Abstract
A proteomic analysis of proteins bound to the osteocalcin OSE2 sequence of the mouse osteocalcin promoter identified TRPS1 as a regulator of osteocalcin transcription. Mutations in the TRPS1 gene are responsible for human tricho-rhino-phalangeal syndrome, which is characterized by skeletal and craniofacial abnormalities. TRPS1 has been shown to bind regulatory promoter sequences containing GATA consensus binding sites and to repress transcription of genes involved in chondrocyte differentiation. Here we show that TRPS1 can directly bind the osteocalcin promoter in the presence or absence of Runx2. TRPS1 binds through a GATA binding sequence in the proximal promoter of the osteocalcin gene. The GATA binding site is conserved in mice, humans, and rats, although its location and orientation are not. Mutation of the mouse or human GATA binding sequence abrogates binding of TRPS1 to the osteocalcin promoter. We show that TRPS1 is expressed in osteosarcoma cells and upon induction of osteoblast differentiation in primary mouse bone marrow stromal cells and that TRPS1 regulates the expression of osteocalcin in both cell types. The expression of TRPS1 modulates mineralized bone matrix formation in differentiating osteoblast cells. These data suggest a role for TRPS1 in osteoblast differentiation, in addition to its previously described role in chondrogenesis.
Introduction
Runx2, previously described as CBFA1, OSF2, AML3, or PEPB2αA, is a runt homology domain transcription factor and plays a key role in driving osteoblast differentiation. Runx2 expression, which occurs in differentiating and mature osteoblasts, is essential for bone formation, as illustrated by the phenotype of Runx2-deficient mice, which lack osteoblasts and have skeletons that are primarily composed of immature chondrocytes (1–3). Haploinsufficiency for RUNX2 in humans results in cleidocranial dysplasia, a syndrome that is characterized by delayed endochondral and intramembranous ossification (4). Runx2 transactivates genes involved in the deposition of bone matrix, e.g. those encoding osteocalcin, collagen IA1, osteopontin, and matrix metalloproteinase 13 (MMP13), and regulation of osteoclastogenesis, e.g. the genes encoding RankL and osteoprotegerin (reviewed in Ref. 5). Although Runx2 is expressed in cells of the osteoblast lineage throughout skeletal development, its levels increase only 1.5–2-fold as preosteoblasts differentiate into mature osteoblasts. Furthermore, the amount of Runx2 bound to target promoters, as measured by chromatin immunoprecipitation, is relatively unchanged throughout differentiation (6), yet the targets it regulates have distinct patterns of expression. These observations suggest that the regulation of Runx2 activity, either by post-translational modification or by the binding of co-regulators, plays a substantial role in defining the Runx2-mediated activation of target genes. Various proteins have been shown to interact with Runx2 and modulate its activity, including Smad3, CREB2 -binding protein (CBP) HES1, Groucho/TLE, and histone deacetylases (reviewed in Ref. 7).
Osteocalcin is one of the most abundant (∼15%) non-collagenous proteins in the bone extracellular matrix and is expressed, secreted, and deposited by osteoblasts, chondroblasts, and odontoblasts during extracellular matrix mineralization. Osteocalcin expression is regulated by Runx2 binding to the OSE2 (osteoblast-specific cis-acting) element in the promoter (8–10). In addition, osteocalcin expression is regulated at the transcriptional level by hormones such as 1,25-dihydroxyvitamin D (vitamin D3) and dexamethasone and by cytokines such as TGF-β.
In this study, we performed an unbiased screen for factors that bind the osteocalcin promoter sequence and identified TRPS1 (tricho-rhino-phalangeal syndrome-1) as a protein that specifically associates with the osteocalcin promoter. Tricho-rhino-phalangeal syndrome is an autosomal dominant disorder that results in craniofacial and skeletal abnormalities (11, 12). Patients have sparse scalp hair, a bulbous nose, short stature, hip abnormalities, brachydactyly, cone-shaped epiphyses in the phalanges, and premature growth plate closure (11, 13, 14). Tricho-rhino-phalangeal syndrome is due to mutations affecting the TRPS1 gene (also known as GC79) and is classified into three variants (15, 16). The TRPS1 protein contains nine zinc finger domains, including one GATA-type zinc finger and two C-terminal zinc fingers with strong similarity to the Ikaros family of lymphoid transcription factors (16, 17). The TRPS1 protein localizes to the nucleus and specifically binds to the GATA consensus motif or its inverse sequence (17). TRPS1 has been reported to act as a transcriptional repressor in vitro, and this activity is dependent on both a highly conserved GATA DNA-binding domain and the Ikaros-like zinc finger motifs (17, 18). Genes that are transcriptionally regulated by TRPS1 include those encoding prostate-specific antigen, STAT3, and PTHrP (19–21).
In the developing mouse embryo at midgestation (embryonic days 12.5–14.5), TRPS1 is expressed in the joints of the limbs, maxilla, mandible, snout, prospective phalanges, and hair follicles (17, 22–24). Mice engineered to express TRPS1 with a GATA-binding domain deletion (Trps1ΔGT mice) have a stronger phenotype in the heterozygous state than mice heterozygous for a null allele of Trps1 (20, 24). Trps1ΔGT homozygote mice die of respiratory failure at birth due to malformation of the thoracic spine and ribs and have delayed endochondral ossification, delayed chondrocyte differentiation, and accelerated mineralization of the perichondrium (25). Trps1−/− knock-out mice die perinatally, with hair follicle abnormalities, an abnormal and fragile rib cage, and shortened maxillary, mandibular, and long bones (20). TRPS1 has been shown to physically and genetically interact with Runx2 (25). Whereas Trps1ΔGT/+ mice exhibit accelerated mineralization and a long growth plate, Runx2+/− mice have delayed mineralization and a shortened growth plate. Double heterozygous mice have a rescued growth plate phenotype, suggesting that TRPS1 regulates Runx2-dependent functions by driving mineralization of the perichondrium during growth plate development. Trps1 was recently identified by a genome scan for candidate genes involved in the regulation of bone mineral density in mice (26).
Osteoblast differentiation depends on the ability of Runx2 to integrate information from co-regulators and chromatin-associated proteins to achieve promoter-specific binding and temporal transcriptional activation of target genes involved in the differentiation process. How distinct promoter complexes are formed and the identities of the proteins involved remain unresolved questions. It is not well understood how Runx2 is able to function as a repressor in some contexts and as an activator in others, although this versatility probably involves the organization of Runx2-binding sites and their proximity to binding sites for other co-regulatory proteins. It has been suggested that TRPS1 may regulate the activity of Runx2 to coordinate chondrocyte hypertrophy and perichondrial mineralization in the developing growth plate (25). In searching for proteins that modulate transcription from the osteocalcin promoter, we found that TRPS1 regulates the Runx2-dependent expression of osteocalcin, a protein expressed by mature osteoblasts and hypertrophic chondrocytes. Our data suggest that TRPS1 may function to coordinate the mineralization of the extracellular matrix by osteoblasts in addition to hypertrophic chondrocytes and thus regulate the development of bones formed by intramembranous or endochondral ossification.
EXPERIMENTAL PROCEDURES
Plasmids
The mouse osteocalcin-Luc plasmid expresses luciferase under the control of the −147 to +13 segment of the mouse osteocalcin gene (27). The p6OSE2-Luc plasmid contains six copies of the OSE2 sequence derived from the mouse osteocalcin promoter followed by a minimal promoter (8). The human osteocalcin-Luc plasmid was generated by PCR amplification of the −212 to +28 region (relative to the transcription start site) of the osteocalcin promoter using genomic DNA isolated from U2OS cells. These regulatory promoter sequences were cloned into the pGL3-Basic luciferase reporter vector (Promega). GATA and Runx binding site mutations were created using the Stratagene QuikChange site-directed mutagenesis kit according to instructions. The TRPS1 expression plasmid was acquired from Open Biosystems. The Runx2 expression plasmid was provided by Philip W. Hinds (Tufts-New England Medical Center). pRK5-βgal expresses β-galactosidase under the control of a cytomegalovirus promoter (28).
Cell Culture
ROS17/2.8 rat osteosarcoma cells were cultured in F-12 medium with 10% fetal bovine serum and 1× penicillin/streptomycin. 293T, HepG2, and U2OS cells were cultured in DMEM, 10% fetal bovine serum, and 1× penicillin/streptomycin. The prostate cancer cell line LNCaP was cultured in RPMI 1640 medium containing 10% fetal bovine serum. Cells were treated with 2 ng/ml TGF-β1 (Peprotech) for 1.5 h, 1 nm synthetic androgen R1881 (NEN Life Science) for 24 h, or 10 nm vitamin D3 (Calbiochem) for 24 h. Bone marrow stromal cells (provided by Tamara Alliston, University of California, San Francisco) were isolated from bone marrow of femora and tibiae of 2.5-month-old C57BL/6 mice. Bone marrow pellets were resuspended and maintained in growth medium (Dulbecco's modified Eagle's medium, 20% fetal bovine serum, non-essential amino acids, vitamin solution, l-glutamine, and 1× penicillin/streptomycin). For osteoblast differentiation, bone marrow stromal cells were plated in growth medium at 3 × 104 cells/well in 12-well plates. At confluence, the cells were switched to medium containing 10% fetal bovine serum, 1× penicillin/streptomycin, 10 mm β-glycerophosphate, and 50 μg/ml l-ascorbate-2-phosphate. Medium was changed every 2 days, and cells and medium were harvested at the indicated time points for RNA isolation, enzyme-linked immunosorbent assay, or alizarin red S staining.
shRNA and Lentiviral Infection
shRNA oligonucleotides against mouse and rat Trps1 were designed using pSicoOligomaker 1.5 (developed by A. Ventura) and checked using Blast. Their sequences are shown in supplemental Table 1. The oligonucleotides were phosphorylated with T4 kinase, annealed, and cloned into pSicoR-puro as described (see the Tyler Jacks Laboratory web site). Lentivirus was produced following co-transfection of 293T cells with the pSicoR-puro-derivative vector and VSV-G and Δ8.9 plasmids (29, 30) using Lipofectamine 2000 (Invitrogen). Viral supernatant was harvested 48–72 h after transfection, passed through a 0.45-μm syringe filter, diluted 2:3 with fresh medium containing 8 μg/ml protamine sulfate, and used to infect target cells in 6-well plates at ∼80% confluence. Selection with puromycin at 2 μg/ml was initiated 48 h after infection. mRNA levels were quantified by qRT-PCR, and protein expression was visualized by immunoblot.
