA of the Swiss Light Source using a PILATUS 6M pixel detector (see Table 1) (14). Raw data were processed with XDS (15). Structures were solved by molecular replacement with the program Phaser (16) using the structure of the complete 12-subunit pol II without nucleic acids (Protein Data Bank code 1Y1W) (9). The molecular replacement solution was subjected to rigid body refinement with CNS Version 1.2 (17). Model building was done with Coot (18) and Moloc (Gerber Molecular Design). The nucleic acids were built stepwise into unbiased Fo − Fc electron density. The register of the nucleic acids was unambiguously defined by bromine labeling as described (10). Refinement was monitored with the free R-factor, calculated from the same set of excluded reflections as in the refinement of the complete pol II complex (19) and the complete pol II EC (9, 10, 13).
TABLE 1.
Crystallographic data and refinement statistics for complete pol II ECs
Diffraction data were collected at beamline PX1 at the Swiss Light Source and were processed with program XDS (15). r.m.s.d., root mean square deviation.
| EC D | EC E | |
|---|---|---|
| Data collection | ||
| Space group | C2221 | C2221 |
| Unit cell axes (Å) | 220.6, 392.0, 281.5 | 221.2, 394.1, 282.3 |
| Wavelength (Å) | 0.9188 | 0.9186 |
| Resolution range (Å) | 50.0–3.7 (3.8–3.7)a | 50.0–3.9 (4.0–3.9)a |
| Unique reflections | 250,702 (19,429) | 216,612 (15,791) |
| Completeness (%) | 99.6 (99.9) | 99.7 (99.8) |
| Rsym (%) | 8.2 (56.7) | 7.8 (86.3) |
| I/σ(I) | 11.9 (2.7) | 13.4 (2.6) |
| Redundancy | 3.8 (3.8) | 3.9 (4.0) |
| Refinement | ||
| Non-hydrogen atoms | 32,348 | 32,298 |
| r.m.s.d. bonds (Å) | 0.008 | 0.008 |
| r.m.s.d. angles | 1.50° | 1.48° |
| Peak in anomalous difference Fourier (σ) | 6.7 | 5.1 |
| Rcryst (%) | 22.5 | 22.8 |
| Rfree (%) | 25.8 | 26.6 |
a Numbers in parentheses correspond to the highest resolution shells.
RESULTS
Yeast pol II Slowly Misincorporates Adenine at 8-oxoG
We reconstituted an S. cerevisiae pol II EC containing 8-oxoG at position +1 of the template strand, opposite the NTP-binding site (Fig. 1b, Scaffold A) (13). Incubation of the resulting EC A with NTPs led to RNA extension and accumulation of the runoff product (Fig. 1c, lanes 1–3; “Experimental Procedures”; and supplemental Fig. 1). Incubation with individual NTPs showed that both cytosine and adenine are incorporated opposite the lesion (Fig. 1c, lanes 4–11). Uridine was also incorporated, but this was also observed for undamaged DNA (supplemental Fig. 1) and is caused by a lesion-independent mechanism called template misalignment (20). Incubation with GTP also resulted in some misincorporation (Fig. 1c, lanes 4–11), and for undamaged DNA, some misincorporation of ATP was also observed (supplemental Fig. 1), but these misincorporations are at a level expected from yeast pol II in the absence of TFIIS (8). To further investigate adenine misincorporation and cytosine incorporation opposite the lesion, we performed a time course analysis with an EC that contained a fully complementary complete scaffold. This revealed that adenine misincorporation was ∼34-fold slower than cytosine incorporation (Fig. 1d and “Experimental Procedures”), suggesting a misincorporation frequency of a few percent, consistent with data for mammalian pol II (7).
