Abstract
The expression of acetylcholinesterase (AChE) in skeletal muscle is regulated by muscle activity; however, the underlying molecular mechanisms are incompletely understood. We show here that the expression of the synaptic collagen-tailed AChE form (ColQ-AChE) in quail muscle cultures can be regulated by muscle activity post-translationally. Inhibition of thiol oxidoreductase activity decreases expression of all active AChE forms. Likewise, primary quail myotubes transfected with protein disulfide isomerase (PDI) short hairpin RNAs showed a significant decrease of both the intracellular pool of all collagen-tailed AChE forms and cell surface AChE clusters. Conversely, overexpression of PDI, endoplasmic reticulum protein 72, or calnexin in muscle cells enhanced expression of all collagen-tailed AChE forms. Overexpression of PDI had the most dramatic effect with a 100% increase in the intracellular ColQ-AChE pool and cell surface enzyme activity. Moreover, the levels of PDI are regulated by muscle activity and correlate with the levels of ColQ-AChE and AChE tetramers. Finally, we demonstrate that PDI interacts directly with AChE intracellularly. These results show that collagen-tailed AChE form levels induced by muscle activity can be regulated by molecular chaperones and suggest that newly synthesized exportable proteins may compete for chaperone assistance during the folding process.
Introduction
Acetylcholinesterase (AChE)3 is an important component of the neuromuscular synapse where it rapidly terminates neurotransmission by hydrolyzing acetylcholine. Two separate genes encode the catalytic (AChE) and collagen tail (ColQ) subunits of the hetero-oligomeric collagen-tailed AChE forms expressed in skeletal muscle (for review, see Refs. 1–3). A single ColQ-AChE molecule consists of three catalytic AChE tetramers attached to a triple-helical collagen-like tail composed of three separate ColQ molecules. In some species and in some muscles, one (A4) or two (A8) tetramers of AChE attached to the triple helical ColQ subunits are observed. Each ColQ strand binds covalently to a tetramer of catalytic subunits, and each tetramer in turn consists of two dimers, with one dimer having its two C-terminal cysteine residues covalently linked to cysteine residues at the N terminus of ColQ. The other two catalytic subunits are also disulfide-linked to each other through the same residues (4, 5). Although many aspects of ColQ-AChE structure, function, and localization have been elucidated (for review, see Refs. 1–3), how this enzyme is regulated by muscle activity remains incompletely understood.
Physiologically the expression of the synaptic ColQ-AChE form is regulated by the incoming nerve and by depolarization of the cell membrane (6–11). Paradoxically, the direction of the regulation appears to be species-specific (for review, see Ref. 3). In rats, for example, muscle paralysis due to denervation or induced pharmacologically leads to dramatic decrease in AChE transcripts and their accumulation at junctional sarcoplasm (12–14). Avian systems behave differently; the addition of veratridine or scorpion venom (ScVn) to cultured avian muscle tissues, maintaining sodium channels in the open state and, thus, chronically depolarizing the membrane, results in increased levels of ColQ-AChE activity and decreased ColQ and AChE mRNAs (15–17). In parallel, there is a large increase in ColQ-AChE as well as an increase in cell surface enzyme activity and clusters of active enzyme molecules. In contrast, when tetrodotoxin (TTX) is added to maintain sodium channels in the closed state, the levels of ColQ-AChE activity decrease and of ColQ and AChE mRNAs increase (15–17). Similar observations have been made in vivo after denervation (18). Yet in the presence of TTX there is a significant decrease in ColQ-AChE expression as well as a decrease in cell surface enzyme activity and clusters of AChE (15, 16, 19). Under both experimental conditions, chronically or blocked membrane depolarization, the levels of AChE catalytic subunit mRNA correlate positively with total AChE activity but not with the levels of catalytically active ColQ-AChE (15, 16). These observations suggest that post-translational regulation occurs at the level of assembly. One possible mechanism to control the expression of ColQ-AChE could be by regulating the rate of assembly of catalytic and collagen tail subunits.
The globular dimeric and tetrameric AChE forms are assembled in the endoplasmic reticulum (ER) and post-translationally processed in the ER and Golgi apparatus (20–23); therefore, ER molecular chaperones are likely to be involved in regulating ColQ-AChE assembly. It has previously been reported that nicotinic acetylcholine receptor biogenesis is regulated in part by the chaperone calnexin (24–27). Presumably this occurs by increasing the efficiency of subunit folding. In addition, AChE catalytic subunits are stabilized by three intrachain disulfide bonds (28, 29); therefore, ER thiol oxidoreductases must be involved in their folding, providing additional candidates for regulating AChE biogenesis. Members of protein disulfide isomerase (PDI) family are the known ER-resident thiol oxidoreductases. They catalyze formation and isomerization of intra- and interchain disulfides bridges. Moreover, PDI is also the β-subunit of prolyl-4-hydroxylase (P4H), an α2/β2 hetero-tetramer localized in the ER that catalyzes co-translationally the hydroxylation of proline residues in Xaa-Pro-Gly sequences on single collagen polypeptide chains, which is an obligatory posttranslational modification before trimerization of the collagen strands (for review, see Ref. 30). Until trimerization occurs, PDI retains unassembled pro-collagen propeptides in the ER (31). Therefore, we hypothesized that PDI is required for ColQ and AChE assembly and maturation and may play a regulatory role controlling collagen-tailed AChE forms expression post-translationally. We now show that inhibition of thiol oxidoreductase activity in primary quail myotubes decreases expression of all intracellular AChE forms. Knockdown of PDI results in a decrease of all collagen-tailed AChE forms and cell surface AChE clusters. Conversely, overexpression of PDI, endoplasmic reticulum protein 72 (Erp72), or calnexin in muscle cells enhances expression of all collagen-tailed AChE forms including the synaptic ColQ-AChE, with PDI having the most dramatic effect with a 100% increase in intracellular ColQ-AChE and cell surface enzyme activity. In addition, we show that levels of PDI are regulated by muscle activity and correlate with the levels of ColQ-AChE. Finally, we demonstrate that PDI interacts directly with AChE during AChE folding. Together our results show that catalytically active ColQ-AChE expression can be regulated by molecular chaperones posttranslationally at the level of individual subunit folding and/or at the level of assembly of catalytic and collagen tail subunits. More importantly, our studies suggest that there may be more competition for ER chaperone-assisted folding than previously thought.
EXPERIMENTAL PROCEDURES
Tissue Culture of Embryonic Muscle
Primary myoblasts were obtained from the pectoral muscles of 10-day-old quail embryos and plated at 5 × 104 cells/ml in Eagle's minimum essential medium (EMEM 210; Invitrogen) supplemented with 2% chicken embryo extract, 10% horse serum (GemCell), and 0.1% gentamicin (Invitrogen). The quail muscle cultures (QMCs) were fed with fresh medium on days three and five after plating, and on day three the medium was supplemented with 10−6 m cytosine-arabinoside (Sigma) to inhibit fibroblast proliferation. Cultures were maintained in a 39 °C in a water-saturated incubator with 5% CO2.
Drug Treatments
To study the newly synthesized AChE, QMCs were treated with 10−4 m diisopropyl fluorophosphate (DFP; Sigma) in phosphate-buffered saline (PBS, pH 7.4) for 10 min at room temperature followed by 3 washes in PBS alone to remove unreacted DFP. The cultures were then returned to the incubator to recover in complete medium with or without additional treatments as described. When used, 6-day-old QMCs were pretreated with 5.4 μm α-amanitin (Fluka) for 30 min and/or 10−4 m DFP for 10 min. The cells were then treated with 20 μm ScVn (Sigma) or 5 μm TTX (Sigma) with or without 5.4 μm α-amanitin for 12 h. In other studies, 6-day-old QMCs were treated with 10−4 m DFP for 10 min and allowed to recover for 24 h in the presence of 1 mm bacitracin, a thiol oxidoreductase activity inhibitor, or incubated for 10 min with 5 mm dithiothreitol (DTT) added directly to the culture medium, washed, and allowed to recover in complete medium for up to 30 min before extraction and analysis.