DNA Pull-down Assays
Biotinylated RVP3 forward primer and GL2 reverse primer (Invitrogen), which flank the multiple cloning site in pGL3-Luc, were used to generate biotinylated double-stranded DNA oligonucleotides from sequences cloned into pGL3-Basic by PCR. PCR products were purified using a Qiagen PCR purification kit and quantified by ethidium bromide staining of an agarose gel using ImageJ. 4 pmol of each biotinylated PCR product were bound to 10 μg of M280 Dynabeads (∼200 pmol/mg; DYNAL). Nuclear extracts were prepared essentially as described (31). Per pull-down assay, 1–2 mg of nuclear lysate was incubated with control DNA (i.e. pGL3-Basic vector without insert) for 1 h at 4 °C. Equal volumes of precleared lysate were bound overnight to the biotinylated DNA sequences in the presence of 5 μg of poly(dI-dC)-poly(dI-dC). The beads were washed four times with incubation buffer (20 mm Tris, pH 7.9, 150 mm NaCl, 5% glycerol, 0.5 mm EDTA, 1 mm MgCl2, 1% Triton X-100). Bound proteins were eluted with 0.25 m NaCl, separated by SDS-PAGE, and stained with Coomassie Blue (Gel Code Blue Stain; Pierce) for analysis by mass spectrometry or transferred to polyvinylidene difluoride (Immobilon-P, Millipore) for immunoblotting.
Transfection and Reporter Assays
ROS17/2.8 and U2OS cells were transfected in 12-well plates using FuGENE 6 or Lipofectamine 2000, respectively. 48 h after transfection, cells were washed with phosphate-buffered saline and lysed in Reporter Lysis Buffer (Promega). Luciferase activities were measured as described by the Marine Genomics Laboratory (see the Marine Genomics Laboratory web site) and normalized against β-galactosidase activity (Tropix). Each assay was performed in triplicate and is representative of at least three independent experiments.
RT-PCR
RNA was isolated using Qiashredder and the RNeasy kit with DNase treatment from Qiagen following the manufacturer's instructions. 1 μg of RNA was used in each reverse transcription reaction using the iScript cDNA synthesis kit (Bio-Rad). Real-time PCR was performed using an Opticon-2 PCR machine (MJ Research/Bio-Rad). cDNA equivalent to 35 ng was amplified in a 40-μl reaction containing a 0.5 μm concentration of each primer, 200 μm dNTPs, 2.5 mm MgCl2, 0.2× SYBR Green (Molecular Probes), and 1.25 units of Taq DNA polymerase (Invitrogen) or Amplitaq Gold DNA Polymerase (Applied Biosystems) for SYBR Green reactions. Amplification was performed as follows: denaturation at 95 °C for 5 min (10 min for Amplitaq Gold), followed by 35 cycles of 95 °C for 30 s, 55 °C for 30 s, 72 °C for 60 s. Melting curve analysis was used to confirm the specificity of the PCR products. For TaqMan, the amplification reaction included a 0.9 μm concentration of each primer, 200 nm probe, 200 μm dNTPs, 3 mm MgCl2, and 1.25 units of Taq DNA Polymerase (Invitrogen). Primer and probe sequences are shown in supplemental Tables 2 and 3. Relative quantification of gene expression was determined using the 2−ΔΔCT method following efficiency testing of primers (32, 33). Samples were run in triplicate and normalized to glyceraldehyde-3-phosphate dehydrogenase or RPL19 mRNA. Each graph is representative of at least three independent experiments.
Immunoblots
Polyvinylidene difluoride membrane (Immobilon-P, Millipore) was blocked in Tris-buffered saline with Tween-20, 5% nonfat dry milk, washed, and incubated with antibodies to TRPS1 (N-18), Runx2 (S-19), Smad3 (FL-245) (Santa Cruz Biotechnology, Inc., Santa Cruz, CA); phospho-Smad3 (95414s, Cell Signaling); or FLAG (M2, Sigma). Horseradish peroxidase-conjugated anti-goat and anti-rabbit secondary antibodies were purchased from Jackson Laboratories. Bands were visualized by ECL (Amersham Biosciences).
Chromatin Immunoprecipitation
Chromatin immunoprecipitations were performed according to the protocol of the chromatin immunoprecipitation assay kit (Upstate Biotechnology, Inc., Lake Placid, NY). Briefly, cells were fixed with 1% formaldehyde for 10 min at room temperature, followed by quenching with glycine (125 mm) for 5 min. The fixed cells were washed with phosphate-buffered saline containing protease inhibitors and lysed in 1% SDS, 10 mm EDTA, 50 mm Tris-HCl, pH 8.1, with protease inhibitors for 10 min on ice prior to sonication, centrifugation, and addition of dilution buffer. 1% of input was removed, and lysates were immunoprecipitated with 2 μg of rabbit anti-FLAG antibody (F7425, Sigma) or species-matched IgG control antibody or 40 μl of anti-FLAG M2 affinity gel suspension (Sigma) for 2 h. Salmon sperm DNA/protein A/G-Sepharose beads were added to the polyclonal FLAG and IgG control immunoprecipitations and incubated overnight. Immune complexes were washed with low salt buffer, high salt buffer, LiCl buffer, and twice with TE sequentially prior to elution in 200 μl of elution buffer (1% SDS, 0.1 m NaHCO3). The eluates were heated at 65 °C for 4 h to reverse cross-linking and treated with RNase A for 30 min at 37 °C, followed by treatment with proteinase K for 1 h at 45 °C to remove RNA and protein. DNA was recovered using a Qiagen PCR purification kit and eluted in 50 μl of Qiagen EB buffer. 1% of input and 10% of the immunoprecipitates were used in PCR analyses using AmpliTaq Gold DNA polymerase (Applied Biosystems) for 35 cycles at 95 °C for 30 s, 51 °C for 30 s, 72 °C for 60 s (after initial denaturation for 10 min at 95 °C). Primers used for chromatin immunoprecipitations are shown in supplemental Table 4.
Alizarin Red S Staining
Cells were washed three times with phosphate-buffered saline and once with distilled H2O before fixation in 100% ethanol for 15 min at room temperature. Cells were rinsed with distilled H2O and stained in 1% alizarin red S (GFS chemicals) for 30–60 min at room temperature. Mineralization was visualized following several rinses with distilled H2O. Quantification of mineralization was performed by dissolving stained mineralized cultures in 10% cetylpyridinium chloride, followed by reading absorbance at 540 nm (34).
Osteocalcin Enzyme-linked Immunosorbent Assay
Medium from differentiating bone marrow stromal cells was collected at day 17. Replicates of 25-μl samples were analyzed for the presence of secreted osteocalcin as per the manufacturer's instructions (BTI Inc., Stoughton, MA).
Mass Spectrometry
In-gel digestion of candidate proteins was performed as described (35). HPLC grade acetonitrile and HPLC water were from Burdick & Jackson; formic acid and trifluoroacetic acid were from Pierce; and sequencing grade modified porcine trypsin was from Promega and used at a final concentration of 12.5 ng/ml.
LC MS/MS
Mixtures of proteolytically generated peptides were analyzed by nano-LC MS/MS utilizing a 2DLC nano-HPLC system (Eksigent) interfaced with the Q-Star XL mass spectrometer (Applied Biosystems/Sciex) equipped with a nanospray II source (Applied Biosystems/Sciex). External calibration was performed in MS/MS mode using fragment ions of Glu-fibrinopeptide as references. An LC Packings Pepmap C18 trap column (300-μm inner diameter, 5-mm length, 300-Å pore size, 5-μm particle size) and a column (75-μm inner diameter, 15-cm length) self-packed with Jupiter Proteo C12 end-capped material (90-Å pore size, 4-μm particle size) were used for desalting and reversed phase peptide separation, respectively. A 20-min linear gradient from 2% B to 50% B was run at a 250 nl/min flow rate, utilizing solvent A (2% acetonitrile, 0.1% formic acid) and solvent B (80% acetonitrile, 0.08% formic acid). Precursor ion selection employed an automated routine (IDA, Analyst QS 1.1, Applied Biosystems/Sciex) that consisted of a series of one survey MS scan (1 s, m/z 400–1700) and two MS/MS scans (2 s, m/z 60–1500); nitrogen served as a collision gas, and collision energy was automatically adjusted, depending on the size and charge state of the precursor ion. Protein identification was accomplished by using the MASCOT 2.0 (Matrix Science) search engine. Mammalia taxonomy was searched within the mass spectrometry data base 20060831 (3,239,079 sequences; 1,079,594,700 residues) utilizing the following settings: precursor ion mass tolerance, 150 ppm; fragment mass tolerance, 0.15 Da; enzyme, trypsin (including cleavages before Pro); number of missed cleavages, three; fixed modifications, S-carboxyamidomethyl; variable modifications, carboxyamidomethyl (N-term 57), deamidation (Asn and Gln), Met-sulfoxide; pyro-Glu (from N-terminal Gln); and phosphorylation of Ser and Thr.
Statistical Analyses
Data are expressed as mean values of at least three independent experiments with 2–3 replicates. Error bars show S.E. Statistical significance was computed using an unpaired two-tailed Student's t test. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
RESULTS
Identification of Proteins Bound Differentially to the Osteocalcin Promoter
Runx2 is thought to act as a scaffold for larger protein complexes. This allows Runx2 to integrate contributions from various signaling pathways and allows for either the activation or repression of Runx2-regulated promoters (7, 36). Whether a particular promoter is activated or repressed at a given point in time is dependent on the intracellular protein context and the combination of binding sites present on the promoter. In an effort to better understand what proteins are involved in the regulation of Runx2-responsive promoters, a DNA pull-down assay was employed to isolate proteins that specifically and differentially bind to a DNA sequence derived from the germ line IgCα promoter (37) and the OSE2 (osteoblast-specific element 2) sequence of the mouse osteocalcin promoter (8) (Fig. 1A). Both sequences contain Runx-binding elements but are activated or repressed in response to different stimuli and in different cell types (38, 39).