Adenine Misincorporation Results in Transcriptional Mutagenesis
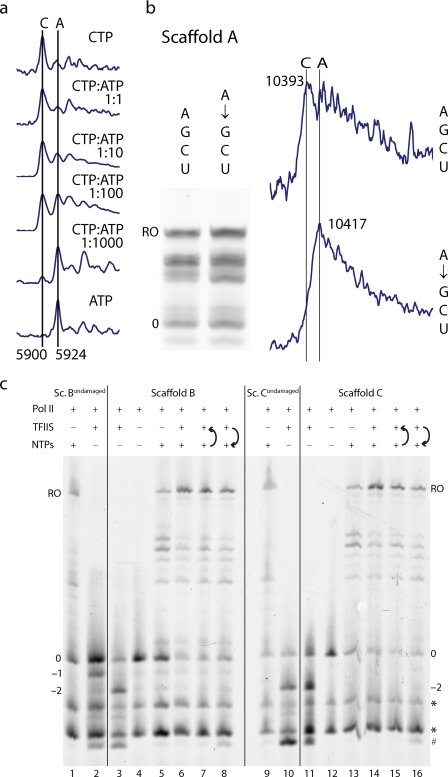
To test whether misincorporation occurs when CTP is present and whether the misincorporated adenine remains in the RNA, we analyzed RNA products by MALDI mass spectrometry. We could distinguish RNAs containing cytosine or adenine, which differ in mass by only 24 Da (Fig. 2). RNA with the misincorporated adenine was detected when an excess of ATP over CTP was used (Fig. 2a). At an equimolar ATP:CTP ratio, misincorporation could not be detected, apparently because the mass of the misincorporation product was indistinguishable from that of the cytosine incorporation product associated with a sodium ion (Fig. 2a). When EC A was incubated with ATP before NTP addition, runoff RNA contained the misincorporated adenine (Fig. 2b). In summary, 8-oxoG can cause adenine misincorporation in the presence of CTP, and the inserted adenine escapes polymerase intrinsic proofreading and remains in the RNA, leading to transcriptional mutagenesis.
FIGURE 2.
Transcriptional mutagenesis. a, adenine misincorporation in EC A in the presence of CTP at different ratios (1 mm ATP, 5-min incubation). RNA products containing adenine (5900 Da) or cytosine (5924 Da) were identified by mass spectrometry (see “Experimental Procedures”). b, misincorporated adenine remains in the runoff (RO) RNA. EC A was incubated with NTPs (1 mm, 60 min) or with ATP (1 mm, 5 min), followed by incubation with NTPs (1 mm, 60 min) as indicated. Mass spectrometry identified runoff RNAs containing cytosine (10,393 Da) or adenine (10,417 Da). c, effect of the cleavage stimulatory factor TFIIS on RNA elongation with ECs B and C. Incubation with TFIIS for 5 min resulted in cleavage products (lanes 3, 10, and 11). The presence of NTPs (1 mm) increased runoff formation (lanes 6 and 14) independent of the order of addition (lanes 7, 8, 15, and 16). * and #, RNA impurities and a cleavage product after backtracking, respectively. Controls included incubations of fluorescent RNA with pol II (lanes 4 and 12) and of lesion-free ECs Bundamaged and Cundamaged with TFIIS (lanes 2 and 10).
TFIIS Removes a Misincorporated Adenine
Because TFIIS enhances intrinsic pol II RNA cleavage (21, 22), we investigated whether TFIIS can stimulate removal of a misincorporated adenine opposite the 8-oxoG lesion. We added recombinant TFIIS to ECs B and C, which contained cytosine or adenine, opposite the lesion (Fig. 1b), and incubated the mixture for 5 min. This led to cleavage of an RNA 3′-dinucleotide, which was not observed for undamaged DNA containing a G-C match (Fig. 2c, Sc. Bundamaged). Dinucleotide cleavage generally occurs in the presence of TFIIS (23) and was also observed for undamaged DNA containing a G-A mismatch (Fig. 2c, Sc. Cundamaged). Upon subsequent incubation with NTPs for 5 min, RNA was elongated (Fig. 2c). MALDI analysis of runoff RNA produced by EC C, incubated with both TFIIS and NTPs for 5 min, indicated replacement of the RNA 3′-adenine by cytosine (data not shown). This showed that TFIIS had stimulated removal of the misincorporated adenine from the RNA prior to elongation. These data suggest that TFIIS suppresses transcriptional mutagenesis by stimulating proofreading, but they do not demonstrate proofreading under rapid elongation conditions. During rapid elongation, proofreading apparently does not occur to a large extent, at least with mammalian pol II (7).