Analysis of AChE Oligomeric Forms and Enzyme Activity Assay
The oligomeric forms of AChE were extracted with borate extraction buffer (20 mm sodium borate, pH 9.0, 0.5% Triton X-100, 5 mm EDTA, 1 m NaCl, 0.5% bovine serum albumin) and the protease inhibitors leupeptin (1 μg/ml) and pepstatin (2 μg/ml). After centrifugation in a microcentrifuge, 150-μl aliquots of crude cell extract pooled from triplicate samples were analyzed by velocity sedimentation in a Beckman SW41ti rotor at 36 × 103 rpm for 16 h on 5–20% sucrose gradients in extraction buffer without bovine serum albumin. To separate G4 and A4, Triton X-100 was replaced by 0.5% Brij-98, and the gradients were run in a Beckman SW41ti rotor at 40 × 103 rpm for 22 h. The fractions were collected and assayed for AChE activity using the Ellman assay (32).
Cloning of Chicken and Quail PDIs and Chicken ERp72 cDNAs
Chicken PDI (chPDI) and Erp72 (chErp72) cDNAs were cloned into pTarget (Promega) by reverse transcription-PCR from total RNA isolated from primary chick embryo fibroblast cultures using primers based on the respective sequences identified as GI:11809988 and GI:57530767, available from the NCBI data base. The cloned cDNAs were confirmed by DNA sequencing and included the endogenous leader sequences at the 5′ end and the ER retention signal at the 3′ end to ensure ER localization of the encoded proteins. A 900-bp quail PDI (qPDI) cDNA fragment was cloned into pcDNA 3.1 (Invitrogen) by reverse transcription-PCR from QMC total RNA using ACCTGCTCGTGGAGTTC as forward and GGCTGCTTGTCCCAGTC as reverse primers. The primers matched conserved regions among human, cow, mouse, rat, and chicken PDI cDNAs, all available from the NCBI data base.
Designing and Cloning of qPDI shRNAs
Based on the sequence of the cloned qPDI cDNA fragment, we designed and cloned in pSilencer 3.0-H1 (Ambion) three different qPDI shRNAs: qPDI shRNA1, CAGCGACCATAGTGATAAT; qPDI shRNA2, AAGGACGTAACAACTTTGA; qPDI shRNA3, CCAGTGAAGTGGTTGTGAT. All experiments carried out here were done using qPDI shRNA2.
Cell Transfections
QMCs were transfected with plasmids encoding quail ColQ (pTarget-qColQ), chPDI (pTarget-chPDI), chErp72 (pTarget-Erp72), canine calnexin (pBK-calnexin) (a generous gift from Dr. Joav Prives, Stony Brook, NY), qPDI shRNA (pSilencer H1 qPDI shRNA), or qColQ (pSilencer H1 qColQ shRNA) using Exgen-500 (Fermentas) following the manufacturer's suggested protocol. In parallel, control groups were transfected with empty pTarget or enhanced green fluorescent protein shRNA (pSilencer H1 EGFPshRNA). For immunofluorescence studies, a plasmid encoding red fluorescent protein pHcRed1-N1 (Clontech) was used to identify transfected cells.
Cell Surface AChE Activity Assay
Primary QMCs were incubated on a shaker platform at 4 °C for 1 h in 1 ml of PBS, pH 7.6, per 35-mm culture dish containing 1 mm acetylcholine and 105 cpm of [3H]acetylcholine iodide (PerkinElmer Life Sciences). At 60 min of incubation, 100 μl of aliquots were removed and counted in a liquid scintillation counter using the two-phase system as previously described (33). Blanks for background subtraction consisted of collagen-coated tissue culture dishes without cells.
DTT Treatment
Six-day-old QMCs and HEK 293 cells stably expressing human AChE catalytic subunit (HEK 293-hAChE) (hAChE cDNA was the generous gift of Dr. Andrew Engel, Mayo Medical School) growing on 35-mm dishes were washed with Hanks' balanced salt solution and incubated with complete medium with or without 5 mm DTT for 20 min at 37 °C in the case of HEK 293-hAChE or 39 °C for the QMCs. After treatment, the DTT was removed by washing twice with Hanks' balanced salt solution, and the cells were allowed to recover for 0, 5, 10, or 15 min in regular medium at the appropriate temperature. Cells were then extracted in RIPA buffer as indicated below, and the total cell extracts were used for coimmunoprecipitation studies.
Coimmunoprecipitations
Six-day-old QMCs or HEK 293-hAChE cells previously treated with DTT as described above were extracted on ice for 20 min in RIPA buffer (0.2% SDS, 1% Triton X-100, 150 mm NaCl, 10 mm Tris-HCl, pH 7.6) containing a protease inhibitor mixture (Pierce) and 20 mm N-ethylmaleimide (Sigma) to preserve PDI-substrate complexes. The lysates were centrifuged for 20 min at maximum speed in a microcentrifuge to remove insoluble material. The RIPA-soluble fractions were incubated with either 4 μl of rabbit anti-PDI (Stressgen), 10 μg of mouse anti-avian AChE 1A2 (21), 10 μg of mouse anti-hAChE AE2 (34), or mouse IgG or rabbit IgG alone for 2 h at 4 °C. Then 10 μl of a 50% slurry of protein A- or protein G-Sepharose beads were added to the mixture, and incubation was continued overnight. The beads were subsequently washed three times with RIPA buffer and one time with PBS containing protease inhibitor mixture followed by resuspension in 2× reducing SDS sample buffer (25 mm Tris base, pH 6.5, 6% SDS, 10% glycerol, 50 mm DTT, bromphenol blue). The samples were then used to detect coimmunoprecipitated AChE or PDI by Western blot analysis. All incubations were done at 4 °C on a rocking platform.
Western Blots
Primary QMCs treated with ScVn and TTX as described above were extracted in RIPA buffer containing protease inhibitor mixture. After centrifugation, samples containing 50 μg of total protein were run in 10% reducing SDS-PAGE. The proteins were electrophoretically transferred onto a nitrocellulose membrane (Whatman) and blocked with 1% powdered skim milk (Carnation) for 1 h, incubated with mouse anti-PDI monoclonal antibody 1D3 (Abcam) (1:1000), washed 4 times with 0.025% Nonidet P-40 in PBS, incubated with goat anti-mouse peroxidase antibody (Jackson Immunochemicals), and developed with West Pico chemiluminescent substrate (Pierce). All immunoprecipitated samples were treated, run, and analyzed under identical conditions. The PDI was detected using anti-PDI antibody (1:5000) and minimal cross-reacting goat anti-rabbit peroxidase (Jackson Immunochemicals) as second antibody. To detect quail AChE, mouse monoclonal antibody 1A2 (1.6 μg/ml) followed by goat anti-mouse peroxidase antibody was used. To detect hAChE, we biotinylated an equimolar mixture of AE2 and AE3 mouse monoclonal antibodies (34) using EZ-Link NHS-PEO Solid Phase Biotinylation kit (Pierce). These biotinylated antibodies were used at a concentration of 0.6 μg/ml followed by incubation with streptavidin-peroxidase 1:20,000 (Pierce). In all cases the positive bands were developed with West Pico chemiluminescent substrate. The integrated density of Western blot bands was determined using ImageJ software from the NIH.