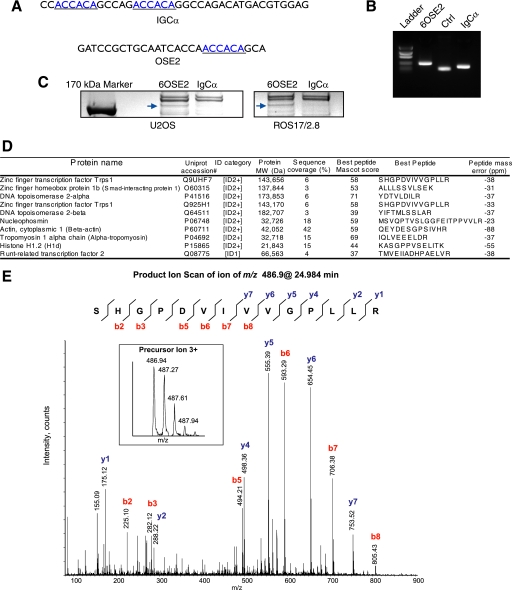
FIGURE 1.
Identification of proteins bound to the 6OSE2 DNA sequence. A, IgCα and OSE2 DNA sequences used in the DNA pull-down assays. Runx2 binding sites are underlined. B, ethidium bromide-stained agarose gel showing biotinylated PCR products. Ctrl, a plasmid containing a DNA sequence without Runx2 binding sites. Bands were quantified using ImageJ. Equal molar amounts of biotinylated oligonucleotides were used in each DNA pull-down assay. C, Coomassie Blue-stained SDS-PAGE of DNA pull-down assays of nuclear extracts from U2OS and ROS17/2.8 cells. The arrows indicate TRPS1 bound to the 6OSE2 sequence but not the IgCα sequence. D, selected proteins bound to the 6OSE2 sequence and identified by mass spectrometry. Protein identification was based on one peptide for the ID1 category and two or more peptides for the ID2+ category. MS/MS spectra of polypeptides matched to a single peptide are shown in supplemental Fig. S2. Unique non-overlapping peptides were used to calculate protein sequence coverage defined as the ratio between the sum of amino acids encompassed by the confidently matched peptides (percentage confidence interval >95%) and the number of amino acids in a polypeptide sequence. Ion score is −10*log(P), where P is the probability that the observed match is a random event. Ion scores above N, the Mascot significance level calculated for p < 0.05, were considered to be statistically non-random. E, product ion scan of the triply charged molecular ion at m/z 486.9 that was authenticated as 535SHGPDVIVVGPLL548 from TRPS1. Inset, spectrum of molecular ion of the precursor. Data were acquired during LC MS/MS analysis at an elution time of 24.984 min. Fragment ions of y- and b-series are annotated in blue and red, respectively. As illustrated by a peptide sequence drawing at the top, the majority of peptide bonds generated discernable fragmentation ions, providing evidence as to the peptide identity. No differentiation between leucine and isoleucine amino acid residues was possible under low energy collision-induced dissociation conditions; their annotation as I and L in the peptide sequence was based on the protein data base and was not established in the course of this analysis.
The IgCα promoter sequence and six tandem copies of the OSE2 sequence, called 6OSE2 (8), were individually cloned into the pGL3-Basic luciferase reporter plasmid. 3′ and biotinylated 5′ primers spanning the cloning site were used to amplify both sequences as well as a control DNA sequence lacking any known DNA binding sites (Fig. 1B). Equal molar amounts of each biotinylated promoter sequence, as visualized by ethidium bromide staining following agarose gel electrophoresis, were used in DNA pull-down assays. The biotinylated DNA sequences were bound to streptavidin-coupled magnetic beads and incubated with nuclear extract from U2OS or ROS 17/2.8 osteosarcoma cells. These cell lines were chosen because they have osteoblast properties yet differ in their osteoinductive potential in vivo and expression of differentiation markers, such as osteocalcin. DNA-bound proteins were separated by SDS-PAGE and visualized by Coomassie Blue staining (Fig. 1C). Proteins bound to the 6OSE2 sequence but not the IgCα sequence were excised from the gel and identified by mass spectrometry. Fig. 1D shows the most prominent proteins identified using MASCOT searches of LC/MS analyses of tryptic peptides from excised bands. MASCOT settings and peptide coverage maps can be found in supplemental Figs. S1 and S2. As a positive control, the band corresponding to Runx2 was excised and identified (supplemental Fig. S3). We focused on the first protein listed in the table in Fig. 1D, TRPS1, which was identified in the DNA pull-down assays from both cell lines. Fig. 1E is an example of a TRPS1 peptide MS/MS spectrum from ROS17/2.8 cells. Indicated are the peptide sequence ions resulting from collision-induced dissociation of the peptide ion in the mass spectrometer.
Binding and Repression of the Osteocalcin Promoter by TRPS1
To examine the binding of TRPS1 to the 6OSE2 sequence, nuclear lysates were prepared from U2OS cells, and DNA pull-down assays were performed to visualize endogenous protein bound to the promoter sequences by immunoblotting (Fig. 2A). Runx2 bound to both the 6OSE2 and IgCα promoters, and TRPS1 only bound to the 6OSE2 promoter. Since TRPS1 has been shown to co-immunoprecipitate with Runx2 (25) (data not shown), we next tested whether binding of TRPS1 to the 6OSE2 sequence was dependent on cell type. Unlike U2OS or ROS17/2.8 cells, HepG2 human epithelial cells did not express endogenous Runx2 or TRPS1, as determined by quantitative RT-PCR (data not shown). When HepG2 cells were transfected to express TRPS1, we found that TRPS1 bound to the 6OSE2 but not the IgCα sequence (Fig. 2B). As a control, Smad3, an effector of TGF-β signaling, bound to the IgCα sequence following TGF-β treatment. Lack of Smad3 binding to the 6OSE2 sequence was probably due to the fact that Smad3 binds the osteocalcin promoter through Runx2 (39), whereas the IgCα sequence contains Smad binding sites. To test this hypothesis, we examined the binding of Smad3 to the 6OSE2 sequence in ROS17/2.8 cells, which express Runx2 (8). Following treatment of ROS17/2.8 cells with TGF-β, Runx2 and Smad3 bound both sequences. However, TRPS1 bound only to the 6OSE2 sequence (Fig. 2C), whereas the addition of TGF-β did not affect the binding of TRPS1 to this sequence in ROS17/2.8 or HepG2 cells (data not shown). Furthermore, ectopic expression of TRPS1 did not result in nonspecific binding to the IgCα sequence (Fig. 2B). Together, these data demonstrate that TRPS1 directly and specifically interacts with the 6OSE2 sequence and suggest that TRPS1 binding to the DNA does not require interaction with Runx2 or Smad3.
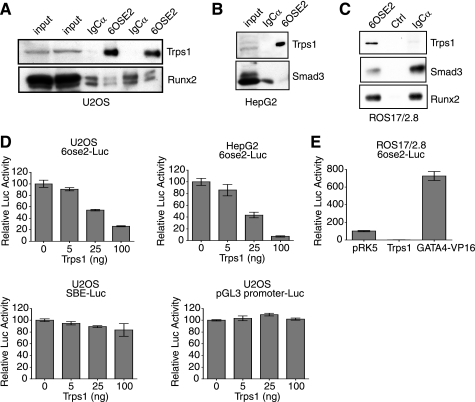
FIGURE 2.
Binding of TRPS1 to the 6OSE2 sequence results in transcriptional repression. A, nuclear lysates of U2OS cells were incubated with 6OSE2 or IgCα DNA, and bound proteins were immunoblotted for endogenous TRPS1 and Runx2. TRPS1 bound only to the 6OSE2 sequence (shown in duplicate). Runx2 bound to both the 6OSE2 and IgCα sequences. The left two lanes show input nuclear lysate. B, similar DNA pull-down assays as in A, except using nuclear lysates from TGF-β-treated HepG2 cells, which do not express Runx2. Immunoblots are shown for transfected TRPS1 and Smad3. TRPS1 bound the 6OSE2 sequence, but Smad3 did not. C, DNA pull-down assays as in A, using TGF-β-treated ROS17/2.8 cells, followed by immunoblot for endogenous TRPS1, Smad3, and Runx2. TRPS1 bound the 6OSE2 sequence, whereas Runx2 and Smad3 bound to both the 6OSE2 and IgCα sequences but not to a control sequence. D, effects of increasing levels of Trps1 on luciferase expression from the Runx2-controlled 6OSE2 promoter. Values are shown relative to the transcription activity in the absence of transfected Trps1. The luciferase assays were performed in U2OS and HepG2 cells. TRPS1 did not affect the transcription from the pGL3- and SBE-Luc reporters. E, TRPS1 represses transcription from the 6OSE2-Luc reporter, whereas GATA4-VP16, a fusion protein that binds GATA sequences, activates transcription. Activity is shown relative to control pRK5 vector.
To determine if the binding of TRPS1 to the 6OSE2 sequence regulates transcription, we co-expressed Runx2 with increasing amounts of TRPS1 into U2OS or HepG2 cells and scored the luciferase activity driven by the 6OSE2 sequence (Fig. 2D). TRPS1 repressed the 6OSE2 promoter in a dose-dependent manner in both cell types, indicating that osteoblast-specific proteins other than Runx2 were not required for repression of transcription. TRPS1 did not repress basal transcription from the pGL3 promoter or SBE-Luc, a Smad-responsive reporter, supporting the specificity of repression by exogenous TRPS1 (Fig. 2D).
Because TRPS1 is a GATA-binding transcription factor, we evaluated whether transcription from the 6OSE2 sequence could be regulated by other GATA family transcription factors. We therefore tested the ability of a GATA4-VP16 fusion protein to regulate the 6OSE2 reporter in ROS17/2.8 cells using a luciferase assay (Fig. 2E). GATA4-VP16 activated the 6OSE2 reporter, indicating that GATA DNA binding can regulate transcription from the 6OSE2 sequence.
