Abstract
The regulation of lipid homeostasis by insulin is mediated in part by the enhanced transcription of the gene encoding sterol regulatory element-binding protein-1c (SREBP-1c). The nascent SREBP-1c is embedded in the endoplasmic reticulum (ER) and must be transported to the Golgi where two sequential cleavages generate its NH2-terminal fragment, nSREBP-1c. We have shown recently that in primary cultures of rat hepatocytes, insulin rapidly and selectively stimulates proteolytic processing of the nascent SREBP-1c by enhancing the affinity of the SREBP cleavage-activating protein (SCAP)·SREBP-1c complex for coatomer protein complex II (COPII) vesicles. The SCAP·SREBP complex is retained in the ER by Insig proteins. We report here that insulin persistently stimulates controlled proteolysis of the nascent SREBP-1c by selectively reducing the level of Insig-2a protein via accelerated degradation of its cognate mRNA. Insulin enhanced the rate of turnover of Insig-2a mRNA via its 3′-untranslated region. Insulin-induced depletion of Insig-2a promotes association of the SCAP·SREBP-1c complex with COPII vesicles and subsequent migration to the Golgi where site-1 and site-2 proteases process the nascent SREBP-1c. Consistent with this mechanism, experimental knockdown of Insig-2a expression with small interfering RNA mimicked insulin-induced proteolysis of the nascent SREBP-1c, whereas exogenous expression of Insig-2a in hepatocytes led to reduced intramembrane proteolysis of the newly synthesized SREBP-1c. The action of insulin on the processing of the nascent SREBP-1c via Insig-2a was highly selective, as proteolysis of the newly synthesized SREBP-2 remained unchanged under identical conditions. On the basis of these data, we propose that the stimulation of SREBP-1c processing by insulin is mediated by a selective depletion of Insig-2a protein by promoting decay of its cognate mRNA. Thus, insulin-induced reduction in Insig-2a protein leads to an enhanced export of the SCAP·SREBP-1c complex from ER to the Golgi.
Introduction
Sterol-regulatory element-binding proteins (SREBPs),3 represented by SREBP-1a, SREBP-1c, and SREBP-2, are key transcription factors that control a suite of genes involved in the homeostatic regulation of cholesterol and lipid metabolism (1–7). SREBP-1a and SREBP-1c, which differ only in their NH2-terminal sequences, are derived from a single gene through the use of alternative promoters, and SREBP-2 is encoded by a separate gene (8). Despite their structural similarities, the three SREBPs regulate different metabolic pathways (5). SREBP-2 is constitutively expressed in most tissues; in contrast, whereas the SREBP-1a gene is preferentially expressed in transformed cell lines, SREBP-1c is the predominant isoform expressed in the liver and adipose tissue (9). The tissue-specific differences in the expression of SREBP-1a, SREBP-1c, and SREBP-2 reflect their unique functions as regulators of cell-specific gene expression.
As key drivers of lipid homeostasis, the genes encoding SREBPs are regulated in response to a number of nutritional and hormonal stimuli by both transcriptional and posttranscriptional mechanisms of gene expression. The newly synthesized SREBPs are embedded in the endoplasmic reticulum (ER) via their two trans-membrane domains, whereas the NH2- and COOH-terminal segments of the nascent SREBPs are exposed to the cytoplasm. Within the ER, SREBPs are associated with two integral membrane proteins, SCAP (SREBP cleavage-activating protein) and Insig-1 (insulin-induced gene product-1) or Insig-2 (1, 2). Following their transport from the ER to the Golgi, the nascent SREBPs undergo intramembrane proteolysis, which liberates their transcriptionally active NH2-terminal fragments. To maintain intracellular levels of sterols in a narrow range, the ER-to-Golgi transit and processing of the cholesterol-regulating isoform SREBP-2 are dynamically controlled by sterols that finely modulate the molecular interactions of ER-loaded nascent SREBP-2 with Insig proteins and SCAP. For example, when cholesterol levels are high, SCAP is tightly associated with Insig-1, causing retention of the SCAP·SREBP-2 complex in the ER and thus preventing proteolytic processing of the nascent SREBP-2. Conversely, when cholesterol levels are depleted, Insig-1 dissociates from the SCAP·SREBP-2 complex, thus facilitating its transport to the Golgi where full-length SREBP-2 undergoes proteolysis to generate nSREBP-2.
Hepatic expression of the SREBP-1c isoform and its downstream targets has received considerable attention in recent years. There is compelling evidence to suggest that altered expression of SREBP-1c in the liver under hyperinsulinemic conditions may be related causally to the mechanisms of hyperlipidemia associated with type II diabetes and obesity (10, 11). Insulin potently stimulates expression of the SREBP-1c gene by a “feed forward” mechanism of gene expression (11–13). It has also been demonstrated that insulin not only stimulates transcription of the SREBP-1c gene (14, 15) but also has a potent effect on the rate of intramembrane proteolysis of the newly synthesized SREBP-1c. The insulin-induced phosphorylation of the nascent SREBP-1c leads to its more efficient transit from the ER to the Golgi compartment, the site of proteolytic processing (16). However, the molecular details of the putative mechanisms by which insulin enhances the biogenesis of nSREBP-1c via more efficient posttranslational proteolysis are unclear. In addition, although both SREBP-1c and -2 isoforms are associated with the SCAP/Insig complex and similarly undergo ER-to-Golgi transport and proteolytic processing, they are selectively regulated by insulin and sterols, respectively (16). The mechanism of this specificity is not clearly defined.
In this work, we show that persistent stimulation of SREBP-1c processing by insulin in primary cultures of rat hepatocytes is obligatorily dependent on its ability to reduce Insig-2a gene expression. Insulin achieves this outcome by selectively promoting the turnover of Insig-2a mRNA without affecting the steady state level of either Insig-2b or Insig-1 mRNA. Furthermore, we report that insulin mediates depletion of Insig-2a protein by degradation of its cognate mRNA, thus promoting the association between the SCAP·SREBP-1c complex and Sec23·Sec24-containing COPII vesicles and their more rapid transport to the Golgi.