Immunofluorescence
Primary QMCs were blocked for 20 min with 1% bovine serum albumin and then sequentially incubated with mouse anti-avian AChE antibody 1A2 for 1 h followed by 20 μg/ml Alexa 488-donkey anti-mouse IgG (Molecular Probes) with 0.1 μg/ml Hoechst dye to stain the nuclei. The cells were fixed with 4% paraformaldehyde in PBS, pH 7, for 20 min. Between incubations the cells were washed 4 times with Hanks' balanced salt solution. All images were acquired with identical exposure parameters using a Leica DMR-A or DMI 6000 fluorescence microscopes operated by 3i Slidebook 4.0 software.
RESULTS
Synaptic ColQ-AChE Is Regulated by Muscle Activity Post-transcriptionally
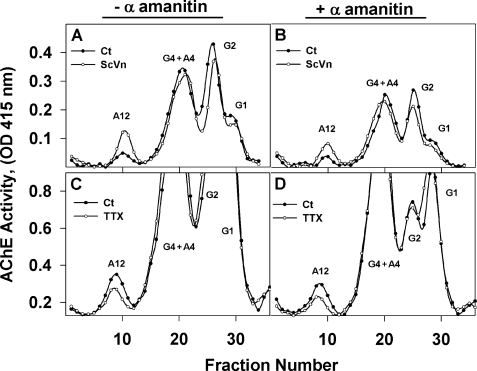
We previously studied the regulation of the structural ColQ subunit at the transcriptional level (17). During QMC development and as a function of muscle activity, we observed a lack of correlation between the levels of total AChE activity on the one hand and ColQ subunit mRNAs as well as the levels of ColQ-AChE activity expressed in QMCs on the other. This was most apparent when muscle activity was up or down-regulated using sodium channel agonists or antagonists (17). To determine whether changes in muscle activity could regulate ColQ-AChE expression at the post-transcriptional level, we treated QMCs with ScVn or TTX in the presence or absence of 4.9 μg/ml α-amanitin to inhibit transcription of mRNA precursors by Pol II. In addition, the QMCs were treated with DFP, a membrane-permeable organophosphate that irreversibly blocks all cell-associated AChE activity. The cells were then allowed to synthesize new AChE molecules for 12 h before extraction and analysis of the expressed AChE forms by velocity sedimentation. As published previously by us and by others (for reviews, see Refs. 1–3), maintaining sodium channels in the open state with ScVn up-regulated expression of synaptic ColQ-AChE (Fig. 1A), whereas blocking membrane depolarization with TTX resulted in a decrease of ColQ-AChE (Fig. 1C). In α-amanitin-treated QMC, total AChE activity was decreased. Nevertheless, the ColQ-AChE form increased (Fig. 1B) or decreased (Fig. 1D) when the cells were treated with ScVn or TTX, respectively. These observations were consistent among three independent experiments in which each experimental group consisted of triplicate cultures. Thus, inhibiting transcription did not affect the manner in which muscle activity regulated ColQ-AChE assembly. As usual, a small percentage of the newly synthesized AChE is assembled into synaptic ColQ-AChE form. The relative proportion of AChE forms in QMCs from Fig. 1, A and B, is different from that in Fig. 1, C and D, because they belong to two separate sets of primary cultures. In general the relative amount of the ColQ-AChE form varies from one set of cultures to the other. There are many factors that affect it including the time of the year, source of quail eggs, age of embryos, and age of cultures as well as the lots of horse serum and chicken embryo extract used to supplement the medium.
FIGURE 1.
Synaptic ColQ-AChE expression is post-transcriptionally regulated. QMCs were pretreated (panels B and D) or not (panels A and C) with 5.4 μm α-amanitin for 30 min followed by 10−4 m DFP in Hanks' balanced salt solution for 10 min and 3 washes. The cells were then allowed to synthesize new AChE in complete medium with (open circles) or without (solid circles) 20 μm ScVn (panels A and B) or 5 μm TTX (panels C and D) in the presence (B and D) or absence (A and C) of 5.4 μm α-amanitin for 12 h. Less than 5% of the total newly synthesized AChE is assembled into collagen-tailed A12 during this short time. The AChE forms were fractionated by velocity sedimentation in 5–20% sucrose gradients. G1, G2, G4, and A4 refer to AChE monomers, dimers, tetramers, and collagen-tailed tetramers, respectively. A12 refers to the ColQ-AChE form. The results in A and B are from a different set of cultures than the results in C and D. Neither the up-regulation of A12 in the presence of ScVn nor its down-regulation by TTX was affected by inhibiting transcription with α-amanitin.
Protein Disulfide Isomerase Is Required for Catalytically Active Collagen-tailed AChE Expression
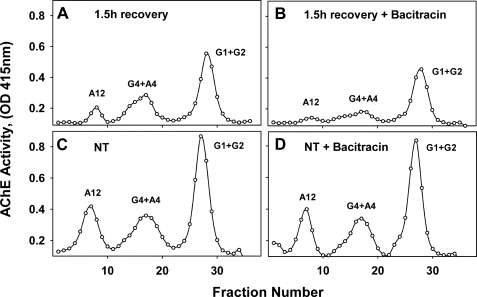
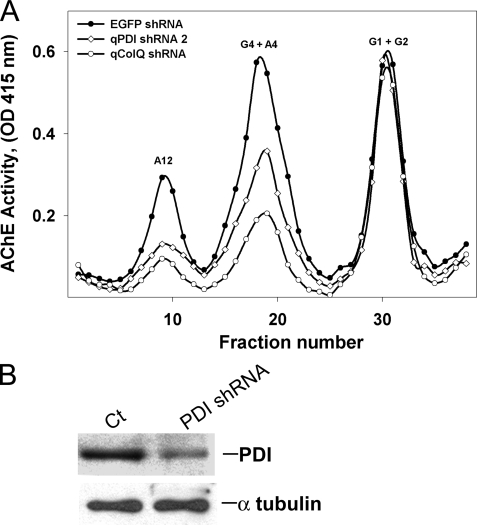
ColQ-AChE is a secreted molecule assembled from 2 different types of subunits, 12 globular catalytic subunits arranged as tetramers and a 3-stranded collagen-like molecule that is covalently attached via disulfide bonds. Based on these structural features of collagen-tailed AChE and the experimental evidence that muscle activity regulates ColQ-AChE expression post-transcriptionally, we postulated that among the multiple ER molecular chaperones, PDI would be one likely candidate to be involved in catalyzing ColQ and AChE assembly. In addition, it could potentially play a regulatory role should the availability of chaperones become limiting. We, therefore, asked whether PDI was required for the expression of collagen-tailed AChE forms. First, we determined the effect of bacitracin, a thiol oxidoreductase activity inhibitor (35, 36), on catalytically active AChE expression in QMCs. We compared both the intracellular steady state and newly synthesized AChE levels in the presence or absence of bacitracin. Inhibition of thiol oxidoreductase activity while the QMCs were recovering from DFP treatment resulted in a decrease of all newly synthesized catalytically active AChE forms, including the ColQ-AChE form (Fig. 2). The percent decrease for the G1+G2, G4+A4, and ColQ-AChE (A12) pool was 18, 36, and 30%, respectively. These results indicate that thiol oxidoreductase activity is required for folding of catalytically active AChE molecules. However, all PDI family members bear thiol oxidoreductase activity, and skeletal muscle fibers appear to express at least three variants (37–39). To determine whether PDI was specifically required for ColQ-AChE expression, we transfected QMCs with a plasmid encoding a qPDI-shRNA to selectively knock down PDI expression without affecting other thiol oxidoreductases. All intracellular collagen-tailed AChE forms were down-regulated when PDI was knocked down by the qPDI shRNA. Nevertheless, the pool of monomers and dimers remained unaffected (Fig. 3A). This experiment is representative of three independent experiments. In this case the percent decrease for the G4+A4 and ColQ-AChE pool was 37 and 55%, respectively.