Identification of a GATA Binding Site in the Mouse Osteocalcin Promoter
Analysis of the OSE2 DNA sequence with the transcription factor binding site identification program TFSEARCH (40) identified a putative GATA binding site, CAATCA (41), upstream of the Runx2 binding site (Fig. 3A). To test if this site mediated TRPS1 binding to the 6OSE2 sequence, we generated two oligonucleotides modeled on the IgCα sequence. One contained two copies of the Runx2 binding site but lacked the GATA site, whereas the other had the same sequence but with a GATA site added 5′ of the Runx2 binding sites (Fig. 3A). In DNA pull-down assays using ROS17/2.8 nuclear lysates, endogenous Runx2 bound efficiently to both sequences, indicating that the GATA binding sequence did not affect Runx2 binding. In contrast, efficient binding of TRPS1 depended on the presence of the GATA sequence CAATCA (Fig. 3B). These data suggest that TRPS1 does not bind through Runx2 but rather requires a GATA site present in OSE2.
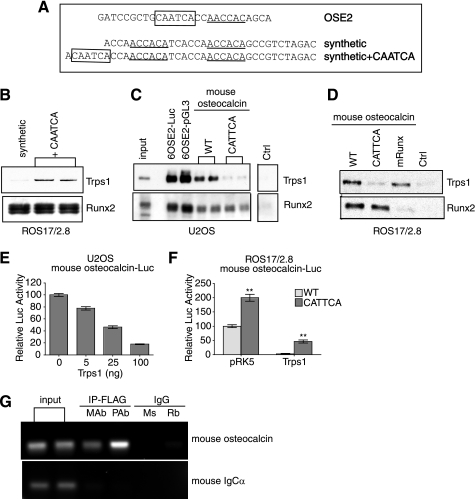
FIGURE 3.
Binding of TRPS1 to the mouse osteocalcin promoter requires a GATA binding sequence. A, the OSE2 sequence and sequences of two synthetic oligonucleotides, each containing two Runx2 sites, one with and one without a putative GATA site (CAATCA). GATA binding sites are boxed, and Runx2 binding sites are underlined. B, immunoblots on the left show the results of DNA pull-down assays from ROS17/2.8 nuclear lysates using the two synthetic oligonucleotides. The presence of a GATA binding sequence (CAATCA) allowed endogenous TRPS1 to bind to the oligonucleotide (shown in duplicate). Runx2 bound equally to both oligonucleotides. C, DNA pull-down assays from lysates of transfected U2OS cells using the 6OSE2 sequence generated by PCR from two different vectors, the −148 to +1 segment of the mouse osteocalcin promoter (wild type; WT) or the same segment with an inactivating point mutation in the GATA-binding sequence (CATTCA). Immunoblotting shows the binding of TRPS1 and Runx2. TRPS1 is bound to the 6OSE2 sequence amplified from either vector and to the wild-type osteocalcin promoter sequence. The CATTCA mutation strongly reduced binding of TRPS1 to the promoter segment as compared with wild type but had no effect on Runx2 binding. D, DNA pull-down assays from lysates of ROS17/2.8 cells. Endogenous TRPS1 bound to the wild-type osteocalcin promoter fragment but not to the CATTCA mutant fragment. Mutation of the Runx binding site (mRunx) had no effect on TRPS1 binding but abolished Runx2 binding. E, increased expression of TRPS1 confers dose-dependent repression of transcription from the mouse osteocalcin promoter in U2OS cells, measured by luciferase activity. Reporter activity is shown relative to the promoter activity in the absence of transfected Trps1. F, transcription activity of the wild-type mouse osteocalcin promoter and CATTCA mutant in ROS17/2.8 cells in the absence or presence of transfected Trps1 or vector control (pRK5). The CATTCA mutant has higher activity than the wild-type promoter segment, and TRPS1 is less efficient at repressing transcription from the mutant promoter segment. Luciferase activity was normalized to the transcription from the wild-type promoter. G, binding of TRPS1 to the mouse osteocalcin promoter in bone marrow stromal cells expressing FLAG-tagged TRPS1, as assessed by chromatin immunoprecipitation using either monoclonal or polyclonal anti-FLAG antibodies and species-matched IgG controls (mouse (Ms) for monoclonal and rabbit (Rb) for polyclonal). The PCR products show that TRPS1 binds the osteocalcin promoter but not the IgCα sequence.
Because the 6OSE2 sequence is a tandem arrangement of sequences derived from the osteocalcin promoter, we tested the binding of TRPS1 to the native −148 to +1 region of the mouse osteocalcin promoter, which contains only one OSE2 sequence. TRPS1 and Runx2 both bound this osteocalcin promoter sequence in U2OS and ROS17/2.8 cells (Fig. 3, C and D). The binding efficiency was reduced compared with the 6OSE2 sequence, which was expected, given that the 6OSE2 sequence contains six tandem repeats of the same sequence. We next made a single base pair mutation in the GATA binding site of the mouse osteocalcin promoter, thus converting CAATCA to CATTCA, which has been shown to disrupt binding (41). Neither ectopic nor endogenous TRPS1 bound the mutated GATA sequence in U2OS or ROS17/2.8 cells. In contrast, Runx2 binding was not affected. Conversely, mutation of the Runx2 binding site in the mouse osteocalcin promoter abolished Runx2 binding but had no effect on the binding of TRPS1 to the promoter sequence, further confirming that TRPS1 does not bind this sequence through Runx2.
Consistent with our findings using the 6OSE2 sequence, TRPS1 repressed transcription from the −148 to +1 region of the mouse osteocalcin promoter in a dose-dependent manner in U2OS cells (Fig. 3E). Point mutation of the GATA binding site resulted in enhanced transcription in the presence of endogenous TRPS1 and a significantly decreased repression of the mouse osteocalcin promoter reporter by overexpressed TRPS1 (Fig. 3F). The ability of TRPS1 to repress the promoter is probably due to the ability of Runx2 and TRPS1 to interact with each other (25) (data not shown) and their overexpression, allowing a low level recruitment of TRPS1 through Runx2.
In vivo binding of TRPS1 to the osteocalcin promoter was tested by chromatin immunoprecipitation. Because antibodies capable of immunoprecipitating endogenous TRPS1 were not available, we infected bone marrow stromal cells with a retroviral vector expressing FLAG-tagged TRPS1. We then performed chromatin immunoprecipitations of FLAG-tagged TRPS1 using monoclonal or polyclonal anti-FLAG antibodies or species-matched nonspecific IgG and amplified the mouse osteocalcin promoter sequence, encompassing the region containing the GATA sequence. Binding of FLAG-tagged TRPS1 to the osteocalcin promoter segment was detected using monoclonal or polyclonal anti-FLAG antibodies, and no reactivity was observed using mouse or rabbit control IgG (Fig. 3G). In control experiments, FLAG-tagged TRPS1 did not bind the IgCα promoter sequence.
Identification of a GATA Binding Site in the Human Osteocalcin Promoter
The identification of a GATA binding site in the mouse osteocalcin promoter at positions −142 to −137 led us to examine the cross-species conservation of this sequence. Fig. 4A shows the sequence alignment of the orthologous rat, mouse, and human osteocalcin promoter sequences. The GATA binding site in the mouse promoter at positions −142 to −137 is not conserved in either the rat or human sequence. However, using the Weeder Motif Locator (MoD tools) (42), we identified a more distal putative GATA binding site that is conserved between the rat (positions −172 to −166) and human (positions −198 to −192) promoter sequences. This GATA binding sequence is in the opposite orientation relative to the transcription start site when compared with the one in the mouse osteocalcin OSE2 element at −142 to −137, although identical in sequence, and is predicted to bind GATA factors. The slightly divergent sequence in the mouse promoter at the corresponding position (Fig. 4A) is also predicted to bind GATA factors, albeit with lower affinity (41). The functionality of this site in the mouse osteocalcin promoter was not tested.
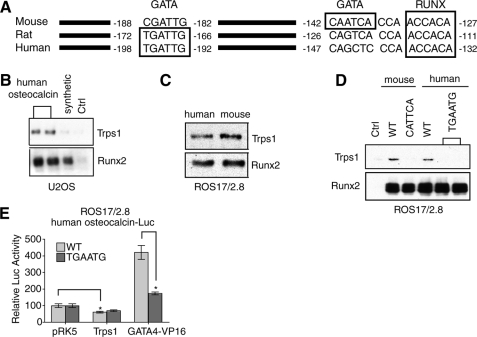
FIGURE 4.
Binding of TRPS1 to the human osteocalcin promoter. A, sequence alignment of mouse, rat, and human osteocalcin promoter sequences. Predicted Runx and GATA binding sites are boxed. The proximal GATA binding site in the mouse promoter is not conserved in the rat and human sequences. A more distal GATA binding site is conserved between the rat and human sequences but not in the mouse sequence. B, DNA pull-down assays using nuclear lysates from transfected U2OS cells. Immunoblotting shows that TRPS1 and Runx2 both bind the human osteocalcin promoter segment. The synthetic oligonucleotide without the GATA binding sequence (Fig. 3A) and a control oligonucleotide were used as negative controls for TRPS1 binding. C, DNA pull-down assays as in B, but analyzing endogenous proteins in ROS17/2.8 cells, show similar binding of TRPS1 and Runx2 to the human and mouse osteocalcin promoter segments. D, DNA pull-down assays from ROS17/2.8 cell lysates, using either wild-type (WT) or mutant mouse or human osteocalcin promoter sequences. Mutation of the GATA binding sites (CATTCA mutation in the mouse sequence and TGAATG mutation in the human sequence) abolishes TRPS1 binding but not Runx2 binding. E, transcription assays in transfected U2OS cells show that the TGAATG mutation in the human osteocalcin promoter slightly reduces the transcription repression by TRPS1 and strongly reduces the activation of the promoter by the GATA4-VP16 fusion. Luciferase activities are normalized to vector control.