MATERIALS AND METHODS
Reagents
Insulin, aprotinin, N-acetyl-leucyl-leucyl-norleucinal (ALLN), dithiothreitol, leupeptin, sodium orthovanadate, sodium deoxycholate, phenylmethylsulfonyl fluoride, compactin, cycloheximide, actinomycin D, and HEPES were purchased from Sigma. The Dual-Luciferase Reporter 1000 assay system, pGL3-basic vector, and pRL-null vector were obtained from Promega Corp. (Madison, WI). Platinum PCR SuperMix and Lipofectamine 2000 transfection reagent were procured from Invitrogen. Protein A/G beads were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Plasmids
As described previously (16), the adenovirus construct Ad-HisSREBP-1cFLAG was designed to express a rat full-length SREBP-1c fused to six tandem copies of His epitope tag at the NH2 terminus and 3× FLAG tag at the COOH terminus under the control of the CMV promoter. Ad-Insig-2aFLAG vector was created by cloning a full-length rat Insig-2a-specific cDNA that was cloned in-frame with a FLAG tag. Insig-2a-specific cDNA was amplified by PCR using 5′-actgtcgacatggcggaagga-3′ (forward) and 5′-cgtgtcgacttcttgatgaga-3′ (reverse) containing an engineered SalI site. The PCR-amplified fragments were ligated into the pShuttle-CMV vector (Stratagene). Then, a fragment encoding a 3× FLAG tag and a stop codon (TGA) was introduced at the COOH terminus of Insig-2a cDNA. The resulting plasmid was linearized with PmeI and transformed into Escherichia coli BJ5183-AD-1. Recombinant adenovirus plasmid DNA was digested with PacI and introduced into AD-293 cells to prepare a stock of FLAG-tagged Insig-2a adenovirus (Ad-Insig-2aFLAG). Recombinant Ad-HisSREBP-1cFLAG, Ad-Insig-2aFLAG, and control Ad-LacZ viruses were propagated in AD-293 cells and purified by CsCl density gradient centrifugation. Recombinant SREBP-1c, Insig-2a, and control adenovirus stocks were stored in PBS containing 10% glycerol at −80 °C until used. Hepatocytes were infected at a multiplicity of infection of 10–30 for 16 to 24 h.
A plasmid vector used to investigate the role of 3′-untranslated regions (3′-UTRs) in the posttranscriptional regulation of Insig-2a mRNA turnover (pLuc/Insig-2a 3′-UTR) and a control vector containing the 3′-UTR sequence of GAPDH (pLuc/GAPDH 3′-UTR) were purchased from SwitchGear Genomics (Menlo Park, CA). Both the Insig-2a and GAPDH constructs were created on the backbone of the pSGG vector. The pLuc/Insig-2a 3′-UTR contains an 1850-bp DNA encompassing the entire 3′-UTR of human Insig-2a gene inserted downstream of the firefly luciferase.
The promoter-luciferase construct pInsig-2a (−2496/+87)-luc was constructed as follows. The rat genomic DNA was isolated from tail biopsy using a Qiagen genomic DNA isolation kit (Valencia, CA). A forward primer containing a KpnI site (5′-gcggtaccctgacattggtg-3′) and a reverse primer containing a BglII site (5′-cgagatctgcagagatgtcagg-3′) were used in PCR to amplify a DNA fragment encompassing 2496 nucleotides upstream of the transcriptional start site (+1) and 87 nucleotides of the first exon from the rat Insig-2a gene. The PCR-amplified DNA fragment was ligated into the multiple cloning region upstream of the luciferase cDNA sequence of the pGL3-basic vector (Promega). The pInsig-2a (−2496/+87)-luc plasmid was verified by DNA sequencing.
Culture of Primary Hepatocytes
Hepatocytes were obtained from livers of male Sprague-Dawley rats (Harlan Laboratories, Indianapolis, IN) by collagenase perfusion as described previously (17). Cells were suspended in Dulbecco's modified Eagle's medium (Invitrogen) containing 5% fetal bovine serum (Sigma), 10 mm glucose, 1 μm dexamethasone, and 100 nm insulin. Each 60-mm-diameter tissue culture dish was coated with rat tail collagen (Collaborative Biochemical Products, Bedford, MA) and seeded with 3 × 106 cells. After 4 h, nonadherent cells were removed, and adherent cells were infected at a multiplicity of infection of 10–30 with Ad-HisSREBP-1cFLAG, Ad-Insig-2aFLAG, or Ad-LacZ virus. After infection of hepatocytes with recombinant adenoviruses, cells were incubated in Dulbecco's modified Eagle's medium with or without insulin (100 nm) for 24 h.
Transient Transfections and Luciferase Reporter Assays
Primary cultures of rat hepatocytes were incubated overnight with 1 μg of pInsig-2a (−2496/+87)-luc or pLuc-Insig-2a 3′-UTR plus 1 μg of pRL-null dispersed with 8 μl of Lipofectin reagent in RPMI 1640 medium. Cells were washed with 1× PBS and incubated for a further 24 h in RPMI 1640 medium containing 100 nm insulin, 20 mm glucose, and 100 nm dexamethasone alone. Cells were lysed, and luciferase activity was quantified using a TD-20/20 luminometer (Turner Designs, Sunnyvale, CA). The Dual-Luciferase Reporter assay system was used to normalize for variation in transfection efficiency, and the data were expressed as the ratio of Photinus to Renilla luciferase activity as outlined in detail elsewhere (14, 15).
Preparation of Cell Total Extracts and Nuclear Extracts
Primary cultures of rat hepatocytes or McA-RH7777 rat hepatoma cells (catalog no. CRL-1601, ATCC, Manassas, VA) were homogenized in cell lysis buffer (Santa Cruz Biotechnology) containing protease and phosphatase inhibitors and then centrifuged at 10,000 × g for 10 min at 4 °C to obtain the supernatants that were used as total cell extracts. Nuclear extracts were prepared according to a published procedure (18) with minor modifications (16). In brief, hepatocytes from five 60-mm plates were washed twice with PBS and collected by centrifugation at 1,000 × g for 5 min. Pelleted cells were then suspended by passage through a 25-gauge needle 10 times in 1 ml of lysis buffer (20 mm HEPES, pH 7.9, 10 mm NaCl, 1.5 mm MgCl2, 0.2 mm EDTA, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, and 0.5% Nonidet P-40) and protease inhibitor mixture (Sigma); the cell suspension was kept on ice for 10 min. Nuclei were pelleted by centrifugation at 500 × g for 10 min at 4 °C and washed once in the lysis buffer. The pelleted nuclei were resuspended in a hypertonic buffer (20 mm HEPES, pH 7.9, 0.42 m NaCl, 1.5 mm MgCl2, 2.5% glycerol, 1 mm EDTA, 1 mm EGTA, 1 mm dithiothreitol, 25 μg/ml ALLN, and protease inhibitor mixture), incubated for 30 min at 4 °C, and centrifuged at 25,000 × g for 30 min at 4 °C; the supernatant containing nuclear proteins was collected.
Preparation of Microsomal Membranes
Microsomal membranes were prepared according to a published procedure (19) with minor modifications as outlined previously (16). Briefly, hepatocytes were washed three times with ice-cold PBS, scraped off the dish with a rubber policeman into 10 mm HEPES-KOH, pH 7.2, 250 mm sorbitol, 10 mm KOAc, and 1.5 mm Mg(OAc)2 plus protease inhibitors, and then broken with 20 passes through a 22-gauge syringe. The broken cells were centrifuged at 500 × g for 10 min at 4 °C, and the supernatant was recentrifuged at 16,000 × g for 20 min at 4 °C. The protein extracted from the pellet containing ER, pre-Golgi, and Golgi membranes was suspended in 20 mm HEPES, pH 7.2, 250 mm sorbitol, 50 mm KOAc, and 1 mm Mg(OAc)2 plus protease inhibitors and subjected to SDS-PAGE and Western blot analysis.