FIGURE 2.
Thiol oxidoreductase activity is required for expression of all catalytically active AChE molecules. 6-Day-old QMCs were DFP-treated (A and B) or not (C and D) to inhibit all AChE molecules and allowed to recover for 1.5 h in the absence (A and C) or presence (B and D) of 1 mm bacitracin. The AChE forms were fractionated by velocity sedimentation in 5–20% sucrose gradients. G1, G2, and G4 refer to AChE monomers, dimers, and tetramers, and A4, A8, and A12 refer to the various collagen-tailed forms consisting of one, two, or three tetramers attached to the ColQ subunit, respectively. A12 refers to the ColQ-AChE form. When cell-associated thiol oxidoreductase activity was blocked, the intracellular levels of all newly synthesized catalytically active AChE forms decreased, including the ColQ-AChE. NT, not treated with DFP.
FIGURE 3.
PDI is required for catalytically active collagen-tailed AChE form expression. A, primary QMCs were transfected with pSilencer qPDI shRNA (open diamonds) or qColQ shRNA (open circles). The AChE forms were fractionated by velocity sedimentation in 5–20% sucrose gradients. G1, G2, and G4 refer to AChE monomers, dimers, and tetramers, and A4, A8, and A12 refer to the various collagen-tailed forms consisting of one, two, or three tetramers attached to the ColQ subunit, respectively. EGFP shRNA (solid circles) was used as negative control. Expression of the synaptic ColQ-AChE and the collagen-tailed A4 forms was decreased in muscle cells when either PDI or ColQ was knocked down specifically by the shRNAs. Panel B, relative PDI content was determined by a Western blot of triplicate QMC crude extracts from cells transfected with either EGFP shRNA (control) or PDI shRNA. Overexpression of PDI shRNA knocks down PDI in QMCs.
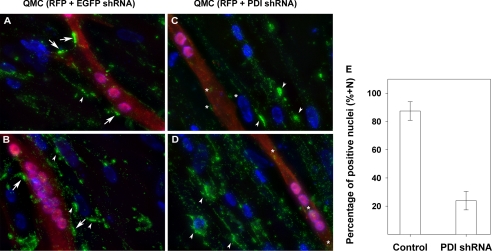
Because the final destination of the synaptic ColQ-AChE molecules is in association with the extracellular matrix on the muscle cell surface, we determined whether PDI knockdown had an effect on the number of AChE cell surface clusters. For this purpose QMCs were transfected with plasmids encoding red fluorescent protein and qPDI shRNA, the red fluorescent protein being used to identify transfected cells. After the cultures, matured cell surface AChE clusters were immunostained using anti-chAChE monoclonal antibody 1A2 as described under “Experimental Procedures” (Fig. 4, A–D). We then counted the total number of nuclei in transfected myotubes and those displaying AChE clusters on the proximal extracellular matrix in triplicate cultures. Using these values we calculated the percentage of nuclei within transfected myotubes exhibiting AChE clusters on the adjacent cell surface and labeled it percentage of positive nuclei. Myotubes transfected with qPDI shRNA showed a dramatic decrease in cell surface AChE clusters (red cells in Fig. 4, C and D). The percentage of positive nuclei was 87 ± 7% for the control group and 24 ± 7% for myotubes transfected with qPDI shRNA (Fig. 4E). Together these results suggest that PDI is required not only for the expression of catalytically active collagen-tailed AChE molecules including synaptic ColQ-AChE but also for the processing that results in a mature enzyme able to localize and cluster on the surface of skeletal muscle cells.
FIGURE 4.
PDI is required for AChE localization on the cell surface. QMCs were co-transfected with red fluorescent protein (RFP) and either an EGFP shRNA as control (panels A and B) or a qPDI shRNA (panels C and D) to knock down PDI. Each group consisted of triplicate cultures, and the images were acquired from a separate culture under the same exposures. The white arrows point at cell surface AChE clusters on transfected cells in control group (panels A and B). The white asterisks highlight the remaining cell surface AChE on qPDI shRNA-transfected cells in the experimental groups (panels C and D). The white arrowheads point at cell surface AChE clusters on non-transfected cells in both control (panels A and B) and experimental group (panels C and D). Panel E, a bar graph shows the percentage of nuclei in multinucleated myotubes displaying AChE clusters on their adjacent cell surface in control and qPDI shRNA-transfected QMCs. All images were acquired using a 40× objective.
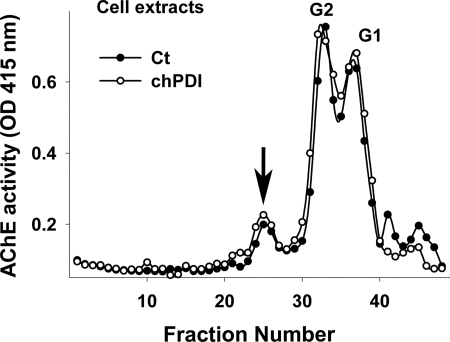
The Levels of Endoplasmic Reticulum Molecular Chaperones PDI, ERp72, and Calnexin Limit Collagen-tailed AChE Expression
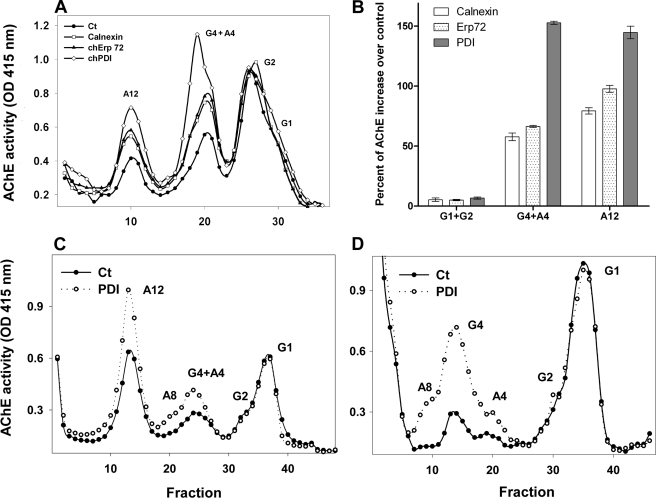
Because the levels of all catalytically active collagen-tailed AChE molecules decrease when PDI is either inhibited pharmacologically or knocked down specifically by the shRNA, we asked whether overexpression of this ER molecular chaperone could up-regulate the expression of these heteromeric AChE forms. The closest available genomic information for quail is the chicken genome; therefore, we cloned full-length chPDI and chErp72 cDNAs from tissue-cultured chicken embryo fibroblast RNA by reverse transcription-PCR and subcloned them into the pcDNA 3.1 vector to test their ability to increase AChE expression in QMCs. The cloned inserts were initially screened by restriction enzyme digestion and confirmed by fully sequencing them in both directions. We cloned chErp72 because it is a member of the PDI family but does not have a known role in collagen synthesis as does PDI. We also tested canine calnexin because it is an ER molecular chaperone unrelated to the PDI family that is known to enhance acetylcholine receptor maturation by promoting subunit folding and assembly (23–26). Overexpression of either of these molecular chaperones consistently increased expression of the tetrameric (G4) and collagen-tailed AChE forms (A12-A4) in three independent experiments (Fig. 5A). Up-regulation of chicken PDI showed the largest increase of more than 100% in the A4-G4 fraction and the ColQ-AChE form (Fig. 5, A and B). When G4 and A4 were separated after PDI overexpression, we found that as the ratio of these two forms changes, they both increase, but G4 increases more than A4 (Fig. 5, C and D). In some experiments chPDI also up-regulated the pool of free AChE monomers and dimers (G1 and G2) although this observation was not consistent across all experiments. Nevertheless, we thought it was important to determine whether PDI could limit the expression of catalytic subunits independently of the presence of ColQ. To this end we overexpressed chPDI in an HEK stable cell line expressing chAChE. PDI overexpression did not affect catalytically active AChE levels in the absence of ColQ (Fig. 6). These results all together suggest that the availability of molecular chaperones PDI, ERp72, and calnexin limits the expression levels of the catalytically active synaptic ColQ-AChE molecules and other AChE collagen-tailed forms in skeletal muscle cells, PDI being the most critical of the three chaperones tested.