To determine if TRPS1 binds the human osteocalcin promoter, we amplified the −212 to +28 region promoter by PCR from genomic DNA of U2OS osteosarcoma cells. We then evaluated the binding of TRPS1 to this sequence in DNA pull-down assays using nuclear extracts from transfected U2OS cells (Fig. 4B) or untransfected ROS17/2.8 cells (Fig. 4C). Ectopically expressed and endogenous Runx2 and TRPS1 bound with similar efficiencies to the human and mouse osteocalcin promoter sequences (Fig. 4C). Mutation of the human GATA binding site (−198 to −192) from TGATTG to TGAATG abolished binding of TRPS1, although the binding of Runx2 was unaffected (Fig. 4D). These data suggest that the distal GATA-binding element in the human osteocalcin promoter is required for TRPS1 binding. In reporter assays using the same promoter fragment, the mutation barely attenuated the ability of TRPS1 to repress Runx2-dependent activation of the human osteocalcin promoter (60% versus 69%) but significantly decreased the activation of the promoter by the GATA4-VP16 fusion protein (Fig. 4E) (420% versus 170%). The weak repression of Runx2-dependent transcription by TRPS1 using the human promoter stands in contrast to the much stronger repression of the mouse promoter (Fig. 3F) and may be due to sequence differences between the two promoters, resulting in the differential binding of other regulatory factors. In addition, it is possible that the interaction of TRPS1 with Runx2 that is required for transcriptional repression at the human sequence depends more on the three-dimensional architecture of the promoter in a chromatin context. Nevertheless, these results, taken together, illustrate that TRPS1 can bind and regulate both the mouse and human osteocalcin promoters through its ability to bind an intact GATA binding site.
Regulation of TRPS1 Expression by Androgen and Vitamin D3
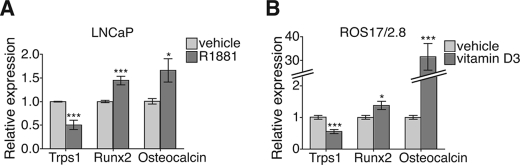
The ability of TRPS1 to repress expression from the osteocalcin promoter led us to evaluate whether TRPS1 expression correlated with osteocalcin gene expression. We used two cell culture systems in which a treatment modulates the expression of either TRPS1 or osteocalcin and examined the regulation of both genes. First, we used a prostate cancer cell model in which it has been shown that TRPS1 mRNA expression is regulated (43). In LNCaP-FGC cells, an androgen-responsive human prostate carcinoma cell line, treatment with the androgen R1881 results in down-regulation of TRPS1 mRNA and protein levels (44). Accordingly, the androgen R1881 down-regulated TRPS1 expression in these cells (Fig. 5A). Although osteocalcin expression has been detected in androgen-independent prostate cancer cells (45), its expression in androgen-responsive cells has not been reported. We showed that treatment of androgen-sensitive LNCaP cells with R1881 results in increased osteocalcin mRNA expression concomitant with down-regulation of TRPS1 mRNA (Fig. 5A). This inverse correlation suggests that a decrease in TRPS1 expression results in an increase in osteocalcin expression.
FIGURE 5.
Effects of androgens and vitamin D3 on Trps1 expression. A and B, effects of the androgen R1881 (A) or vitamin D3 (B) on the expression of Trps1 mRNA in LNCaP prostate cancer cells (A) or ROS17/2.8 osteosarcoma cells (B), assessed by qRT-PCR.
To further evaluate this correlation, we also studied the regulation of Trps1 and osteocalcin expression in ROS17/2.8 osteosarcoma cells. Under normal growth conditions, these cells express low levels of osteocalcin, but its expression can be enhanced by treating the cells with vitamin D3 (46). Vitamin D3 increases the expression of osteocalcin through a vitamin D3-responsive element at positions −465 to −438 in the rat osteocalcin promoter (46). In response to vitamin D3, ROS17/2.8 cells decreased the expression of Trps1 mRNA, measured using quantitative RT-PCR (Fig. 5B). Conversely, osteocalcin mRNA levels were increased in response to vitamin D3. These results, generated using two different cell types, illustrate that two different physiologically relevant treatments result in an increase in osteocalcin and decrease in Trps1 expression, consistent with the control of osteocalcin expression by TRPS1. The results of these experiments prompted us to use a more targeted approach to manipulate TRPS1 levels and to address the role of endogenous TRPS1 in the regulation of osteocalcin transcription.
TRPS1 Regulates Osteocalcin Expression in ROS17/2.8 Cells
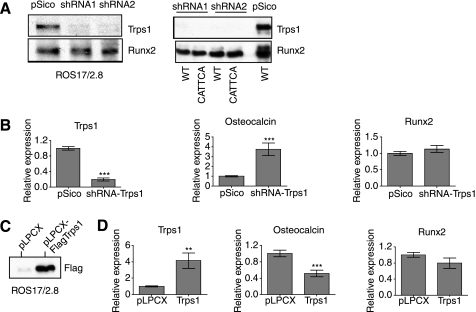
To determine the role of TRPS1 in the regulation of osteocalcin expression, we designed shRNAs targeting the Trps1 transcript and cloned each into a lentiviral vector. ROS17/2.8 cells were infected with lentivirus expressing shRNA to Trps1, and the efficiency of down-regulation of TRPS1 expression was evaluated by immunoblotting. The results for the two most efficient short hairpins are shown in Fig. 6A. In cells expressing either shRNA, no TRPS1 protein was detected in DNA pull-down assays using either the wild-type mouse osteocalcin promoter or the corresponding sequence in which the CAATCA sequence was mutated to CATTCA (Fig. 3, B and C). By quantitative RT-PCR, the level of Trps1 mRNA in cells expressing the Trps1 shRNA was decreased by ∼80% (Fig. 6B). Strikingly, in these cells, the osteocalcin mRNA levels increased reciprocally by 4–5-fold. Because a TRPS1 binding site has been identified in the Runx2 distal promoter at position −1176 (47), we also examined the effect of decreased TRPS1 expression on Runx2 levels. There was a moderate but statistically significant increase in Runx2 mRNA levels when Trps1 expression was reduced.
FIGURE 6.
Effect of TRPS1 levels on endogenous osteocalcin and Runx2 expression in ROS17/2.8 cells. A (left), immunoblot of nuclear lysates from ROS17/2.8 cells infected with a control pSico vector or lentivirus expressing shRNAs targeting Trps1. Right, DNA pull-down assays of the same nuclear lysates showing a lack of TRPS1 binding to wild-type or CATTCA mutant mouse osteocalcin promoter sequences in cells expressing the shRNA. Nuclear lysates of pSico control-infected cells show TRPS1 binding to the wild-type promoter sequence. B, mRNA expression of Trps1, osteocalcin, and Runx2 mRNA in ROS17/2.8 cells expressing shRNA to Trps1 or control-infected cells. Expression levels, assessed by qRT-PCR, are shown relative to vector control-infected cells. C, immunoblot of lysates from cells expressing FLAG-tagged TRPS1 or vector control. D, mRNA expression of Trps1, osteocalcin, and Runx2 mRNA in ROS17/2.8 cells with increased Trps1 expression or control-infected cells. Expression levels, assessed by qRT-PCR, are shown relative to vector control-infected cells.
We next evaluated the effect of increased TRPS1 expression on osteocalcin and Runx2 mRNA levels. ROS17/2.8 cells were infected with retrovirus expressing FLAG-tagged TRPS1 or control virus. Overexpression of TRPS1 was demonstrated by immunoblot (Fig. 6C), and quantitative RT-PCR analysis showed a 2–3-fold increase in Trps1 mRNA levels (Fig. 6D) compared with vector control cells. The increased TRPS1 expression resulted in an ∼50% reduction in the amount of osteocalcin mRNA (Fig. 6D). These data demonstrate that TRPS1 represses transcription of the osteocalcin gene and suggest that TRPS1 expression may affect the transcriptional autoregulation of the Runx2 gene (27).
Trps1 Expression and OSE2 DNA Binding in Differentiating Bone Marrow Stromal Cells
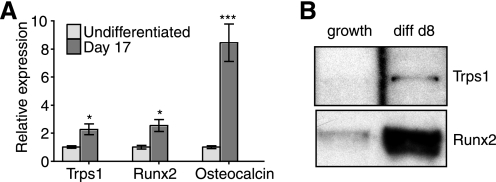
Because ROS17/2.8 cells were derived from an osteosarcoma (48, 49), we wanted to evaluate the role of TRPS1 in the regulation of osteocalcin expression in primary osteoblasts undergoing differentiation. To this end, we used mouse bone marrow stromal cells, which are capable of differentiating into several different lineages, including osteoblasts, when cultured under the appropriate conditions (50, 51). These bone marrow stromal cells were allowed to differentiate at confluence in the presence of β- glycerophosphate and l-ascorbate-2-phosphate. The levels of Trps1 mRNA were examined in undifferentiated cells and at 17 days following the addition of differentiation medium (Fig. 7A). The expressions of Trps1, osteocalcin, and Runx2 mRNA were all increased following the addition of differentiation medium for 17 days. This result is consistent with the possibility that Trps1 plays a role in intramembranous ossification and has the potential to regulate osteoblast differentiation markers, such as osteocalcin.
FIGURE 7.
Expression and DNA binding activity of TRPS1 in differentiated bone marrow stromal cells. A, relative Trps1, osteocalcin, and Runx2 mRNA levels, determined by qRT-PCR, in undifferentiated bone marrow stromal cells and bone marrow stromal cells cultured in differentiation medium for 17 days. Expression levels are expressed relative to mRNA levels in undifferentiated cells. B, DNA pull-down assays reveal binding of endogenous TRPS1 and Runx2 in lysates of differentiated cells to the 6OSE2 sequence. No binding was observed using undifferentiated cells cultured in growth medium.
The level of TRPS1 protein in bone marrow stromal cells that is capable of binding to the 6OSE2 DNA sequence was assayed by DNA pull-down assays. Nuclear lysates were prepared from confluent bone marrow stromal cells maintained in either growth or differentiation medium for 8 days. DNA pull-down assays were performed using the 6OSE2 sequence, and the bound proteins were analyzed by immunoblotting (Fig. 7B). TRPS1 bound to the 6OSE2 sequence only in differentiated cells and not in cells in culture conditions that do not result in detectable Trps1 mRNA expression. Runx2 binding to the 6OSE2 sequence also increased in differentiated cells, consistent with its increased expression when the cells were treated with differentiation medium.