SDS-PAGE and Immunoblot Analysis
Protein concentrations of total cellular and nuclear extracts were measured with a BCA kit (Pierce Biotechnology), and equal amounts of protein were subjected to SDS-PAGE. Proteins were transferred to nitrocellulose membranes (Amersham Biosciences), and immunoblot analysis was carried out with an ECL Plus Western blotting detection kit (Amersham Biosciences) according to the manufacturer's instructions. Images were captured with a phosphorimaging device (Bio-Rad) and quantified with ImageJ software (from NIH Image). The following primary antibodies were used: anti-FLAG (F3040) from Sigma; and anti-His (SC-8036), anti-SREBP1 (SC-13551), anti-SCAP (SC-9675), anti-histone H1 (SC-8030), anti-Insig-1 (SC-25124), anti-Insig-2 (SC-34821), anti-Sec23 (SC-12107), anti-calnexin (SC-46669), and anti-β-actin (SC-1616) from Santa Cruz Biotechnology.
Immunoprecipitation
Microsomal extracts containing 500–1000 μg of protein from control and insulin-treated hepatocytes were immunoprecipitated with 2 μg of anti-Sec23, anti-Insig-2, or anti-Insig-1 antibodies overnight at 4 °C. The immunoprecipitated proteins were collected by incubation with 40 μl of 50% (w/v) protein A/G-Sepharose beads followed by serial washings with PBS and lysis buffer. The protein A/G-Sepharose beads containing immunocomplexed protein were suspended in 40 μl of Laemmli sample buffer, heated for 10 min in boiling water, resolved by SDS-PAGE, and subjected to Western blot analysis as described previously (16).
Isolation of RNA and Its Quantification by Real-time PCR
Total RNA was isolated from primary cultures of rat hepatocytes by using an RNeasy Mini Kit (Qiagen). Twenty microliters of RNA was initially treated with DNase I (Ambion, Austin, TX) at 37 °C for 1 h, and the RNA in the samples were quantified by measuring absorbance at 260 nm. Equal amounts of DNA-free RNA were used for the first-strand cDNA synthesis that was done by using a SuperScript III first-strand synthesis kit (Invitrogen). Up to 1 μg of RNA was mixed with 1 μl of dNTP mix (10 mm) and 1 μl of random hexamers (50 ng/μl). Each sample was incubated at 70 °C for 10 min and then placed on ice for at least 2 min. Next, 2 μl of 10× reverse transcriptase buffer, 4.5 μl of 25 mm MgCl2, 2 μl of 0.1 m dithiothreitol, and 1 μl of RNaseOUT recombinant ribonuclease inhibitor were added to each tube. After incubation at room temperature for 2 min, 1 μl of SuperScript III reverse transcriptase was added to each sample. The samples were incubated sequentially at room temperature for 10 min, at 42 °C for 1 h, and at 70 °C for 15 min. RNase H was then added to each tube, and each tube was incubated at 37 °C for 20 min. Synthesized cDNA was mixed subsequently with 2× SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) and a set of forward and reverse primers and quantified by real-time PCR analysis. The primers used were: Insig-2a, 5′-cgctcttggttgccatgta-3′ (forward) and 5′-tgatgagatttttgagcaataacttt-3′ (reverse); Insig-1, 5′-atgtatcgcggtgtttgttg-3′ (forward) and 5′-tcgatcaaacgtccacca-3′ (reverse); and 18 S rRNA, 5′-cggctaccacatccaaggaa-3′ (forward) and 5′-ttttcgtcactacctccccg-3′ (reverse). The parameters for real-time PCR were 95 °C for 11 min followed by 40 cycles of 95 °C for 30 s and 60 °C for 1 min; parameters for the melt curve were 100 cycles of 10 s from 60 to 100 °C in increments of 0.4 °C. All reactions were performed in triplicate. The relative amounts of mRNAs were calculated by using the comparative Ct method.
Statistics
All experiments were repeated at least three times; data are presented as mean ± S.E. The data were analyzed by unpaired Student's t test using Prism 4.0 (GraphPad Software, San Diego, CA). Quantifications with p values of <0.05 were considered statistically significant.
RESULTS
Insulin Stimulates Processing of SREBP-1c in Rat Hepatocytes
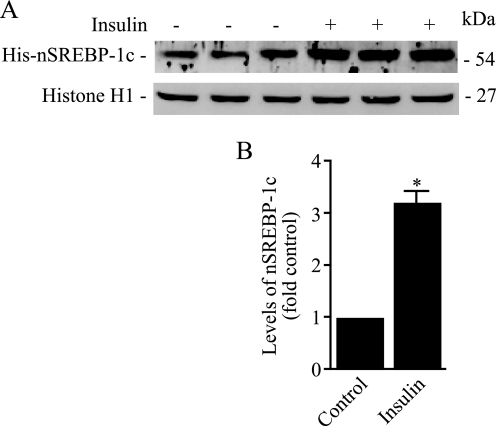
Using adenovirus vectors designed to express the full-length SREBP-1c in rat hepatocytes, we recently demonstrated that insulin potently and selectively stimulates proteolytic processing of the nascent SREBP-1c (16). We used a similar strategy here to extend these observations. Specifically, we carried out experiments to test the hypothesis that insulin-induced signals enhance the intramembrane proteolysis of SREBP-1c by altering its association with ER-bound Insig and/or COPII proteins. To accomplish this objective, we infected hepatocytes with Ad-HisSREBP-1cFLAG followed by incubation of these cells in medium with or without 100 nm insulin for an additional 24 h. To prevent degradation of the newly generated nSREBP-1c, we incubated adenovirus-infected cells with the proteasomal inhibitor ALLN (25 μg/ml) in the final 6 h before harvesting. We fractionated nuclear protein extracts by SDS-PAGE, transferred the proteins to nylon membranes, and reacted these blots with antibodies against His tag to measure the accumulation of His-tagged nSREBP-1c. To measure equivalencies of protein loading across various lanes, blots were subsequently probed with a histone H1-specific antibody. Representative Western blots from three independent experiments and quantification of the resulting data are shown in Fig. 1. Consistent with previously published data, nuclear extracts prepared from insulin-treated hepatocytes were more highly enriched in nSREBP-1c compared with extracts from untreated cells; in insulin-treated hepatocytes, the nuclear SREBP-1c was increased by more than 3-fold (Fig. 1).
FIGURE 1.