8-oxoG and the Misincorporated Adenine Form a Hoogsteen Pair
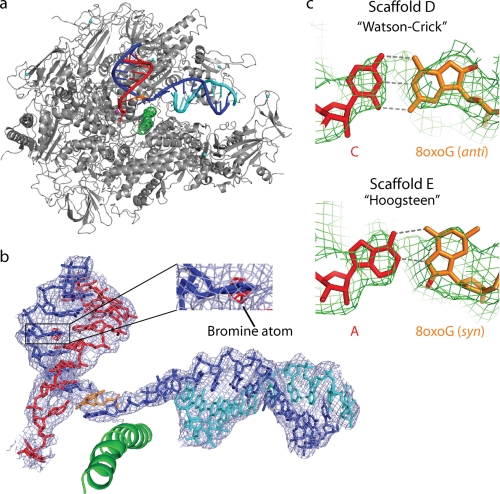
To investigate the molecular basis for 8-oxoG transcription, we solved crystal structures of pol II ECs containing 8-oxoG (see “Experimental Procedures”). An interpretable electron density map was obtained with an 8-oxoG-C bp at the penultimate position in the hybrid (Fig. 1b, Scaffold D). The structure was solved at 3.7 Å resolution (Fig. 3 and Table 1). Bromine labeling of the DNA revealed that the EC was not post-translocated but adopted a state with a frayed RNA 3′-end at register +1. This RNA fraying does not influence our analysis, as it occurred also in a control structure with undamaged DNA (supplemental Fig. 2). We recently published a detailed structural analysis of RNA fraying and showed that it occurs with different types of terminal RNA nucleotides and that frayed nucleotides can be stabilized by a neighboring mismatch pair (8).
FIGURE 3.
Structures of 8-oxoG-containing pol II ECs. a, overview of the EC D structure. silver, pol II; green, bridge helix; orange, 8-oxoG. b, structure and 2Fo − Fc electron density (blue, contoured at 1.0σ) of nucleic acids in EC E. The anomalous difference Fourier map (red, contoured at 3.5σ) reveals the bromine atom. c, unbiased difference electron density (green, contoured at 2.7σ) for 8-oxoG-containing bp in ECs D (upper) and E (lower).
The 8-oxoG-C bp adopted Watson-Crick geometry at register −1, consistent with normal cytosine incorporation (Fig. 3c). An additional structure at 3.9 Å (Fig. 3b) contained an adenine opposite 8-oxoG (Fig. 1b, Scaffold E; and Table 1). The unbiased difference electron density for the bp at −1 revealed the 8-oxoG in syn-conformation, forming a Hoogsteen bp with adenine in the RNA (Fig. 3c).
DISCUSSION
We have shown that yeast pol II can misincorporate adenine at an 8-oxoG lesion in the DNA template strand, that the misincorporated adenine forms a Hoogsteen bp with the lesion at the polymerase active center, and that the misincorporated adenine escapes polymerase intrinsic proofreading and remains in the RNA as a product of transcriptional mutagenesis. How may misincorporation and proofreading occur? In incoming DNA, 8-oxoG likely adopts a standard anti-conformation within a Watson-Crick bp, but its base must rotate by 180° to the syn-conformation in the 8-oxoG-A DNA-RNA Hoogsteen bp at the active center. Modeling shows that base rotation in the templating position at register +1 would result in a clash with the bridge helix and the DNA base at register −1. We therefore propose that base rotation occurs during 8-oxoG translocation from the downstream position +2 to the templating position +1 in the recently characterized pre-templating position (24), where the 8-oxoG base would be unpaired and free to rotate from anti to syn before being loaded into the templating position. In the templating site, 8-oxoG likely forms a Hoogsteen bp with an incoming ATP, leading to adenine misincorporation. TFIIS induces backtracking of 8-oxoG from the active center to downstream DNA. When repeated translocation preserves the anti-conformation, correct cytosine incorporation occurs, and RNA is elongated error-free.