TRPS1 Regulates Osteocalcin Expression in Differentiating Bone Marrow Stromal Cells
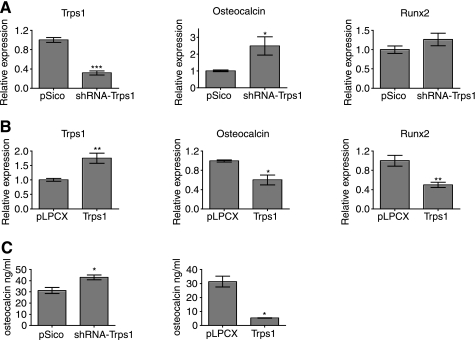
To test the effect of modulation of Trps1 levels on osteocalcin expression in differentiating cells, bone marrow stromal cells were infected with lentivirus expressing short hairpin RNAs targeting Trps1 or retrovirus expressing Trps1. Following selection, the cells were induced to undergo osteoblast differentiation. Reduction of Trps1 mRNA levels resulted in increased osteocalcin gene expression (Fig. 8A). Runx2 mRNA levels were not affected by Trps1 down-regulation but were decreased by increased Trps1 expression (Fig. 8B). Consistent with the repression of osteocalcin expression by TRPS1, increased expression of Trps1 strongly repressed the level of osteocalcin mRNA. These data are in agreement with our experiments in ROS17/2.8 cells. We next quantified the osteocalcin protein levels in the medium by enzyme-linked immunosorbent assay (Fig. 8C). Increased Trps1 expression resulted in greatly decreased levels of osteocalcin in the medium, whereas reduction of Trps1 expression by lentiviral shRNA expression increased the level of secreted osteocalcin.
FIGURE 8.
Effect of decreased or increased TRPS1 levels on osteocalcin and Runx2 expression in differentiated bone marrow stromal cells. A and B, Trps1, osteocalcin, and Runx2 mRNA levels in differentiated cells expressing Trps1 shRNA (A) or increased Trps1 levels (B). The mRNA levels, determined by qRT-PCR, are shown relative to vector control-infected cells. C, secreted osteocalcin levels determined by enzyme-linked immunosorbent assay from bone marrow stromal cells with increased or down-regulated Trps1 expression or infected with the corresponding vector controls.
TRPS1 Expression Defines the Level of Osteoblast Differentiation and Mineralization
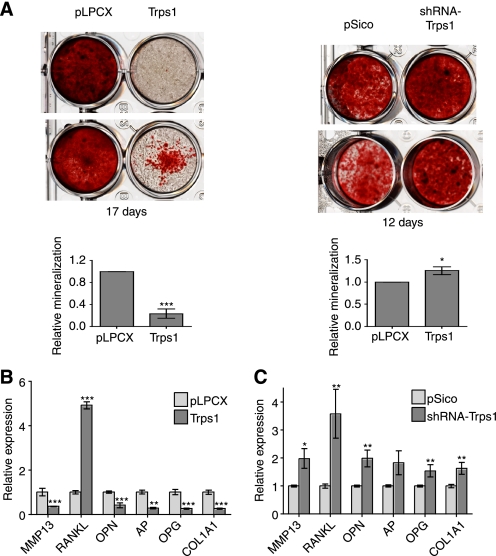
Osteocalcin expression in osteoblasts occurs at the onset of mineralization and increases with mineral deposition (52, 53). To evaluate the role of TRPS1 in osteoblast mineralization, we infected bone marrow stromal cells with retrovirus expressing Trps1 or lentivirus expressing shRNA targeting Trps1 and allowed the cells to differentiate. By quantifying the calcium deposition using alizarin red S staining, we found that, after 12–17 days, cells with increased Trps1 expression had decreased mineralization, whereas reduction of Trps1 expression slightly increased mineralization (Fig. 9A). These results suggest that TRPS1 regulates not only the expression of osteocalcin during osteoblast differentiation but also the deposition of mineralized matrix by differentiating osteoblasts.
FIGURE 9.
Effect of TRPS1 on osteoblast differentiation. A, matrix mineralization by cells with increased Trps1 expression or expression of shRNA to Trps1, visualized by alizarin red S staining after differentiation for 17 and 12 days, respectively (shown in duplicate for each). The quantification of alizarin red S stain is shown compared with vector control. B and C, relative mRNA quantification of osteoblast marker gene expression by qRT-PCR in differentiated cells expressing Trps1 (B) or Trps1 shRNA (C) or corresponding vector controls. mRNA levels of MMP13, RankL, osteopontin (OPN), alkaline phosphatase (AP), osteoprotegerin (OPG), and collagen 1A1 (Col1A1) are shown relative to vector control-infected cells.
To further characterize the regulation of osteoblast differentiation by TRPS1, we assessed the expression of other osteoblast genes, in addition to the osteocalcin gene, in differentiated cells. This set of genes includes those encoding collagen 1A1, the major protein constituent of bone (54); alkaline phosphatase, a non-collagenous matrix protein involved in matrix stabilization (55); osteopontin, which is associated with matrix formation and maturation (52); MMP13, an enzyme important for bone metabolism and homeostasis (56); RankL, a stimulator of osteoclastogenesis (57); and osteoprotegerin, an inhibitor of RankL (58). Increased Trps1 expression resulted in decreased expression of the genes encoding MMP13, osteopontin, alkaline phosphatase, osteoprotegerin, and collagen 1A1 (Fig. 9B). In contrast, RankL expression was strongly enhanced in cells with increased Trps1 expression. Using lentiviral expression of Trps1-specific shRNA, we found that decreased expression of Trps1 resulted in a reciprocal expression pattern for the MMP13, osteopontin, alkaline phosphatase, osteoprotegerin and collagen 1A1 genes, demonstrating that TRPS1 represses the expression of these genes, either directly or indirectly (Fig. 9C).
MMP13, osteopontin, and osteocalcin are late markers of osteoblast differentiation, and their expression occurs concomitantly with mineralization. In contrast, alkaline phosphatase is an early marker of osteoblast differentiation and is the only gene tested that does not contain a Runx2 binding site in its promoter (5). Using the Weeder program (MoD tools) (42) to locate conserved motifs in co-regulated genes of the same species, we identified potential GATA-binding sequences within 400 bp of the transcription start site in the promoters of all genes tested. Whether or not these binding sites are functional and are able to bind TRPS1 remains to be investigated. However, our data suggest that TRPS1 may be able to regulate the onset of mineralization in both chondrocytes and osteoblasts.
DISCUSSION
In this study, we sought to identify modulators of Runx2-dependent gene expression. We took a proteomics approach using DNA segments from two differentially regulated Runx-responsive promoters to isolate and identify proteins that could regulate transcription. Proteins differentially bound to the two DNA segments were isolated from two different osteosarcoma cell lines and identified by mass spectrometry. One of the proteins that we identified was TRPS1, which is known to be a GATA-binding transcription repressor and is encoded by a gene that is responsible for the human skeletal disease known as tricho-rhino-phalangeal syndrome.
We initially confirmed the interaction of TRPS1 with both a short DNA segment (OSE2) and the proximal region of the mouse osteocalcin promoter that encompasses the OSE2 sequence. TRPS1 repressed the luciferase expression from a reporter containing the OSE2 sequence in a dose-dependent manner. Importantly, TRPS1 bound to the osteocalcin promoter through a previously unidentified GATA binding site rather than through protein interactions with Runx2. TRPS1 bound the OSE2 sequence of the osteocalcin promoter in either the presence or absence of Runx2. These results explain why the GATA domain of TRPS1 is required for the repression of Runx2-dependent transcription (25). Further, mutation of the Runx2 binding sequence does not affect the binding of TRPS1 to the osteocalcin promoter, and TRPS1 binding does not affect the ability of Runx2 to bind the DNA. However, TRPS1 binding and transcriptional repression was affected by mutating the GATA binding site of the mouse osteocalcin promoter, and activation of the osteocalcin promoter by GATA4-VP16 was also abolished by mutation of the GATA binding site. Although TRPS1 was identified by mass spectrometry and chromatin immunoprecipitation as a transcription factor binding to the short mouse osteocalcin promoter sequence that contains the GATA binding site, we cannot exclude the possibility that other GATA factors might also be involved in the regulation of osteocalcin gene transcription.
In the mouse promoter, the GATA site is located at −142 to −137 relative to the transcription start site, whereas it is found in the opposite orientation at −172 to −166 in the rat and −198 to −192 in the human osteocalcin promoter sequences. We have shown that TRPS1 can bind the GATA motif in both the mouse and human osteocalcin promoter sequences, and it has been documented that TRPS1 can bind the GATA sequence in either orientation (17). Although many regulatory sites are spatially conserved between orthologs, this is not always the case, and the positions of highly conserved motifs can deviate by over 100 bp (59). In fact, the PTHrP gene also has differences between the position and number of GATA binding sites in promoters of the mouse and human orthologs. The mouse PTHrP promoter has GATA sites at −442 and −422, both of which are inverted, and the human PTHrP promoter has GATA sites at −422, −173, and −119, with only the last one inverted (60). This result highlights a limitation of many regulatory sequence prediction algorithms that use comparative sequence analysis with the assumption that transcription factor binding sites between orthologs are spatially conserved.
The mechanism by which TRPS1 is able to repress transcription remains unclear. The repression domain of TRPS1 maps to the Ikaros-like zinc fingers in the C terminus of the protein (17). This region of TRPS1 has been shown to interact with two proteins, LC8a (dynein light chain 8) (61) and RNF4 (RING finger protein 4) (62), both of which suppress TRPS1-mediated transcriptional repression of GATA-responsive promoters by an unknown mechanism. TRPS1 is also sumoylated at two lysines within the C-terminal repression domain (63). Sumoylation enhances repression by TRPS1, in contrast to Ikaros sumoylation, which disrupts both histone deacetylase-dependent and -independent repression (64). Further study is required to determine which proteins and post-translational modifications regulate TRPS1 and the nature of the transcription complexes in which it participates.