FIGURE 5.
Overexpression of calnexin, Erp72, and PDI increase catalytically active collagen-tailed AChE form expression. A, primary QMCs were transfected with expression vectors encoding calnexin (open squares), chErp72 (closed triangles), and chPDI (open diamonds), the AChE forms were separated by velocity sedimentation, and the enzymatic activity was measured. The control consisted of transfection with an empty vector alone (solid circles). B, averaged percent increase over control of the different AChE forms after calnexin, Erp72, and PDI overexpression in QMC from three independent experiments. The error bars are the S.E. C and D, primary QMCs were transfected with expression vector encoding chPDI (open circles), and the AChE forms were separated by velocity sedimentation in the presence of Triton X-100 (C) or Brij-98 (D) to resolve the G4 and A4 forms. G1, G2, G4, and A4 refer to AChE monomers, dimers, tetramers, and collagen-tailed tetramers, respectively. A12 refers to the ColQ-AChE form.
FIGURE 6.
Overexpression of PDI does not affect the levels of catalytically active AChE monomers and dimers. HEK 293 stable cells expressing chAChE were transfected with expression vector encoding chPDI (open circles). The control consisted of transfection with an empty vector alone (solid circles). The different AChE forms expressed in total cell extracts were fractionated by velocity sedimentation, and their enzymatic activity was measured. G1 and G2 refer to AChE monomers and dimers. The arrow indicates most likely AChE tetramers.
Protein Disulfide Isomerase and qColQ Interact Synergistically to Increase Collagen-tailed AChE Expression
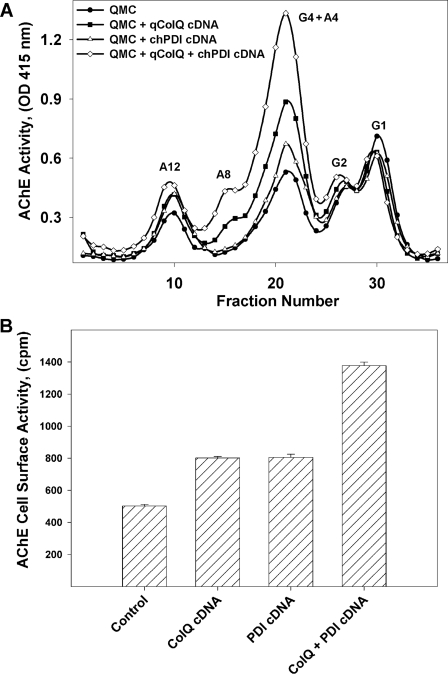
Based on the evidence that PDI was required for ColQ-AChE expression, we reasoned that PDI could mediate, but not necessarily limit, at least three processes during AChE biogenesis; that is, the folding of the catalytic AChE subunit, the processing of single collagen strands into triple helical structures, or their subsequent assembly with the catalytic AChE subunits to form the ColQ-AChE molecules. Knowing that overexpression of additional AChE collagen tail subunits in QMCs was sufficient to increase ColQ-AChE expression as well as a fraction of collagen tailed tetramers (A4) (17), we tested whether this ability of the collagen subunit could be limited by the availability of PDI. To test this, we overexpressed qColQ and chPDI either alone or together in QMCs. Intracellular A4, A8, and A12 forms were increased by up-regulation of either qColQ or chPDI (Fig. 7A). Interestingly, when the two proteins were coexpressed, expression of all collagen-tailed AChE forms, including the ColQ-AChE, was up-regulated more dramatically (Fig. 7A). In parallel cultures we measured cell surface AChE activity, which is the most stringent test for ColQ-AChE expression, because we are quantifying the active ColQ-AChE in its final physiological location. Cell surface AChE activity was increased ∼50% when either qColQ or PDI was expressed, but when they were coexpressed, cell surface AChE activity increased 175% (Fig. 7B). These results support our previous observation that PDI levels limit the rate of ColQ-AChE assembly. More importantly, they show that ColQ levels limit the extent in which PDI increases ColQ-AChE expression, thus suggesting that the PDI limiting role can take place at the level of assembly of catalytic and collagen tail subunits by mediating the formation of proper interstrand disulfide bonds or at the level of ColQ co- and post-translational modifications.
FIGURE 7.
PDI and ColQ act synergistically to increase catalytically active collagen-tailed AChE form expression. Primary QMCs were transfected with qColQ (solid squares), chPDI (open triangles), or both together (open diamonds). The control consisted of transfection with an empty vector alone (solid circles). A, the AChE forms in total cell extracts from the different treatment groups were fractionated by velocity sedimentation, and their enzymatic activity was measured. G1, G2, G4, and A4 refer to AChE monomers, dimers, tetramers, and collagen-tailed tetramers, respectively. A12 refers to the ColQ-AChE form. B, cell surface AChE activity was measured in a parallel set of cultures. Overexpression of either or both PDI and ColQ results in a significant increase in ColQ-AChE expression and deposition of AChE on the cell surface. Transfection with ColQ- or PDI-expressing plasmids increased cell surface AChE by ∼50%, whereas transfection with both together resulted in more than twice that amount (175%).
Muscle Activity Regulates PDI, Calnexin, and Calreticulin Levels
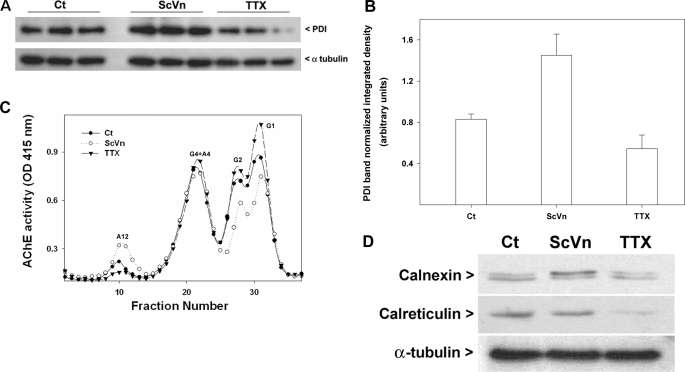
Because the expression of synaptic ColQ-AChE is increased by depolarization of the muscle cell membrane and decreased when membrane depolarization is blocked (40–43), we reasoned that this could at least in part be due to an effect of muscle activity on PDI expression. To determine whether PDI levels were affected by muscle activity, QMCs were treated for 48 h with ScVn or TTX. The distribution of the different AChE forms was then analyzed by velocity sedimentation. Indeed, ScVn increased ColQ-AChE expression, whereas TTX decreased it (Fig. 8C). The expression of PDI was then analyzed by Western blot. Primary QMCs treated with ScVn showed increased PDI levels, whereas those cells treated with TTX exhibited a decrease in PDI levels relative to control cells (Fig. 8, A and B). Under the same conditions we determined the levels of calnexin and calreticulin. We found that calnexin appears as a doublet, possibly due to phosphorylation of its cytosolic domain (44, 45), and the higher molecular weight band increased when QMCs were treated with ScVn, whereas no change is detected in the presence of TTX (Fig. 8D). Calreticulin levels decreased dramatically when QMCs were treated with TTX and did not change in the presence of ScVn (Fig. 8D). These observations were consistent across three independent experiments. These studies demonstrate that the levels of ER chaperones PDI calnexin and calreticulin are regulated by muscle activity and, unlike the levels of ColQ and AChE mRNA and protein (17), correlate positively with the levels of catalytically active ColQ-AChE induced.