Insulin stimulates processing of SREBP-1c at 24 h. Hepatocytes were infected with Ad-HisSREBP-1cFLAG and incubated in medium with or without insulin (100 nm). ALLN (25 μg/ml) was added for the final 6 h of incubation, and cells were harvested. Nuclear extracts were fractionated by SDS-PAGE, and immunoblots were developed using anti-His or anti-histone antibodies. Representative blots of triplicate samples of three experiments are shown. B, densitometric quantification of nSREBP-1c bands shown in A. The data are expressed as a percentage of control optical density of nSREBP-1c-specific band. *, p < 0.05 versus control.
Insulin Down-regulates Insig-2a Gene Expression
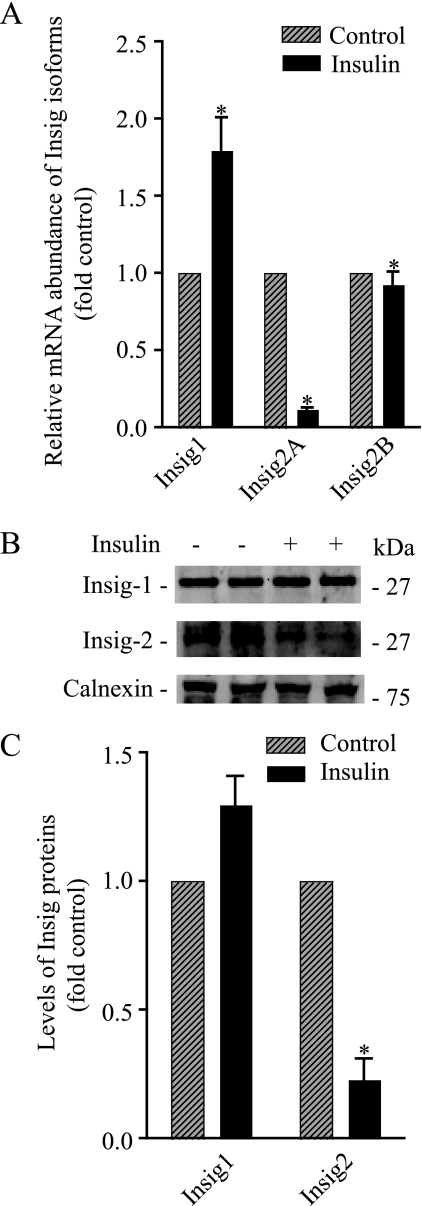
Two highly conserved integral ER proteins, Insig-1 and Insig-2, are critically involved in the ER-to-Golgi transport of the newly synthesized SREBPs and in their intramembrane proteolysis. Insig proteins bind to SCAP, thereby promoting its retention in the ER and consequently preventing SCAP from escorting SREBPs from the ER to the Golgi complex. It has been documented that sterols reduce the affinity of Insig-1 for the SCAP·SREBP-2 complex, thus facilitating the transit of this complex from the ER to the Golgi, where processing of SREBP-2 occurs. Under these conditions, Insig-1 is degraded by the ubiquitin pathway (20, 21). In light of these observations, we reasoned that insulin might also regulate the ER-to-Golgi transport of the newly synthesized SREBP-1c by its ability to alter interactions among Insig-1/Insig-2, SCAP·SREBP-1c, and COPII proteins (i.e. the Sec23·Sec24 protein complex) in the ER. Unlike Insig-1, which is expressed ubiquitously, Insig-2a is expressed specifically in the liver, and it is known that insulin reduces the Insig-2 mRNA in rat hepatocytes (22, 23). On the basis of these observations, we hypothesized that a selective diminution in the Insig-2a gene expression might be involved mechanistically in the action of insulin on the processing of SREBP-1c. To test this hypothesis experimentally, we began by measuring the steady state levels of Insig-1, Insig-2a, and Insig-2b mRNAs in control and insulin-treated primary cultures of rat hepatocytes by real-time PCR analysis. As shown in Fig. 2A, insulin reduced the level of Insig-2a mRNA by more than 5-fold. The effect of insulin on Insig-2a mRNA was selective, as the levels of Insig-1 and Insig-2b mRNAs in insulin-treated hepatocytes remained unaltered.
FIGURE 2.
Insulin selectively reduces the levels of Insig2A mRNA and protein. A, primary cultures of rat hepatocytes were treated with and without insulin (100 nm) for 24 h, and RNA was isolated. The mRNA abundance of Insig-1, Insig-2a, and Insig-2b was measured by real-time PCR analysis as described under “Materials and Methods” using 18 S rRNA as the control. The data are expressed as -fold induction of control Insig isoform(s) mRNA abundance. B, hepatocytes were incubated with and without insulin (100 nm) for 24 h, and microsomes were isolated. Microsomal membranes were fractionated by SDS-PAGE and immunoblotted using Insig-1, Insig-2, and calnexin antibodies. Representative blots are shown. C, densitometric quantification of Insig bands from duplicates of three independent experiments as shown in B. The data are expressed as -fold control of Insig specific bands. *, p < 0.05 versus control.
To determine whether the insulin-mediated decline in Insig-2a mRNA leads to a depletion of Insig-2a protein, we extracted proteins from microsomal membranes prepared from insulin-treated and untreated hepatocytes and probed these proteins by Western blot using Insig-specific antibodies. These blots were also probed with an antibody to calnexin, an ER marker protein, to assess protein loading. As seen in a representative Western blot (Fig. 2B), microsomes from insulin-treated hepatocytes contained a lower amount of Insig-2 protein. Quantification of these blots revealed that insulin treatment resulted in an ∼3-fold reduction in the level of Insig-2 protein without significantly affecting Insig-1 protein (Fig. 2C). Because the Insig-2a transcript is the predominant species in the liver, we interpreted these data to mean that insulin selectively reduced the expression of the Insig-2a isoform. However, our interpretation should be viewed with the caveat that our antibodies did not distinguish between Insig-2a and Insig-2b proteins.
Insulin Selectively Alters the Rate of Turnover of Insig-2a mRNA
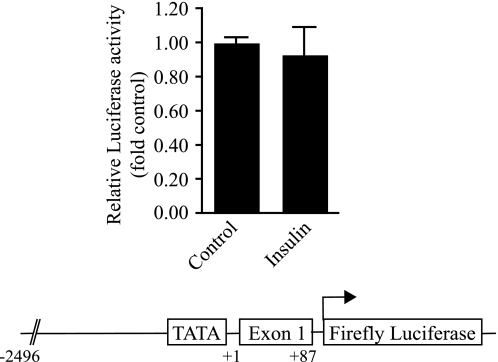
We next examined whether a reduction in the steady state levels of Insig-2a mRNA in response to insulin was because of its ability to repress transcription of the Insig-2a gene or whether insulin achieved this outcome via selectively accelerating the degradation of Insig-2a mRNA. To assess the effect of insulin on the transcription of the Insig-2a gene, we cloned a 2.5-kb DNA fragment representing the 5′-flanking region of the Insig-2a gene and tested its potential for regulation by insulin in primary cultures of rat hepatocytes. Cells were transfected with a DNA construct in which expression of the luciferase reporter was driven by the 5′-flanking region of the Insig-2a gene. Luciferase expression in insulin-treated and untreated hepatocytes was quantified as outlined previously (15). As demonstrated in Fig. 3, insulin exerted no measurable effect on the activity of the Insig-2a promoter-luciferase reporter. On the basis of these analyses, we reasoned that a reduction in the steady state of Insig-2a mRNA by insulin was most likely because of its effect on the rate of turnover of Insig-2a mRNA.