Transcriptional mutagenesis as demonstrated here in vitro may be a threat in vivo because a yeast strain lacking the gene encoding TFIIS is sensitive to oxidative stress (25, 26). This in vivo effect may result at least in part from the accumulation of mutant RNAs that give rise to mutant proteins with impaired function. Transcriptional mutagenesis does not occur at dinucleotide lesions in the template, although these also cause misincorporation (10, 13). At a cyclobutane pyrimidine dimer, misincorporation triggers pol II stalling (10), whereas misincorporation is a result of stalling at a cisplatin cross-link lesion (13).
Like RNA polymerases (6, 27–29), DNA polymerases incorporate cytosine or adenine opposite 8-oxoG to various extents (2, 30–32). High fidelity DNA polymerases accommodate and bypass an 8-oxoG-C bp (2, 33–35). One structure of a high fidelity DNA polymerase revealed an 8-oxoG-A Hoogsteen bp (2). This suggests that the same mechanism of adenine misincorporation at 8-oxoG occurs during replication and transcription and that stable Hoogsteen pairing can dominate nucleic acid arrangements, although the active centers are complementary to structurally very different B- and A-form template-product duplexes in these structurally unrelated DNA and RNA polymerases, respectively.
Supplementary Material
Acknowledgments
We thank Dmitry Vassylyev, Thomas Fröhlich, Georg Arnold, and members of the Cramer laboratory.
This work was supported in part by grants from the Deutsche Forschungsgemeinschaft, Sonderforschungsbereich SFB646, and Transregio 5. Part of this work was performed at the Swiss Light Source at the Paul Scherrer Institut (Villigen, Switzerland).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
The atomic coordinates and structure factors (codes 3I4M and 3I4N) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- 8-oxoG
- 8-oxoguanine
- pol
- polymerase
- TFIIS
- transcription factor IIS
- EC
- elongation complex
- MALDI
- matrix-assisted laser desorption ionization.
REFERENCES
- 1.Lindahl T. (1993) Nature 362, 709–715 [DOI] [PubMed] [Google Scholar]
- 2.Hsu G. W., Ober M., Carell T., Beese L. S. (2004) Nature 431, 217–221 [DOI] [PubMed] [Google Scholar]
- 3.Kuraoka I., Endou M., Yamaguchi Y., Wada T., Handa H., Tanaka K. (2003) J. Biol. Chem. 278, 7294–7299 [DOI] [PubMed] [Google Scholar]
- 4.Tornaletti S., Maeda L. S., Kolodner R. D., Hanawalt P. C. (2004) DNA Repair 3, 483–494 [DOI] [PubMed] [Google Scholar]
- 5.Kathe S. D., Shen G. P., Wallace S. S. (2004) J. Biol. Chem. 279, 18511–18520 [DOI] [PubMed] [Google Scholar]
- 6.Kuraoka I., Suzuki K., Ito S., Hayashida M., Kwei J. S., Ikegami T., Handa H., Nakabeppu Y., Tanaka K. (2007) DNA Repair 6, 841–851 [DOI] [PubMed] [Google Scholar]
- 7.Charlet-Berguerand N., Feuerhahn S., Kong S. E., Ziserman H., Conaway J. W., Conaway R., Egly J. M. (2006) EMBO J. 25, 5481–5491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sydow J. F., Brueckner F., Cheung A. C., Damsma G. E., Dengl S., Lehmann E., Vassylyev D., Cramer P. (2009) Mol. Cell 34, 710–721 [DOI] [PubMed] [Google Scholar]
- 9.Kettenberger H., Armache K. J., Cramer P. (2004) Mol. Cell 16, 955–965 [DOI] [PubMed] [Google Scholar]