FIGURE 8.
PDI levels correlate positively with the levels of synaptic ColQ-AChE under different conditions of muscle activity. Sets of QMCs were treated for 48 h with ScVn or TTX. A, relative PDI content was determined by a Western blot of triplicate samples analyzed by SDS-PAGE and immunoblotted with anti-PDI and anti-α-tubulin antibodies. B, the integrated density of the PDI bands in A was determined and normalized versus the integrated density of the α-tubulin bands. C, the different AChE forms contained in the same volume of total cell extracts were fractionated by velocity sedimentation, and their enzymatic activity was assayed; solid circles, open circles, and solid triangles refer to control and ScVn- and TTX-treated groups, respectively. G1, G2, G4, A4, and A12 refer to AChE monomers, dimers, tetramers, collagen-tailed tetramers, and the ColQ-AChE form, respectively. D, relative calnexin and calreticulin content was determined by Western blot of pooled triplicate samples analyzed by SDS-PAGE and immunoblotted with anti-calnexin, anti-calreticulin, and anti-α-tubulin antibodies.
Protein Disulfide Isomerase Forms an Intermediate Complex with AChE Catalytic Subunit
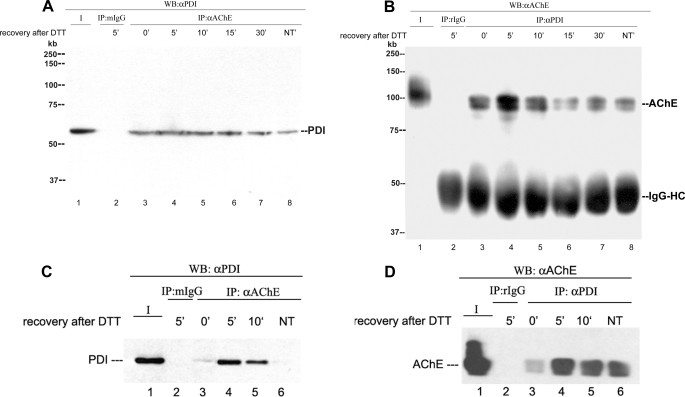
The results presented above suggest that PDI directly or indirectly facilitates ColQ-AChE biogenesis. To determine whether PDI and AChE interact directly, we took advantage of the formation of transient covalently bound PDI-substrate complexes during PDI-assisted disulfide bond formation (46, 47) to determine whether PDI and AChE could be coimmunoprecipitated. Using monoclonal antibody 1A2, which recognizes all AChE forms (21), we were able to coimmunoprecipitate qPDI from QMC total cell extracts (Fig. 9A). When QMCs were treated with 5 mm DTT and allowed to recover for up to 30 min in complete medium, the amount of coimmunoprecipitated PDI increased, reaching a maximum at 5 min after DTT treatment. From the same cells under the same conditions, we were able to coimmunoprecipitate qAChE using anti-PDI antibody (Fig. 9B). To determine whether these interactions could occur independently of ColQ, we coimmunoprecipitated endogenous PDI using anti-hAChE monoclonal antibodies AE2 and AE3 (34) from a HEK 293 cell line stably expressing human AChE previously created in our laboratory (Fig. 9C) and hAChE using anti-PDI antibody (Fig. 9D). In both primary cultures and cell lines, PDI and AChE interact directly during the AChE folding process.
FIGURE 9.
PDI and AChE catalytic subunits form a complex during AChE folding. Lane 1 in each blot is a positive control consisting of an aliquot of the protein of interest. Lane 2 in each blot is the total cell extract from cells treated with DTT that were allowed to recover for 5 min and immunoprecipitated with the corresponding nonspecific IgG. Lanes 3–7 (A and B) and lanes 3–5 (C and D) are total cell extracts from cells treated with DTT that were allowed to recover from the treatment for 0, 5, 10, 15, or 30 min. Lanes 8 (A and B) and 6 (C and D) are total cell extracts from control cells not treated with DTT. A, QMC cell extracts were blotted directly with anti-PDI (lane 1) or after immunoprecipitation (IP) with anti-AChE (lane 3–8). WB, Western blot. B, parallel cell extracts were blotted directly with anti-AChE (lane 1) or after immunoprecipitation with anti-PDI (lane 3–8). C, HEK 293-hAChE cell extracts were blotted directly with anti-PDI (lane 1) or after immunoprecipitation with anti-AChE (lane 3–6). PDI levels are low in cells at 0 min after DTT treatment and in cells not DTT-treated. D, parallel cell extracts were blotted directly with anti-AChE (lane 1) or after immunoprecipitation with anti-PDI (lane 3–6). The higher apparent molecular size of the control protein in panel B, lane 1, is due to “smiling” of the gel. NT, not treated with DFP.
DISCUSSION
In the present studies we show that inhibition of skeletal muscle RNA Pol II does not alter the regulation of synaptic ColQ-AChE by muscle activity for at least the first 12 h under conditions of chronic membrane depolarization or sodium channel block (Fig. 1). Longer time periods could not be studied due to the toxicity caused by inhibiting transcription. In these studies we used 4.9 μg/ml α-amanitin to completely inhibit RNA Pol II. At this concentration, RNA Pol I and III remain active (48, 49). It is unlikely that the AChE activity detected corresponds to the remaining AChE mRNA transcription by RNA Pol I or III because only RNA Pol II transcribes mRNA precursors (50, 51). There is a dramatic effect in total AChE activity in the presence of α-amanitin (Fig. 1, B and D) due to AChE mRNA decay after Pol II inhibition. These results together with the studies of the regulation of AChE and ColQ mRNA and protein levels by muscle activity (17) are experimental evidence unambiguously suggesting that the synaptic ColQ-AChE form is regulated by muscle activity at least in part through post-translational controls. The sequence of post-translational events during ColQ-AChE biogenesis is not completely understood. The catalytic and ColQ subunit assembly process involves the formation of inter- and intrachain disulfide bonds (4, 5, 52), a reaction catalyzed by members of the PDI family. Indeed, we show that PDI interacts directly with the AChE catalytic subunits (Fig. 9), suggesting that PDI can facilitate the formation of intrastrand disulfide bonds during catalytic subunit folding; however, changes in PDI levels do not affect the levels of catalytically active AChE monomers and dimers as shown in (Figs. 3 and 5–7), suggesting that it is not limiting during folding of the AChE catalytic subunit. Nevertheless, PDI levels do limit the expression of all catalytically active collagen-tailed AChE forms including synaptic ColQ-AChE and the AChE tetramers G4, as shown by the overexpression and down-regulation studies (Figs. 3, 5, and 7). It is not known whether G4, A4, and A8 are precursors or degradation products during A12 biogenesis in muscle cells. Therefore, we cannot conclude that G4, A4, or A8 increases after PDI or ColQ overexpression contribute positively toward synaptic ColQ-AChE synthesis.