FIGURE 3.
Insulin does not activate the rat Insig-2a promoter. Primary cultures of rat hepatocytes were transfected with pInsig-2a (−2496/+87)-luc and pRL-null vector designed to express Renilla luciferase. Cells were then incubated with or without insulin for 24 h. Cells were lysed, and luciferase activity was quantified following normalization of transfection efficiency. The data shown represent three independent experiments done in triplicate. The Insig-2a-luc construct contains a −2496/+87 Insig-2a-specific genomic DNA ligated upstream of the firefly luciferase in the pGL3-basic vector.
To test this hypothesis, we carried out experiments to determine the rates of turnover of Insig-2a mRNA in untreated and insulin-treated hepatocytes. Thus, primary cultures of rat hepatocytes were preincubated for 8 h in the presence or absence of insulin followed by incubation of these cells in growth medium supplemented with α-amanitin (5 μg/ml) to block transcription. At the end of this incubation, the steady state levels of Insig-2a mRNA were quantified by real-time PCR analysis. As shown in Fig. 4A, in insulin-treated hepatocytes the half-life of the Insig-2a mRNA was shortened from 9.0 to 4.0 h. A similar reduction in the half-life of Insig-2a mRNA was observed when the duration of insulin treatment was extended to 24 h (data not shown). Because insulin had an insignificant effect on the turnover of Insig-1 mRNA in rat hepatocytes exposed to an identical experimental regimen (Fig. 4B), we believe that insulin selectively promotes the rate of turnover of Insig-2a mRNA.
FIGURE 4.
Insulin accelerates the turnover rate of Insig-2a mRNA. Hepatocytes were treated with and without insulin (100 nm) for 8 h, at which time cells were treated with 5 μm α-amanitin (time 0), and then RNA was isolated at the indicated times. Insig-2a mRNA (A) and Insig-1 (B) mRNA abundance was determined by real-time PCR analysis as described under “Materials and Methods”; the results were normalized with respect to time 0 values. The first order decay rate constants were derived and used to calculate half-life values. C, schematic diagram of the pLuc/Insig-2a 3′-UTR reporter plasmid (upper panel). Hepatocytes were transiently transfected with pRL-null and either the pLuc/GAPDH 3′-UTR or pLuc/Insig-2a 3′-UTR construct and then incubated with or without insulin (100 nm) for 24 h. Luciferase activity was measured as described under “Materials and Methods.” The resulting firefly luciferase activity was normalized against the corresponding Renilla luciferase activity. Data are mean ± S.E. of luciferase activity relative to untreated (Control) hepatocytes transfected with pLuc/GAPDH 3′-UTR construct (n = 3 determinations from three separate hepatocyte preparations). BP, basal promoter; MCS, multicloning site. *, p < 0.05 versus control.
The AU-rich sequences in the 3′-UTRs of mRNAs are thought to determine their unique rates of turnover, and in principle, a given 3′-UTR can confer its regulatory properties on a heterologous reporter mRNA (24). Guided by these observations, we investigated whether the insulin-mediated enhanced turnover of Insig-2a mRNA could be regulated via its cognate 3′-UTR. We transfected parallel cultures of hepatocytes with either pLuc/Insig-2a 3′-UTR DNA or a control vector, pLuc/GAPDH 3′-UTR. Cells were harvested 24–48 h after transfection, and cell extracts were used to quantify expression of the luciferase reporter. As shown in Fig. 4C, luciferase activity was 60% lower in insulin-treated hepatocytes compared with their untreated counterparts. It is apparent that the destabilizing effect of insulin on the luciferase mRNA was mediated by the Insig-2a-specific 3′-UTR because the expression of luciferase in hepatocytes transfected with pLuc/GAPDH 3′-UTR DNA was similar regardless of the presence or absence of insulin in the medium (Fig. 4C). These studies show that insulin treatment specifically accelerates the rate of degradation of Insig-2a mRNA via its 3′-UTR sequences. The molecular mechanisms involved in selective insulin-induced decay of Insig-2a mRNA remain unresolved. Conceivably, insulin could regulate the synthesis of a putative AU-rich 3′-UTR-interacting protein(s) that promotes mRNA degradation (24). Therefore, we measured the steady state levels of Insig-2a mRNA in hepatocytes treated with cycloheximide to inhibit de novo protein synthesis. We discovered that blocking protein synthesis by cycloheximide led to a complete elimination of the effect of insulin on Insig-2a mRNA turnover (data not shown).
Reduced Expression of Insig-2a Mimics the Effect of Insulin on Processing of Nascent SREBP-1c
To investigate the role of Insig-2a in the processing of SREBP-1c, we knocked down the expression of Insig-2a in rat hepatoma cells with Insig-2a siRNA according to published procedures (25). As shown in Fig. 5A, real-time PCR analysis of mRNA abundance showed that in cells transfected with a pool of three Insig-2a-specific siRNAs (Ambion), the Insig-2a mRNA was knocked down by about 75%. The effect of Insig-2a siRNA knockdown was isoform-specific, because the expression of Insig-1 or Insig-2b mRNA remained unchanged under these conditions (Fig. 5A).
FIGURE 5.
Knockdown of Insig-2a expression by Insig-2a siRNA selectively promotes SREBP-1c processing. A, rat hepatoma cells were transfected with or without Insig-2a siRNA or control siRNA targeting GAPDH, and total RNA was isolated. The mRNA abundance of Insig-2a, Insig-2b, and Insig-1 was measured by real-time PCR analysis as described under “Materials and Methods”; 18 S rRNA was used as the control. The data are expressed as -fold change in the expression of Insig mRNA compared with mRNA levels in untransfected cells. B, rat hepatoma cells were co-transfected with pHisSREBP-1cFLAG plus either control siRNA for GAPDH or Insig-2a siRNA for 48 h, and cells were harvested. Nuclear extracts were separated by SDS-PAGE and immunoblotted for anti-His or anti-histone H1 antibodies. A representative blot of microsomal proteins fractionated by SDS-PAGE and immunoblotted using anti-Insig-2 antibodies is shown. The slowly migrating band above His-nSREBP-1c in the right two lanes represents the phosphorylated form of His-nSREBP-1c.