Previous studies have revealed a role for TRPS1 in the differentiation of chondrocytes (19, 20, 25, 60), but no studies have proposed a role in osteoblasts. Such a role is, however, consistent with the observation that Trps1 is expressed in the mandible (17), a bone that is made by intramembranous ossification. We found that TRPS1 is functionally expressed at early and late stages of osteoblast differentiation of bone marrow stromal cells in culture. Furthermore, using cells with silenced or increased Trps1 expression, we demonstrated that TRPS1 negatively regulates late stage osteoblast differentiation, as apparent from its effects on osteocalcin expression and mineralization, and the expression of other osteoblast markers, including osteoprotegerin, osteopontin, alkaline phosphatase, matrix metalloproteinase 13, collagen 1A1, and RankL. Putative GATA binding sites exist in the proximal 400 bp of the Runx2-responsive promoters of the mouse genes encoding matrix metalloprotease 13, osteoprotegerin, osteopontin, and collagen 1A1,3 and GATA binding sites were also detected in the mouse RankL and alkaline phosphatase promoter sequences. The results for RankL are confusing; however, there are conflicting reports regarding the role of Runx2 in the expression of RankL as well as the potential for RankL expression by extraskeletal fibroblasts (65). The possibility of a role for TRPS1 in osteoclast regulation requires further study. Given that TRPS1 levels affect the expression of all genes surveyed, it is tempting to propose that TRPS1 may coordinate the expression of a set of osteoblast genes through interaction with GATA binding sites in their promoters.
Mice that lack functional TRPS1 expression show accelerated mineralization of the perichondrium, and it has been suggested that TRPS1 acts to synchronize chondrocyte mineralization with chondrocyte differentiation in the growth plate (25). Osteocalcin is expressed in mature osteoblasts and hypertrophic chondrocytes undergoing mineralization, which also express osteopontin and bone sialoprotein (66). It is interesting to speculate that TRPS1 may repress Runx2-regulated genes that are involved in mineralization of both osteoblasts and chondrocytes until the onset of terminal differentiation, at which time the transcriptional repression by TRPS1 might be relieved, thus allowing the expression of terminal differentiation markers to occur in a coordinated fashion. Perhaps TRPS1 can function in two stages, to suppress Runx2 expression in non-osteoblastic cells and to repress Runx2-dependent expression of particular genes at some stages of osteoblast or chondrocyte differentiation.
TRPS1 expression is reduced in LNCaP cells treated with the synthetic androgen R1881 (43) (present work). This repression is mediated by the binding of the androgen receptor to a hormonal response element in the TRPS1 promoter (67). Our data indicate that the treatment of LNCaP cells with R1881 results in increased osteocalcin gene expression. Interestingly, high levels of Runx2 and bone matrix proteins, such as osteocalcin, bone sialoprotein, and osteopontin, have been detected in the majority of primary and metastatic prostate cancers but not in normal prostate tissue (68). Osteocalcin expression is high in androgen-insensitive cell lines and low in androgen-sensitive cells like LNCaP cells (69). Further, TRPS1 expression shows an inverse correlation, with high expression in androgen-sensitive cell lines and low expression in insensitive cells (43). These results suggest that TRPS1 may play a role in suppressing the expression of bone-specific markers, such as osteocalcin, in prostate carcinomas and other non-mineralizing cells.
Finally, we found that vitamin D3, a hormone that increases the expression of osteocalcin mRNA in ROS17/2.8 cells (70), induced a reduction in Trps1 expression. We were unable to identify a putative vitamin D3 receptor binding site in the genomic sequence preceding the Trps1 coding sequence but did find a number of other hormone receptor binding sites, including those for glucocorticoid, androgen, estrogen, and progesterone. Moreover, increased TRPS1 expression has been detected in breast cancers (71). These correlations between TRPS1 expression, hormone regulation, and osteoblast marker expression in different cell and tissue contexts are intriguing and will hopefully be explored to determine whether the relationship is causal and could be used as a target for therapy or predictive and could be used as a useful marker of disease progression.
Supplementary Material
Acknowledgments
We thank Tamara Alliston, Lisa Choy, and David M. Thomas for critically reading the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant CA63101 (to R. D.) and a National Institutes of Health Institutional Research Service Award (to D. P.). The UCSF Sandler-Moore Mass Spectrometry Core Facility was supported by the Sandler Family Foundation, the Gordon and Betty Moore Foundation, and National Institutes of Health, NCI, Grant P30 CA82103.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1–4 and Figs. S1–S3.
D. M. Piscopo and R. Derynck, unpublished results.
- CREB
- cAMP-response element-binding protein
- TGF
- transforming growth factor
- shRNA
- short hairpin RNA
- RT
- reverse transcription
- qRT
- quantitative reverse transcription
- HPLC
- high pressure liquid chromatography
- LC
- liquid chromatography
- MS
- mass spectrometry
- PTHrP
- parathyroid hormone-related peptide.
REFERENCES
- 1.Ducy P., Zhang R., Geoffroy V., Ridall A. L., Karsenty G. (1997) Cell 89, 747–754 [DOI] [PubMed] [Google Scholar]
- 2.Komori T., Yagi H., Nomura S., Yamaguchi A., Sasaki K., Deguchi K., Shimizu Y., Bronson R. T., Gao Y. H., Inada M., Sato M., Okamoto R., Kitamura Y., Yoshiki S., Kishimoto T. (1997) Cell 89, 755–764 [DOI] [PubMed] [Google Scholar]
- 3.Otto F., Thornell A. P., Crompton T., Denzel A., Gilmour K. C., Rosewell I. R., Stamp G. W., Beddington R. S., Mundlos S., Olsen B. R., Selby P. B., Owen M. J. (1997) Cell 89, 765–771 [DOI] [PubMed] [Google Scholar]
- 4.Mundlos S., Otto F., Mundlos C., Mulliken J. B., Aylsworth A. S., Albright S., Lindhout D., Cole W. G., Henn W., Knoll J. H., Owen M. J., Mertelsmann R., Zabel B. U., Olsen B. R. (1997) Cell 89, 773–779 [DOI] [PubMed] [Google Scholar]
- 5.Otto F., Lübbert M., Stock M. (2003) J. Cell Biochem. 89, 9–18 [DOI] [PubMed] [Google Scholar]
- 6.Blyth K., Terry A., Mackay N., Vaillant F., Bell M., Cameron E. R., Neil J. C., Stewart M. (2001) Oncogene 20, 295–302 [DOI] [PubMed] [Google Scholar]
- 7.Schroeder T. M., Jensen E. D., Westendorf J. J. (2005) Birth Defects Res. C Embryo Today 75, 213–225 [DOI] [PubMed] [Google Scholar]
- 8.Ducy P., Karsenty G. (1995) Mol. Cell Biol. 15, 1858–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geoffroy V., Ducy P., Karsenty G. (1995) J. Biol. Chem. 270, 30973–30979 [DOI] [PubMed] [Google Scholar]
- 10.Merriman H. L., van Wijnen A. J., Hiebert S., Bidwell J. P., Fey E., Lian J., Stein J., Stein G. S. (1995) Biochemistry 34, 13125–13132 [DOI] [PubMed] [Google Scholar]
- 11.Giedion A., Burdea M., Fruchter Z., Meloni T., Trosc V. (1973) Helv. Paediatr. Acta 28, 249–259 [PubMed] [Google Scholar]
- 12.Niikawa N., Kamei T. (1986) Am. J. Med. Genet. 24, 759–760 [DOI] [PubMed] [Google Scholar]
- 13.Felman A. H., Frias J. L. (1977) AJR Am. J. Roentgenol. 129, 631–638 [DOI] [PubMed] [Google Scholar]
- 14.Giedion A. (1966) Helv. Paediatr. Acta 21, 475–485 [PubMed] [Google Scholar]
- 15.Lüdecke H. J., Schaper J., Meinecke P., Momeni P., Gross S., von Holtum D., Hirche H., Abramowicz M. J., Albrecht B., Apacik C., Christen H. J., Claussen U., Devriendt K., Fastnacht E., Forderer A., Friedrich U., Goodship T. H., Greiwe M., Hamm H., Hennekam R. C., Hinkel G. K., Hoeltzenbein M., Kayserili H., Majewski F., Mathieu M., McLeod R., Midro A. T., Moog U., Nagai T., Niikawa N., Orstavik K. H., Plöchl E., Seitz C., Schmidtke J., Tranebjaerg L., Tsukahara M., Wittwer B., Zabel B., Gillessen-Kaesbach G., Horsthemke B. (2001) Am. J. Hum. Genet. 68, 81–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Momeni P., Glöckner G., Schmidt O., von Holtum D., Albrecht B., Gillessen-Kaesbach G., Hennekam R., Meinecke P., Zabel B., Rosenthal A., Horsthemke B., Lüdecke H. J. (2000) Nat. Genet. 24, 71–74 [DOI] [PubMed] [Google Scholar]
- 17.Malik T. H., Shoichet S. A., Latham P., Kroll T. G., Peters L. L., Shivdasani R. A. (2001) EMBO J. 20, 1715–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang G. T., van den Bemd G. J., Jhamai M., Brinkmann A. O. (2002) Apoptosis 7, 13–21 [DOI] [PubMed] [Google Scholar]