In avian muscle cells ∼70–80% of the newly synthesized AChE molecules are rapidly degraded without ever becoming catalytically active or leaving the endoplasmic reticulum (22). However, once AChE monomers are organized into catalytically active oligomers and acquire endoglycosidase H resistance, they become stable (23), and when ColQ is overexpressed in muscle cells, it can induce the assembly of additional catalytically active AChE heterooligomeric forms (17). Our results suggest that by mediating catalytic AChE tetramerization and its assembly with the collagen tail subunit and/or ColQ-processing, PDI levels can limit the numbers of catalytically active tetramers and collagen-tailed AChE molecules expressed. In addition, ColQ levels most likely determine the extent to which PDI overexpression increases the levels of catalytically active collagen-tailed AChE forms including ColQ-AChE (Fig. 7).
It has been shown that heteromeric association of AChE and ColQ does not require the cysteines of either AChE tryptophan amphiphilic tetramerization or ColQ proline-rich attachment domain (53). In these studies the rate of ColQ and AChE assembly in the presence or absence of the cysteines mentioned above was not determined. Therefore, our results do not contradict these findings; instead, they suggest that the cysteines in AChE tryptophan amphiphilic tetramerization and ColQ proline-rich attachment domain may allow the cells to have an additional level of regulation over the expression of AChE-tailed forms by having PDI-catalyzing AChE and ColQ assembly.
One possibility is that PDI, in addition to catalyzing ColQ-AChE assembly, also controls ColQ maturation independently through its interaction with P4H α-subunit (P4Hα). cis-Hydroxyproline, an inhibitor of collagen synthesis, blocks the assembly of synaptic ColQ-AChE and its precursors (54). Moreover, P4Hα mRNA and P4H activity levels decrease in rat soleus and gastrocnemius after muscle immobilization (55), PDI mRNA decreases after gastronemius and plantaris muscle immobilization (56), and in all cases type I and III collagen mRNA levels decreased. These results are not only consistent with our observation that PDI controls catalytically active collagen-tailed AChE expression; they also emphasize the possibility that the posttranslational regulation of the collagen tail subunit could be limiting during AChE biogenesis. Therefore, we should not underestimate the possible role of P4Hα in the regulation of synaptic ColQ-AChE by catalyzing ColQs proline residue hydroxylation. Nevertheless, PDI or ColQ overexpression alone in QMCs is sufficient to increase the expression of collagen-tailed AChE forms, suggesting that the P4Hα subunit is abundant and not a limiting enzyme during ColQ maturation and synaptic AChE biogenesis. However, this remains to be determined.
We show here for the first time that muscle activity in the form of membrane depolarization regulates the levels of PDI, calnexin, and calreticulin, and they correlate positively with the levels of catalytically active ColQ-AChE. It would be interesting to study the levels of expression of other muscle specific proteins under the same experimental conditions. In Caenorhabditis elegans decreased γ-aminobutyric acid signaling or increased acetylcholine signaling causes increased premature aggregation of chimerical polyglutamine-yellow fluorescent protein in body wall muscle cells (57). Defective γ-aminobutyric acid signaling is responsible for the exposure of a misfolding defect of a temperature-sensitive para-myosin mutant at permissive temperatures in the same muscle cells (57). These studies indeed demonstrated that cell-cell communication has an important role in controlling protein homeostasis (57). In addition, they suggest that regulation of muscle activity through γ-aminobutyric acid and acetylcholine signaling may control the levels of molecular chaperones that are responsible for the observed accumulation of misfolded proteins in muscle cells in C. elegans.
Another important finding that comes from our studies is that calnexin, ERp72, and PDI overexpression in muscle cells results in increased catalytically active AChE levels; both the tetrameric and the hetero-oligomeric ColQ-associated forms increase. The extent to which either of these ER molecular chaperones increases synaptic ColQ-AChE expression, PDI being the one responsible for the most dramatic increase, could be determined by many factors. ERp72 for example, has a less efficient capacity for disulfide bond formation and isomerization compared with PDI (58–60). Furthermore, it remains to be determined which is the chaperone(s) assisting rate-limiting step(s) during AChE folding and assembly. Our observations suggest that exportable molecules may compete for chaperone folding assistance in the endoplasmic reticulum; therefore, the relative distribution of the different ER molecular chaperones from cell to cell could provide the molecular basis for a new level of control over the pattern of expression of mature exportable proteins. These hypotheses and how the ordering of these competing interactions could be prioritized by the cell remain open questions for future studies.
Acknowledgments
We thank Emilio Marrero for excellent technical help in cloning the full-length quail ColQ cDNA and Dr. Joav Prives (SUNY, Stony Brook) for the calnexin-expressing plasmid. We also thank Dr. Pedro Salas, Emilio Marrero, and Gustavo Rubio for helpful discussions and comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants AG005917 and NS057994 (to R. L. R.).
- AChE
- acetylcholinesterase
- ColQ
- acetylcholinesterase collagen tail subunit
- PDI
- protein disulfide isomerase
- P4Hα
- prolyl-4-hydroxylase
- Erp72
- endoplasmic reticulum protein 72
- shRNA
- short hairpin RNA
- ScVn
- scorpion venom
- TTX
- tetrodotoxin
- ER
- endoplasmic reticulum
- DFP
- diisopropyl fluorophosphate
- PBS
- phosphate-buffered saline
- DTT
- dithiothreitol
- Pol
- polymerase
- q-
- quail
- ch-
- chicken
- h-
- human
- RIPA
- radioimmune precipitation assay buffer
- EGFP
- enhanced green fluorescent protein
- QMC
- quail muscle culture.