To investigate whether the knockdown of Insig-2a mRNA and protein affected the rate of proteolytic processing of the nascent SREBP-1c, rat hepatoma cells were co-transfected with Insig-2a siRNA and His-SREBP-1cFLAG plasmids. The microsomal membranes from cells expressing exogenous Insig-2a siRNA and His-SREBP-1c-FLAG were assessed for the presence of Insig-2a protein; concomitantly, the nuclear extracts were also processed to quantify the nSREBP-1c by His-antibodies. As shown in Fig. 5B, Insig-2a-specific siRNA reduced the levels of Insig-2 protein; these cells also elicited an ∼50% greater accumulation of His-nSREBP-1c in their nuclei. The upper band seen in the last two lanes (Fig. 5B) depicting His-nSREBP-1c is phosphorylated His-nSREBP-1c; the appearance of the phosphorylated nSREBP-1c band varied somewhat from experiment to experiment.4 In striking contrast, a nonspecific siRNA (GAPDH) did not affect the SREBP-1c processing (Fig. 5B).
Insig-2a Selectively Associates with the SCAP·SREBP-1c Complex
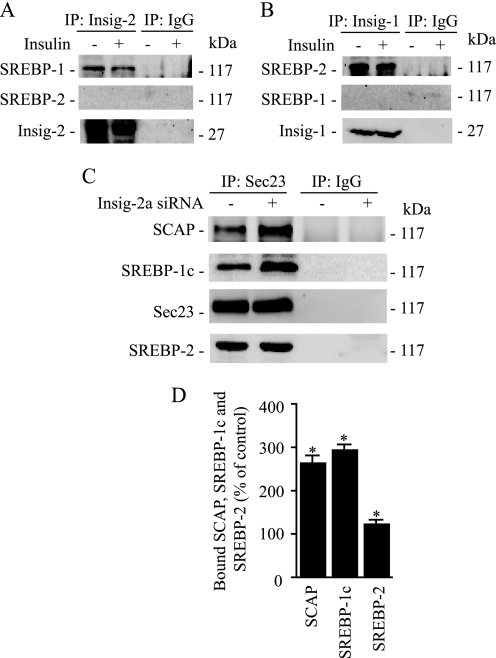
To undergo proteolytic cleavage, the nascent SREBP-1c must be exported from the ER to the Golgi, prior to which the SCAP·SREBP-1c complex must dissociate from the ER retention protein, Insig. Therefore, we sought to determine whether decreased expression of Insig-2a in response to insulin correlated with a reduced binding of the SCAP·SREBP-1c complex to Insig. We immunoprecipitated microsomal proteins with Insig-1- or Insig-2-specific antibodies from control and insulin-treated hepatocytes and assayed these extracts for the presence of SREBP-1, SREBP-2, or Insig-2. As shown in Fig. 6A, SREBP-1c was coimmunoprecipitated with Insig-2a; this association was greatly decreased in hepatocytes treated with insulin. We then tested whether the association/dissociation of Insig-2a was specific for SREBP-1c. Consistent with the published data showing that the SREBP-2 isoform is not subject to insulin-mediated regulation (16), we found no detectable association of SREBP-2 with Insig-2a in immunoprecipitates derived from either control or insulin-treated hepatocytes (Fig. 6A, middle panel). These findings indicate that Insig-2a selectively associates with SREBP-1c and that insulin decreases this association. To further corroborate the selectivity of association between Insig-2a and SREBP-1c, we immunoprecipitated the sterol-regulated isoform Insig-1 (26) from control and insulin-treated hepatocytes and assayed the immunoblots for the presence of SREBP-1c and -2. As expected, SREBP-2 was associated with Insig-1 (Fig. 6B). In contrast, SREBP-1c was not detected in Insig-1 immunoprecipitates derived from either untreated or insulin-treated hepatocytes (Fig. 6B, middle panel). The presence of Insig-1 protein from the immunoprecipitated samples was confirmed by reprobing the Western blots for Insig-1 (Fig. 6B, bottom panel). Overall, these data support the conclusions that 1) Insig-2a selectively associates with the SREBP-1c complex and 2) the effect of insulin to enhance SREBP-1c processing results from the dissociation of the Insig-2a·SREBP-1c complex.
FIGURE 6.
Insig-1 and Insig-2a selectively associate with the SCAP·SREBP-2 and SCAP·SREBP-1c complexes, respectively. Representative blots are shown in A–C. A, hepatocytes were incubated with and without insulin for 24 h (100 nm), and microsomes were isolated. Microsomal membranes were immunoprecipitated with anti-Insig-2 or preimmune IgG, fractionated by SDS-PAGE, and immunoblotted for SREBP-1c or SREBP-2. B, following incubation of hepatocytes with or without insulin for 24 h, microsomes were prepared. Microsomal membranes were immunoprecipitated (IP) using anti-Insig-1 or preimmune IgG and analyzed by immunoblotting using SREBP-1c or SREBP-2 antibodies. C, hepatocytes were transfected with Insig-2a siRNA, and microsomes were isolated. Microsomal membranes were immunoprecipitated with anti-Sec23 or preimmune IgG, fractionated by SDS-PAGE, and immunoblotted for SCAP, SREBP-1c, or SREBP-2. D, densitometric quantification of the amount of bound SCAP, SREBP-1c, and SREBP-2. Data are expressed as a percentage of change over control optical density values of SCAP, SREBP-1c, and SREBP-2 bands. *, p < 0.05 versus control.
Depletion of Insig-2a Increases the Association of the SCAP·SREBP-1c Complex with COPII Proteins
The insulin-mediated regulation of transport of SREBP-1c is initiated by binding of the SCAP·SREBP-1c complex to COPII proteins (Sec23·Sec24 protein complex) that bud from the ER. To determine whether depletion of Insig-2a promotes binding of the SREBP-1c complex to COPII proteins, we knocked down the expression of Insig-2a using siRNA and analyzed the association of SREBP-1c with Sec23 in microsomes isolated from Insig-2a-depleted hepatocytes. The Insig-2a-depleted microsomes were then immunoprecipitated with Sec23 followed by immunoblotting with monospecific antibodies to detect the presence of SCAP and SREBP-1c. Representative blots shown in Fig. 6C illustrate that in Insig-2a-depleted microsomes the amount of SCAP and SREBP-1c associated with Sec23 was significantly increased. To examine whether depletion of Insig-2a specifically regulated the SREBP-1c·SCAP complex, we also probed these blots for the presence of SREBP-2. In contrast to its effect on SREBP-1c, Insig-2a depletion had no effect on the association of SREBP-2 and Sec23 (Fig. 6C, bottom panel). Quantification of the Western blots from three independent experiments shown in Fig. 6D demonstrated a 2–3-fold greater presence of SREBP-1c and SCAP in Insig-2a-depleted microsomal membranes. These data indicate that Insig-2a depletion leads to selective enrichment of SCAP and SREBP-1c proteins in COPII vesicles.