- 10.Brueckner F., Hennecke U., Carell T., Cramer P. (2007) Science 315, 859–862 [DOI] [PubMed] [Google Scholar]
- 11.Komissarova N., Kireeva M. L., Becker J., Sidorenkov I., Kashlev M. (2003) Methods Enzymol. 371, 233–251 [DOI] [PubMed] [Google Scholar]
- 12.Dengl S., Cramer P. (2009) J. Biol. Chem. 284, 21270–21279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Damsma G. E., Alt A., Brueckner F., Carell T., Cramer P. (2007) Nat. Struct. Mol. Biol. 14, 1127–1133 [DOI] [PubMed] [Google Scholar]
- 14.Broennimann Ch., Eikenberry E. F., Henrich B., Horisberger R., Huelsen G., Pohl E., Schmitt B., Schulze-Briese C., Suzuki M., Tomizaki T., Toyokawa H., Wagner A. (2006) J. Synchrotron Radiat. 13, 120–130 [DOI] [PubMed] [Google Scholar]
- 15.Kabsch W. (1993) J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
- 16.McCoy A. J., Grosse-Kunstleve R. W., Storoni L. C., Read R. J. (2005) Acta Crystallogr. 61, 458–464 [DOI] [PubMed] [Google Scholar]
- 17.Brunger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 18.Emsley P., Cowtan K. (2004) Acta Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 19.Armache K. J., Mitterweger S., Meinhart A., Cramer P. (2005) J. Biol. Chem. 280, 7131–7134 [DOI] [PubMed] [Google Scholar]
- 20.Kashkina E., Anikin M., Brueckner F., Pomerantz R. T., McAllister W. T., Cramer P., Temiakov D. (2006) Mol. Cell 24, 257–266 [DOI] [PubMed] [Google Scholar]
- 21.Jeon C., Agarwal K. (1996) Proc. Natl. Acad. Sci. U. S. A. 93, 13677–13682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas M. J., Platas A. A., Hawley D. K. (1998) Cell 93, 627–637 [DOI] [PubMed] [Google Scholar]
- 23.Izban M. G., Luse D. S. (1993) J. Biol. Chem. 268, 12864–12873 [PubMed] [Google Scholar]
- 24.Brueckner F., Cramer P. (2008) Nat. Struct. Mol. Biol. 15, 811–818 [DOI] [PubMed] [Google Scholar]
- 25.Koyama H., Ito T., Nakanishi T., Sekimizu K. (2007) Genes Cells 12, 547–559 [DOI] [PubMed] [Google Scholar]
- 26.Koyama H., Ito T., Nakanishi T., Kawamura N., Sekimizu K. (2003) Genes Cells 8, 779–788 [DOI] [PubMed] [Google Scholar]
- 27.Yamaguchi Y., Mura T., Chanarat S., Okamoto S., Handa H. (2007) Genes Cells 12, 863–875 [DOI] [PubMed] [Google Scholar]
- 28.Viswanathan A., Doetsch P. W. (1998) J. Biol. Chem. 273, 21276–21281 [DOI] [PubMed] [Google Scholar]
- 29.Chen Y. H., Bogenhagen D. F. (1993) J. Biol. Chem. 268, 5849–5855 [PubMed] [Google Scholar]
- 30.Maga G., Villani G., Crespan E., Wimmer U., Ferrari E., Bertocci B., Hübscher U. (2007) Nature 447, 606–608 [DOI] [PubMed] [Google Scholar]
- 31.Broyde S., Wang L., Rechkoblit O., Geacintov N. E., Patel D. J. (2008) Trends Biochem. Sci. 33, 209–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rechkoblit O., Malinina L., Cheng Y., Kuryavyi V., Broyde S., Geacintov N. E., Patel D. J. (2006) PLoS Biol. 4, e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krahn J. M., Beard W. A., Miller H., Grollman A. P., Wilson S. H. (2003) Structure 11, 121–127 [DOI] [PubMed] [Google Scholar]
- 34.Freisinger E., Grollman A. P., Miller H., Kisker C. (2004) EMBO J. 23, 1494–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brieba L. G., Eichman B. F., Kokoska R. J., Doublié S., Kunkel T. A., Ellenberger T. (2004) EMBO J. 23, 3452–3461 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.