Abstract
Laminin-2 promotes basement membrane assembly and peripheral myelinogenesis; however, a receptor-binding motif within laminin-2 and the downstream signaling pathways for motif-mediated cell adhesion have not been fully established. The human laminin-2 α2 chain cDNAs cloned from human keratinocytes and fibroblasts correspond to the laminin α2 chain variant sequence from the human brain. Individually expressed recombinant large globular (LG) 1 protein promotes cell adhesion and has heparin binding activities. Studies with synthetic peptides delineate the DLTIDDSYWYRI motif (Ln2-P3) within the LG1 as a major site for both heparin and cell binding. Cell adhesion to LG1 and Ln2-P3 is inhibited by treatment of heparitinase I and chondroitinase ABC. Syndecan-1 from PC12 cells binds to LG1 and Ln2-P3 and colocalizes with both molecules. Suppression of syndecan-1 with RNA interference inhibits cell adhesion to LG1 and Ln2-P3. The binding of syndecan-1 with LG1 and Ln2-P3 induces the recruitment of protein kinase Cδ (PKCδ) into the membrane and stimulates its tyrosine phosphorylation. A decrease in PKCδ activity significantly reduces cell adhesion to LG1 and Ln2-P3. Taken together, these results indicate that the Ln2-P3 motif and LG1 domain, containing the motif, within the human laminin-2 α2 chain are major ligands for syndecan-1, which mediates cell adhesion through the PKCδ signaling pathway.
Introduction
Laminin is a heterotrimeric glycoprotein specific to the basement membrane and has many biological functions, including cell adhesion, migration, cell proliferation, differentiation, neurite outgrowth, angiogenesis, and tumor invasion (1). Laminins are composed of α, β, and γ chains, which assemble into a cross-shaped heterotrimer (αβγ) through a coiled-coil interaction at the long arm of the cross (2). At least 15 laminin isoforms have been identified with 11 genetically distinct chains: five α chains (α1–α5), three β chains (β1-β3), and three γ chains (γ1–γ3) (3). The laminin α2 chain, a component of laminin-2 (α2β1γ1), laminin-4 (α2β2γ1), and laminin-12 (α2β1γ3), is expressed in skeletal and cardiac muscle, peripheral nerves, the brain, and placenta (4).
Mutations in the laminin α2 chain gene cause merosin-deficient congenital muscular dystrophy in both humans and mice (5, 6). The phenotype of dystrophic mice is characterized by muscular dystrophy, defective basement membranes in muscles and nerves, and peripheral nerve dysmyelination (7–9). Laminin-2 expression is absent in both the peripheral nerve and skeletal muscle of dystrophic mice (7). It is important to note that laminin-2 is critical for basement membrane assembly and peripheral myelinogenesis.
The laminin α2 chain contains a large globular (LG)3 domain at the C terminus, which consists of a tandem repeat of five homologous LG domains (LG1 to LG5), each domain containing an ∼200-amino acid residue autonomous folding unit (10). The LG domains of laminin α chains have been shown to bind integrins, α-dystroglycan, and heparin/heparan sulfate proteoglycans (3) and are implicated as active regions for various biological functions. Mouse laminin α2 chain LG4–5 domain contains binding sites for heparin/sulfatides and α-dystroglycan (11, 12). The laminin α2 chain LG1–3 domain promotes cell binding activity via several integrins, such as α3β1, α6β1, and α7β1, and this domain is required for acetylcholine receptor clustering (3, 13). Several synthetic peptides derived from the mouse laminin α2 chain LG domains promote cell adhesion, heparin binding, neurite outgrowth, and acinar formation (14–16). For example, MG-73 peptide (KNRLTIELEVRT, amino acids 2780–2791) derived from the mouse laminin α2 chain LG4 domain promotes cell adhesion and neurite outgrowth and binds to syndecan-1, a cell surface heparan sulfate proteoglycan (15, 16). Similarly, the EF-2 peptide (DFGTVQLRNGFPFFSYDLG, amino acids 2808–2826), which is located on the connecting loop region of the mouse laminin α2 chain LG4 domain, shows cell adhesion and syndecan-2 binding (17). These results indicate that the mouse laminin α2 chain LG4 domain contains two heparin-binding sites and has multiple biological functions.