Insig-2a Overexpression Attenuates Insulin-enhanced SREBP-1c Processing
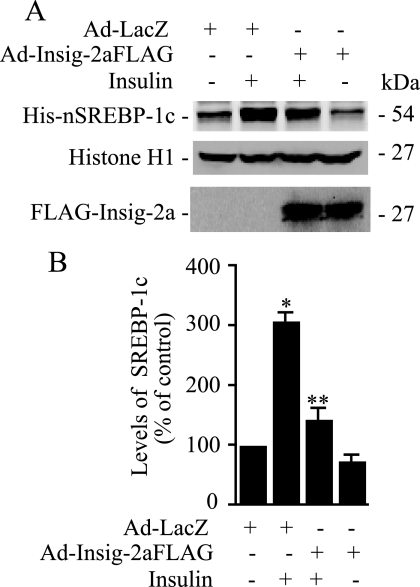
Another prediction of our postulated mechanism of the action of insulin on the posttranslational processing of SREBP-1c would be that if depletion of Insig-2a by insulin leads to increased migration of the newly synthesized SCAP·SREBP-1c complex from the ER to the Golgi, then overexpression of Insig-2a would lead to an opposite outcome. To test this scenario, we infected hepatocytes with AdHisSREBP-1cFLAG with or without co-infection of FLAG-tagged Insig-2a (AdCMV-Insig-2aFLAG) and then assessed the effect of insulin on the processing of the nascent SREBP-1c. As shown in Fig. 7, insulin enhanced the processing of SREBP-1c; this effect was markedly attenuated in hepatocytes co-expressing exogenous Insig-2a. These results are consistent with the interpretation that under conditions of high insulin, reduced Insig-2a mediates enhanced SREBP-1c processing.
FIGURE 7.
Exogenous expression of Insig-2a in primary cultures of rat hepatocytes inhibits SREBP-1c processing by insulin. A, hepatocytes co-infected with Ad-HisSREBP-1cFLAG and Ad-Insig2aFLAG or Ad-LacZ were incubated in medium containing insulin (100 nm) or without additions for 24 h. ALLN (25 μg/ml) was added during the final 6 h of incubation, and cells were harvested. Nuclear extracts were fractionated by SDS-PAGE, and immunoblots were developed using anti-His or anti-histone antibodies. Microsomal fraction was immunoblotted with anti-FLAG antibodies to assess the expression of Insig-2a (bottom panel). B, densitometric quantification of nSREBP-1c bands from three independent blots as shown in A; the data are expressed as a percentage of control optical density of the nSREBP-1c-specific band. *, p < 0.05 versus control; **, p < 0.05 versus insulin treatment.
DISCUSSION
SREBP-1c is the primary regulator of insulin-induced hepatic lipogenesis. Insulin induces expression of SREBP-1c mRNA and its encoded protein (11–14, 27), which must be transported to the Golgi for regulated proteolysis to release the transcriptionally active NH2 terminus (nSREBP). We reported previously that insulin selectively enhances ER-to-Golgi transport and controls proteolytic cleavage of nascent SREBP-1c by promoting phosphorylation-dependent binding of SCAP·SREBP-1c to COPII proteins (16). However, the SCAP·SREBP-1c complex must also dissociate from the ER retention protein, Insig, for ER-to-Golgi transport to occur. We now report that an additional effect of insulin is to promote the ER-to-Golgi transport of SCAP·SREBP-1c via depletion of Insig-2a. We have further demonstrated that insulin selectively enhances the rate of turnover of Insig-2a mRNA and leads to the depletion of Insig-2a protein. The resulting reduced availability of Insig-2a protein allows association of the SREBP-1c-SCAP complex with COPII proteins, thereby facilitating ER-to-Golgi transport and proteolytic processing of nascent SREBP-1c.
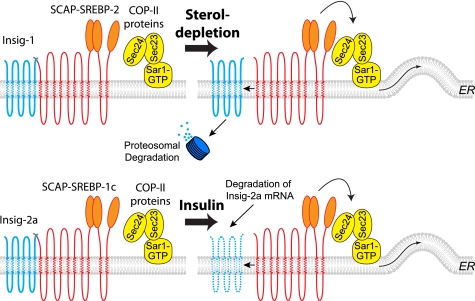
There are several possible mechanisms by which insulin could enhance the processing of SREBP-1c. Previous studies have elegantly defined the role of Insig-1 in regulating the posttranslational processing of the SREBP-2 isoform by cholesterol (21, 26). On the basis of the preliminary observation of rapid decline in levels of Insig-2a mRNA following insulin administration, we (23) and others (22) have suggested that reduced expression of Insig-2a protein is involved in insulin-mediated enhanced proteolytic processing of nascent full-length SREBP-1c. By assessing the contrasting effects of suppression and overexpression of Insig-2a on the COPII-dependent ER-to-Golgi transport of the SREBP-1c-SCAP complex, the current study provides strong mechanistic evidence in support of this hypothesis. Our data further show that Insig-2a protein associates selectively with the SREBP-1c-SCAP complex and that insulin preferentially represses Insig-2a, thereby forming a mechanistic basis for the selectivity of the effect of insulin on SREBP-1c processing. In contrast, we demonstrated that Insig-1 was specifically associated with the SCAP·SREBP-2 complex. ON the basis of a number of previous observations (28–30) and the data presented here, we present a mechanistic scheme that depicts dif-ferential posttranscriptional regula-tion of SREBP-1c and SREBP-2 pro-teins by insulin and cholesterol, respectively (Fig. 8).
FIGURE 8.
Proposed model of isoform-specific regulation of ER-to-Golgi export of SREBPs in response to sterol depletion and insulin. Under the conditions of sterol depletion, Insig-1 dissociates from the SCAP·SREBP-2 and SCAP·SREBP-1a complexes and is targeted for proteasomal degradation. Concomitantly, the SCAP·SREBP-2 and SCAP·SREBP-1a complexes are transported via COPII proteins from the ER to the Golgi, where proteolytic processing occurs (upper panel). SCAP·SREBP-1c associates preferentially with Insig-2a isoform in the liver and is retained in the ER membrane. Insulin treatment selectively degrades Insig-2a mRNA leading to depletion of Insig-2a protein (depicted as a discontinuous line in the lipid bilayer). This in turns leads to enhanced association of SCAP·SREBP-1c with COPII proteins and their export from the ER to the Golgi and enhanced generation of nSREBP-1c (lower panel). These models are based on results from earlier publications (16, 28, 29, 30) and the data contained in this paper.
The present study provides impor-tant insights into the mechanisms by which insulin reduces Insig-2a expression. We have shown that the promoter of rat Insig-2a is unresponsive to insulin in transfected hepatocytes. Although we cannot completely exclude transcriptional repression, the weight of our experimental evidence points to reduced mRNA stability as the major mechanism by which insulin reduces Insig-2a mRNA levels. Insulin is known to destabilize the insulin-responsive mRNAs encoding for phosphoenolpyruvate carboxykinase, glucose transporter 4, and glycogen synthase (31), but the pathways by which the stability of these mRNAs is controlled by insulin remain unknown. Further studies are needed to unravel the cis-acting element and trans-acting factors involved in the destabilization of Insig-2a mRNA in response to insulin. Identification of the precise 3′-UTR elements and proteins that bind to these sequences may reveal the molecular basis of insulin regulation not only for Insig-2a expression but also for other genes that are posttranscriptionally regulated by insulin.