- 19.Itoh S., Kanno S., Gai Z., Suemoto H., Kawakatsu M., Tanishima H., Morimoto Y., Nishioka K., Hatamura I., Yoshida M., Muragaki Y. (2008) Genes Cells 13, 355–363 [DOI] [PubMed] [Google Scholar]
- 20.Suemoto H., Muragaki Y., Nishioka K., Sato M., Ooshima A., Itoh S., Hatamura I., Ozaki M., Braun A., Gustafsson E., Fässler R. (2007) Dev. Biol. 312, 572–581 [DOI] [PubMed] [Google Scholar]
- 21.van den Bemd G. J., Jhamai M., Brinkmann A. O., Chang G. T. (2003) Biochem. Biophys. Res. Commun. 312, 578–584 [DOI] [PubMed] [Google Scholar]
- 22.Fantauzzo K. A., Bazzi H., Jahoda C. A., Christiano A. M. (2008) Gene Expr. Patterns 8, 51–57 [DOI] [PubMed] [Google Scholar]
- 23.Kunath M., Lüdecke H. J., Vortkamp A. (2002) Mech. Dev. 119, Suppl. 1, S117–S120 [DOI] [PubMed] [Google Scholar]
- 24.Malik T. H., Von Stechow D., Bronson R. T., Shivdasani R. A. (2002) Mol. Cell Biol. 22, 8592–8600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Napierala D., Sam K., Morello R., Zheng Q., Munivez E., Shivdasani R. A., Lee B. (2008) Hum. Mol. Genet. 17, 2244–2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishimori N., Stylianou I. M., Korstanje R., Marion M. A., Li R., Donahue L. R., Rosen C. J., Beamer W. G., Paigen B., Churchill G. A. (2008) J. Bone Miner Res. 23, 1529–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ducy P., Starbuck M., Priemel M., Shen J., Pinero G., Geoffroy V., Amling M., Karsenty G. (1999) Genes Dev. 13, 1025–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng X. H., Filvaroff E. H., Derynck R. (1995) J. Biol. Chem. 270, 24237–24245 [DOI] [PubMed] [Google Scholar]
- 29.Naldini L., Blömer U., Gallay P., Ory D., Mulligan R., Gage F. H., Verma I. M., Trono D. (1996) Science 272, 263–267 [DOI] [PubMed] [Google Scholar]
- 30.Yee J. K., Friedmann T., Burns J. C. (1994) Methods Cell Biol. 43, 99–112 [DOI] [PubMed] [Google Scholar]
- 31.Dignam J. D., Lebovitz R. M., Roeder R. G. (1983) Nucleic Acids Res. 11, 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nolan T., Hands R. E., Bustin S. A. (2006) Nat. Protoc. 1, 1559–1582 [DOI] [PubMed] [Google Scholar]
- 33.Schmittgen T. D., Livak K. J. (2008) Nat. Protoc. 3, 1101–1108 [DOI] [PubMed] [Google Scholar]
- 34.Stanford C. M., Jacobson P. A., Eanes E. D., Lembke L. A., Midura R. J. (1995) J. Biol. Chem. 270, 9420–9428 [DOI] [PubMed] [Google Scholar]
- 35.Jimenez C. R., Huang L., Qiu Y., Burlingame A. L. (1998) Current Protocols in Protein Science, pp. 16.4.1–16.4.5Wiley & Sons, Inc., New York [Google Scholar]
- 36.Lian J. B., Stein J. L., Stein G. S., van Wijnen A. J., Montecino M., Javed A., Gutierrez S., Shen J., Zaidi S. K., Drissi H. (2003) Connect. Tissue Res. 44, Suppl. 1, 141–148 [PubMed] [Google Scholar]
- 37.Shi M. J., Stavnezer J. (1998) J. Immunol. 161, 6751–6760 [PubMed] [Google Scholar]
- 38.Zhang Y., Derynck R. (2000) J. Biol. Chem. 275, 16979–16985 [DOI] [PubMed] [Google Scholar]
- 39.Alliston T., Choy L., Ducy P., Karsenty G., Derynck R. (2001) EMBO J. 20, 2254–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heinemeyer T., Wingender E., Reuter I., Hermjakob H., Kel A. E., Kel O. V., Ignatieva E. V., Ananko E. A., Podkolodnaya O. A., Kolpakov F. A., Podkolodny N. L., Kolchanov N. A. (1998) Nucleic Acids Res. 26, 362–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Merika M., Orkin S. H. (1993) Mol. Cell Biol. 13, 3999–4010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pavesi G., Mereghetti P., Zambelli F., Stefani M., Mauri G., Pesole G. (2006) Nucleic Acids Res. 34, W566–W570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang G. T., Blok L. J., Steenbeek M., Veldscholte J., van Weerden W. M., van Steenbrugge G. J., Brinkmann A. O. (1997) Cancer Res. 57, 4075–4081 [PubMed] [Google Scholar]
- 44.Chang G. T., Jhamai M., van Weerden W. M., Jenster G., Brinkmann A. O. (2004) Endocr. Relat. Cancer 11, 815–822 [DOI] [PubMed] [Google Scholar]
- 45.Yeung F., Law W. K., Yeh C. H., Westendorf J. J., Zhang Y., Wang R., Kao C., Chung L. W. (2002) J. Biol. Chem. 277, 2468–2476 [DOI] [PubMed] [Google Scholar]
- 46.Bortell R., Owen T. A., Shalhoub V., Heinrichs A., Aronow M. A., Rochette-Egly C., Lutz Y., Stein J. L., Lian J. B., Stein G. S. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 2300–2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Napierala D., Garcia-Rojas X., Sam K., Wakui K., Chen C., Mendoza-Londono R., Zhou G., Zheng Q., Lee B. (2005) Mol. Genet. Metab. 86, 257–268 [DOI] [PubMed] [Google Scholar]
- 48.Majeska R. J., Rodan S. B., Rodan G. A. (1980) Endocrinology 107, 1494–1503 [DOI] [PubMed] [Google Scholar]
- 49.McCabe L. R., Last T. J., Lynch M., Lian J., Stein J., Stein G. (1994) J. Cell Biochem. 56, 274–282 [DOI] [PubMed] [Google Scholar]
- 50.Dennis J. E., Merriam A., Awadallah A., Yoo J. U., Johnstone B., Caplan A. I. (1999) J. Bone Miner Res. 14, 700–709 [DOI] [PubMed] [Google Scholar]
- 51.Pittenger M. F., Mackay A. M., Beck S. C., Jaiswal R. K., Douglas R., Mosca J. D., Moorman M. A., Simonetti D. W., Craig S., Marshak D. R. (1999) Science 284, 143–147 [DOI] [PubMed] [Google Scholar]
- 52.Owen T. A., Aronow M., Shalhoub V., Barone L. M., Wilming L., Tassinari M. S., Kennedy M. B., Pockwinse S., Lian J. B., Stein G. S. (1990) J. Cell Physiol. 143, 420–430 [DOI] [PubMed] [Google Scholar]
- 53.Pockwinse S. M., Wilming L. G., Conlon D. M., Stein G. S., Lian J. B. (1992) J. Cell Biochem. 49, 310–323 [DOI] [PubMed] [Google Scholar]
- 54.Kern B., Shen J., Starbuck M., Karsenty G. (2001) J. Biol. Chem. 276, 7101–7107 [DOI] [PubMed] [Google Scholar]
- 55.Stein G. S., Lian J. B. (1993) Endocr. Rev. 14, 424–442 [DOI] [PubMed] [Google Scholar]
- 56.Stickens D., Behonick D. J., Ortega N., Heyer B., Hartenstein B., Yu Y., Fosang A. J., Schorpp-Kistner M., Angel P., Werb Z. (2004) Development 131, 5883–5895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yasuda H., Shima N., Nakagawa N., Yamaguchi K., Kinosaki M., Mochizuki S., Tomoyasu A., Yano K., Goto M., Murakami A., Tsuda E., Morinaga T., Higashio K., Udagawa N., Takahashi N., Suda T. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 3597–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lacey D. L., Timms E., Tan H. L., Kelley M. J., Dunstan C. R., Burgess T., Elliott R., Colombero A., Elliott G., Scully S., Hsu H., Sullivan J., Hawkins N., Davy E., Capparelli C., Eli A., Qian Y. X., Kaufman S., Sarosi I., Shalhoub V., Senaldi G., Guo J., Delaney J., Boyle W. J. (1998) Cell 93, 165–176 [DOI] [PubMed] [Google Scholar]
- 59.Pritsker M., Liu Y. C., Beer M. A., Tavazoie S. (2004) Genome Res. 14, 99–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nishioka K., Itoh S., Suemoto H., Kanno S., Gai Z., Kawakatsu M., Tanishima H., Morimoto Y., Hatamura I., Yoshida M., Muragaki Y. (2008) Bone 43, 64–71 [DOI] [PubMed] [Google Scholar]
- 61.Kaiser F. J., Tavassoli K., Van den Bemd G. J., Chang G. T., Horsthemke B., Möröy T., Lüdecke H. J. (2003) Hum. Mol. Genet. 12, 1349–1358 [DOI] [PubMed] [Google Scholar]
- 62.Kaiser F. J., Möröy T., Chang G. T., Horsthemke B., Lüdecke H. J. (2003) J. Biol. Chem. 278, 38780–38785 [DOI] [PubMed] [Google Scholar]
- 63.Kaiser F. J., Lüdecke H. J., Weger S. (2007) Biol. Chem. 388, 381–390 [DOI] [PubMed] [Google Scholar]
- 64.Gómez-del Arco P., Koipally J., Georgopoulos K. (2005) Mol. Cell Biol. 25, 2688–2697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Galli C., Fu Q., Wang W., Olsen B. R., Manolagas S. C., Jilka R. L., O'Brien C. A. (2009) J. Biol. Chem. 284, 12654–12662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mueller M. B., Tuan R. S. (2008) Arthritis Rheum. 58, 1377–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chang G. T., Steenbeek M., Schippers E., Blok L. J., van Weerden W. M., van Alewijk D. C., Eussen B. H., van Steenbrugge G. J., Brinkmann A. O. (2000) J. Natl. Cancer Inst. 92, 1414–1421 [DOI] [PubMed] [Google Scholar]
- 68.Pratap J., Lian J. B., Javed A., Barnes G. L., van Wijnen A. J., Stein J. L., Stein G. S. (2006) Cancer Metastasis Rev. 25, 589–600 [DOI] [PubMed] [Google Scholar]
- 69.Huang W. C., Xie Z., Konaka H., Sodek J., Zhau H. E., Chung L. W. (2005) Cancer Res. 65, 2303–2313 [DOI] [PubMed] [Google Scholar]
- 70.Drissi H., Pouliot A., Koolloos C., Stein J. L., Lian J. B., Stein G. S., van Wijnen A. J. (2002) Exp. Cell Res. 274, 323–333 [DOI] [PubMed] [Google Scholar]
- 71.Radvanyi L., Singh-Sandhu D., Gallichan S., Lovitt C., Pedyczak A., Mallo G., Gish K., Kwok K., Hanna W., Zubovits J., Armes J., Venter D., Hakimi J., Shortreed J., Donovan M., Parrington M., Dunn P., Oomen R., Tartaglia J., Berinstein N. L. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 11005–11010 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.