REFERENCES
- 1.Massoulié J., Pezzementi L., Bon S., Krejci E., Vallette F. M. (1993) Prog. Neurobiol. 41, 31–91 [DOI] [PubMed] [Google Scholar]
- 2.Legay C. (2000) Microsc. Res. Tech. 49, 56–72 [DOI] [PubMed] [Google Scholar]
- 3.Rotundo R. L. (2003) J. Neurocytol. 32, 743–766 [DOI] [PubMed] [Google Scholar]
- 4.Rosenberry T. L., Richardson J. M. (1977) Biochemistry 16, 3550–3558 [DOI] [PubMed] [Google Scholar]
- 5.Anglister L., Silman I. (1978) J. Mol. Biol. 125, 293–311 [DOI] [PubMed] [Google Scholar]
- 6.Vigny M., Di Giamberardino L., Couraud J. Y., Rieger F., Koenig J. (1976) FEBS Lett. 69, 277–280 [DOI] [PubMed] [Google Scholar]
- 7.Vigny M., Koenig J., Rieger F. (1976) J. Neurochem. 27, 1347–1353 [DOI] [PubMed] [Google Scholar]
- 8.Koenig J., Vigny M. (1978) Nature 271, 75–77 [DOI] [PubMed] [Google Scholar]
- 9.Rieger F., Powell J. A., Pinçon-Raymond M. (1984) Dev. Biol. 101, 181–191 [DOI] [PubMed] [Google Scholar]
- 10.Hall Z. W. (1973) J. Neurobiol. 4, 343–361 [DOI] [PubMed] [Google Scholar]
- 11.Weinberg C. B., Hall Z. W. (1979) Dev. Biol. 68, 631–635 [DOI] [PubMed] [Google Scholar]
- 12.Michel R. N., Vu C. Q., Tetzlaff W., Jasmin B. J. (1994) J. Cell Biol. 127, 1061–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hubatsch D. A., Jasmin B. J. (1997) Am. J. Physiol. 273, C2002–C2009 [DOI] [PubMed] [Google Scholar]
- 14.Cresnar B., Crne-Finderle N., Breskvar K., Sketelj J. (1994) J. Neurosci. Res. 38, 294–299 [DOI] [PubMed] [Google Scholar]
- 15.Rossi S. G., Vazquez A. E., Rotundo R. L. (2000) J. Neurosci. 20, 919–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rossi S. G., Katz S., Rotundo R. L. (1998) J. Physiol. 92, 486–487 [Google Scholar]
- 17.Ruiz C. A., Rotundo R. L. (2009) J. Biol. Chem. 284, 21488–21495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rimer M., Randall W. R. (1999) Biochem. Biophys. Res. Commun. 260, 251–255 [DOI] [PubMed] [Google Scholar]
- 19.Rubin L. L. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 7121–7125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rotundo R. L. (1984) Proc. Natl. Acad. Sci. U.S.A. 81, 479–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rotundo R. L. (1984) J. Biol. Chem. 259, 13186–13194 [PubMed] [Google Scholar]
- 22.Rotundo R. L. (1988) J. Biol. Chem. 263, 19398–19406 [PubMed] [Google Scholar]
- 23.Rotundo R. L., Thomas K., Porter-Jordan K., Benson R. J., Fernandez-Valle C., Fine R. E. (1989) J. Biol. Chem. 264, 3146–3152 [PubMed] [Google Scholar]
- 24.Chang W., Gelman M. S., Prives J. M. (1997) J. Biol. Chem. 272, 28925–28932 [DOI] [PubMed] [Google Scholar]
- 25.Gelman M. S., Chang W., Thomas D. Y., Bergeron J. J., Prives J. M. (1995) J. Biol. Chem. 270, 15085–15092 [DOI] [PubMed] [Google Scholar]
- 26.Keller S. H., Lindstrom J., Taylor P. (1996) J. Biol. Chem. 271, 22871–22877 [DOI] [PubMed] [Google Scholar]
- 27.Keller S. H., Lindstrom J., Taylor P. (1998) J. Biol. Chem. 273, 17064–17072 [DOI] [PubMed] [Google Scholar]
- 28.Sussman J. L., Harel M., Frolow F., Oefner C., Goldman A., Toker L., Silman I. (1991) Science 253, 872–879 [DOI] [PubMed] [Google Scholar]
- 29.Bourne Y., Taylor P., Bougis P. E., Marchot P. (1999) J. Biol. Chem. 274, 2963–2970 [DOI] [PubMed] [Google Scholar]
- 30.Wilkinson B., Gilbert H. F. (2004) Biochim. Biophys. Acta 1699, 35–44 [DOI] [PubMed] [Google Scholar]
- 31.Bottomley M. J., Batten M. R., Lumb R. A., Bulleid N. J. (2001) Curr. Biol. 11, 1114–1118 [DOI] [PubMed] [Google Scholar]
- 32.Ellman G. L., Courtney K. D., Andres V., Jr., Feather-stone R. M. (1961) Biochem. Pharmacol. 7, 88–95 [DOI] [PubMed] [Google Scholar]
- 33.Rotundo R. L., Fambrough D. M. (1980) Cell 22, 583–594 [DOI] [PubMed] [Google Scholar]
- 34.Fambrough D. M., Engel A. G., Rosenberry T. L. (1982) Proc. Natl. Acad. Sci. U.S.A. 79, 1078–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clive D. R., Greene J. J. (1994) Exp. Cell Res. 214, 139–144 [DOI] [PubMed] [Google Scholar]
- 36.Osborne C. K., Monaco M. E., Lippman M. E., Kahn C. R. (1978) Cancer Res. 38, 94–102 [PubMed] [Google Scholar]
- 37.Haugstetter J., Blicher T., Ellgaard L. (2005) J. Biol. Chem. 280, 8371–8380 [DOI] [PubMed] [Google Scholar]
- 38.Knoblach B., Keller B. O., Groenendyk J., Aldred S., Zheng J., Lemire B. D., Li L., Michalak M. (2003) Mol. Cell. Proteomics 2, 1104–1119 [DOI] [PubMed] [Google Scholar]
- 39.Vattemi G., Engel W. K., McFerrin J., Askanas V. (2004) Am. J. Pathol. 164, 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De La Porte S., Vigny M., Massoulié J., Koenig J. (1984) Dev. Biol. 106, 450–456 [DOI] [PubMed] [Google Scholar]
- 41.Brockman S. K., Younkin L. H., Younkin S. G. (1984) J. Neurosci. 4, 131–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rieger F., Koenig J., Vigny M. (1980) Dev. Biol. 76, 358–365 [DOI] [PubMed] [Google Scholar]
- 43.Fernandez-Valle C., Rotundo R. L. (1989) J. Biol. Chem. 264, 14043–14049 [PubMed] [Google Scholar]
- 44.Wada I., Rindress D., Cameron P. H., Ou W. J., Doherty J. J., 2nd, Louvard D., Bell A. W., Dignard D., Thomas D. Y., Bergeron J. J. (1991) J. Biol. Chem. 266, 19599–19610 [PubMed] [Google Scholar]
- 45.Ou W. J., Bergeron J. J., Li Y., Kang C. Y., Thomas D. Y. (1995) J. Biol. Chem. 270, 18051–18059 [DOI] [PubMed] [Google Scholar]
- 46.Ye J. M., Key C. J., Wolfe J. L. (1996) Biochem. Biophys. Res. Commun. 223, 153–159 [DOI] [PubMed] [Google Scholar]
- 47.Lumb R. A., Bulleid N. J. (2002) EMBO J. 21, 6763–6770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weinmann R., Raskas H. J., Roeder R. G. (1974) Proc. Natl. Acad. Sci. U.S.A. 71, 3426–3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weinmann R., Roeder R. G. (1974) Proc. Natl. Acad. Sci. U.S.A. 71, 1790–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zylber E. A., Penman S. (1971) Proc. Natl. Acad. Sci. U.S.A. 68, 2861–2865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reeder R. H., Roeder R. G. (1972) J. Mol. Biol. 67, 433–441 [DOI] [PubMed] [Google Scholar]
- 52.MacPhee-Quigley K., Vedvick T. S., Taylor P., Taylor S. S. (1986) J. Biol. Chem. 261, 13565–13570 [PubMed] [Google Scholar]
- 53.Simon S., Krejci E., Massoulié J. (1998) EMBO J. 17, 6178–6187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vallette F. M., De la Porte S., Koenig J., Massoulié J., Vigny M. (1990) J. Neurochem. 54, 915–923 [DOI] [PubMed] [Google Scholar]
- 55.Ahtikoski A. M., Koskinen S. O., Virtanen P., Kovanen V., Takala T. E. (2001) Acta Physiol. Scand. 172, 131–140 [DOI] [PubMed] [Google Scholar]
- 56.Han X. Y., Wang W., Myllylä R., Virtanen P., Karpakka J., Takala T. E. (1999) J. Appl. Physiol 87, 90–96 [DOI] [PubMed] [Google Scholar]
- 57.Garcia S. M., Casanueva M. O., Silva M. C., Amaral M. D., Morimoto R. I. (2007) Genes Dev. 21, 3006–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Satoh M., Shimada A., Kashiwai A., Saga S., Hosokawa M. (2005) Cell Stress Chaperones 10, 211–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Füllekrug J., Sönnichsen B., Wünsch U., Arseven K., Nguyen Van P., Söling H. D., Mieskes G. (1994) J. Cell Sci. 107, 2719–2727 [DOI] [PubMed] [Google Scholar]
- 60.Rupp K., Birnbach U., Lundström J., Van P. N., Söling H. D. (1994) J. Biol. Chem. 269, 2501–2507 [PubMed] [Google Scholar]