We have previously shown that short term (1 h) insulin treatment rapidly enhances SREBP-1c processing via phosphorylation of the nascent SREBP-1c (16). The current study presents an additional and complementary mechanism of insulin-induced Insig-2a repression whereby longer term regulation of SREBP-1c processing is effected. Furthermore, we speculate that phosphorylation of SREBP-1c by insulin may also reduce the association of Insig-2a with the SCAP·SREBP-1c complex, as dissociation of Insig-2a is a requisite step for ER-to-Golgi transport of the nascent SREBP-1c prior to its proteolysis.
Acknowledgment
We thank Poonam Kumar for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1 DK075504 (to M. B. E., R. R., and C. R. Y.) and RO1 DK059368 (to E. A. P.) from the NIDDK. This work was also supported by a postdoctoral fellowship grant from the American Heart Association (to C. R. Y.), a grant-in-aid from the American Heart Association, Southeastern Affiliate (to M. B. E. and C. R. Y.), and the Office of Research and Development, Department of Veterans Affairs (to M. B. E.).
C. R. Yellaturu, unpublished observations.
- SREBP
- sterol regulatory element-binding protein
- nSREBP
- NH2-terminal fragment of SREBP
- SCAP
- SREBP cleavage-activating protein
- COPII
- coatomer protein complex II
- ER
- endoplasmic reticulum
- ALLN
- N-acetyl-leucyl-leucyl-norleucinal
- CMV
- cytomegalovirus
- PBS
- phosphate-buffered saline
- UTR
- untranslated region
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- siRNA
- small interfering RNA.
REFERENCES
- 1.Brown M. S., Goldstein J. L. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 11041–11048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Edwards P. A., Tabor D., Kast H. R., Venkateswaran A. (2000) Biochim. Biophys. Acta 1529, 103–113 [DOI] [PubMed] [Google Scholar]
- 3.Eberlé D., Hegarty B., Bossard P., Ferré P., Foufelle F. (2004) Biochimie 86, 839–848 [DOI] [PubMed] [Google Scholar]
- 4.McPherson R., Gauthier A. (2004) Biochem. Cell Biol. 82, 201–211 [DOI] [PubMed] [Google Scholar]
- 5.Espenshade P. J., Hughes A. L. (2007) Annu. Rev. Genet. 41, 401–427 [DOI] [PubMed] [Google Scholar]
- 6.Espenshade P. J. (2006) J. Cell Sci. 119, 973–976 [DOI] [PubMed] [Google Scholar]
- 7.Raghow R., Yellaturu C., Deng X., Park E. A., Elam M. B. (2008) Trends Endocrinol. Metab. 19, 65–73 [DOI] [PubMed] [Google Scholar]
- 8.Hua X., Wu J., Goldstein J. L., Brown M. S., Hobbs H. H. (1995) Genomics 25, 667–673 [DOI] [PubMed] [Google Scholar]
- 9.Shimomura I., Shimano H., Horton J. D., Goldstein J. L., Brown M. S. (1997) J. Clin. Invest. 99, 838–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shimomura I., Bashmakov Y., Horton J. D. (1999) J. Biol. Chem. 274, 30028–30032 [DOI] [PubMed] [Google Scholar]
- 11.Foretz M., Guichard C., Ferré P., Foufelle F. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 12737–12742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Azzout-Marniche D., Becard D., Guichard C., Foretz M., Ferre P., Foufelle F. (2000) Biochem. J. 350, 389–393 [PMC free article] [PubMed] [Google Scholar]
- 13.Shimomura I., Bashmakov Y., Ikemoto S., Horton J. D., Brown M. S., Goldstein J. L. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 13656–13661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cagen L. M., Deng X., Wilcox H. G., Park E. A., Raghow R., Elam M. B. (2005) Biochem. J. 385, 207–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng X., Yellaturu C., Cagen L., Wilcox H. G., Park E. A., Raghow R., Elam M. B. (2007) J. Biol. Chem. 282, 17517–17529 [DOI] [PubMed] [Google Scholar]
- 16.Yellaturu C. R., Deng X., Cagen L. M., Wilcox H. G., Mansbach C. M., 2nd, Siddiqi S. A., Park E. A., Raghow R., Elam M. B. (2009) J. Biol. Chem. 284, 7518–7532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thorngate F. E., Raghow R., Wilcox H. G., Werner C. S., Heimberg M., Elam M. B. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 5392–5396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dignam J. D., Lebovitz R. M., Roeder R. G. (1983) Nucleic Acids Res. 11, 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rowe T., Aridor M., McCaffery J. M., Plutner H., Nuoffer C., Balch W. E. (1996) J. Cell Biol. 135, 895–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang T., Espenshade P. J., Wright M. E., Yabe D., Gong Y., Aebersold R., Goldstein J. L., Brown M. S. (2002) Cell 110, 489–500 [DOI] [PubMed] [Google Scholar]
- 21.Sun L. P., Li L., Goldstein J. L., Brown M. S. (2005) J. Biol. Chem. 280, 26483–26490 [DOI] [PubMed] [Google Scholar]
- 22.Yabe D., Komuro R., Liang G., Goldstein J. L., Brown M. S. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 3155–3160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yellaturu C. R., Deng X., Cagen L. M., Wilcox H. G., Park E. A., Raghow R., Elam M. B. (2005) Biochem. Biophys. Res. Commun. 332, 174–180 [DOI] [PubMed] [Google Scholar]
- 24.von Roretz C., Gallouzi I. E. (2008) J. Cell Biol. 181, 189–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elbashir S. M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. (2001) Nature 411, 494–498 [DOI] [PubMed] [Google Scholar]
- 26.Goldstein J. L., DeBose-Boyd R. A., Brown M. S. (2006) Cell 124, 35–46 [DOI] [PubMed] [Google Scholar]
- 27.Foretz M., Pacot C., Dugail I., Lemarchand P., Guichard C., Le Lièpvre X., Berthelier-Lubrano C., Spiegelman B., Kim J. B., Ferré P., Foufelle F. (1999) Mol. Cell. Biol. 19, 3760–3768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Radhakrishnan A., Ikeda Y., Kwon H. J., Brown M. S., Goldstein J. L. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 6511–6518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun L. P., Seemann J., Goldstein J. L., Brown M. S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 6519–6526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Radhakrishnan A., Goldstein J. L., McDonald J. G., Brown M. S. (2008) Cell Metab. 8, 512–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Brien R. M., Granner D. K. (1996) Physiol. Rev. 76, 1109–1161 [DOI] [PubMed] [Google Scholar]