However, little is known regarding the biological functions of the human laminin α2 LG domains and their cellular receptors and downstream signaling pathways. Here, we individually express three human laminin α2 LG domains, such as LG1, LG2, and LG3, as monomeric, soluble fusion proteins and examine their biological functions and signaling. More significantly, we identify a biologically active motif that is crucial for LG1 function within the human laminin α2 LG1 domain. The findings herein demonstrate that the DLTIDDSYWYRI motif (amino acids 2221–2232; Ln2-P3) and the LG1 domain, containing the motif, within the human laminin α2 chain promote cell adhesion and heparin binding and bind to syndecan-1. Human laminin α2 LG1 domain/syndecan-1-mediated cell adhesion is achieved through the membrane localization and tyrosine phosphorylation of the protein kinase C (PKC) δ.
EXPERIMENTAL PROCEDURES
Cells and Peptides
The PC12 cell line from transplantable rat pheochromocytoma was cultured in RPMI 1640 medium (BioWhittaker Cambrex, Walkersville, MD) containing 10% fetal bovine serum. Normal human epidermal keratinocytes and normal human dermal fibroblasts were prepared and maintained as described previously (18). The mouse embryo fibroblast cell line NIH/3T3 and normal African green monkey kidney fibroblast cell line CV-1 were purchased from the American Type Culture Collection (Manassas, VA) and cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. All peptides were synthesized by the Fmoc (9-fluorenylmethoxycarbonyl)-based solid-phase methods with a C-terminal amide using a Pioneer peptide synthesizer (Applied Biosystems, Foster City, CA), purified, and characterized at the Korea Basic Science Institute.
Construction, Expression, and Purification of Human Laminin α2 LG Domains and Mutant LG1
The human laminin α2 cDNA was cloned using a reverse transcriptase-PCR with Superscript II reverse transcriptase (Invitrogen) under the conditions recommended by the manufacturer using mRNA isolated from human keratinocytes and fibroblasts. Three C-terminal LG cDNA fragments (LG1 to LG3) of the laminin α2 chain were amplified by PCR using the laminin α2 cDNA as a template and were ligated into the pGEM-T Easy vector (Promega, Madison, WI). The PCR primers used were as follows: LG1--1, 5′-AAACAAGCCAATTCTATCAA-3′ (sense) and 5′-ATTTCCATTTCCCATCATT-3′ (antisense); LG1, 5′-GCCACTCGAGCAGGAGGTGACTG-3′ (sense) and 5′-GCCACCATGGTCAACTGACAGTGCATCC-3′ (antisense); LG2, 5′-GCCACTCGAGCAGTCCTCAGGTG-3′ (sense) and 5′-GCCACCATGGTCACTCCACAAAACCAGGCTTA-3′ (antisense); and LG3, 5′-GCCACTCGAGTGTGGAGCTCTCCCCTGT-3′ (sense) and 5′-GCCACCATGGTCAAACTGGGGTGGGCGTAGGA-3′ (antisense). LG1 was amplified by a nested PCR using two primer combinations as follows: first with the LG1-1 sense and antisense primers, and then the product was amplified with the LG1 sense and antisense primers. The nucleotide sequences of all of the plasmid constructs were confirmed by sequence analysis. The pGEM-T Easy vector containing LG cDNA fragments was digested with XhoI and NcoI. These cDNA fragments were subsequently cloned into corresponding sites of the mammalian expression plasmid vector pRSET (Invitrogen). Correct orientation of the inserts was verified by sequence analysis. The LG1 clone was used to generate three mutants using the Muta-DirectTM site-directed mutagenesis kit (iNtRON Biotechnology, Korea). The PCR primers used for specific mutations were as follows: LG1-M1 (DLTIDDSYWYRI → ALTIAASYWYRI), 5′-GTGTAGAGTACCCAGCTTTGACTATTGCTGCCTCATATTGGTACC-3′ (sense) and 5′-GGTACCAATATGAGGCAGCAATAGTCAAAGCTGGGTACTCTACAC (antisense); and LG1-M2 (DLTIDDSYWYRI → DLTIDDSYWYAI), 5′-GATGACTCATATTGGTACGCTATCGTAGCATCAAGAACT-3′ (sense) and 5′-AGTTCTTGATGCTACGATAGCGTACCAATATGAGTCATC-3′ (antisense). LG1-M3 (DLTIDDSYWYRI → ALTIAASYWYAI) was amplified with LG1-M2 primers using the LG1-M1 plasmid DNA as a template. PCR and enzyme treatment were carried out according to the manufacturer's instructions. Each clone was verified by sequence analysis.
The expression and purification of recombinant LG (rLG) proteins were performed as reported previously (19). Briefly, rLG proteins were induced in Escherichia coli strain BL21 grown to the mid-log phase in Luria-Bertani medium using 1 mm isopropyl β-d-thiogalactopyranoside (Promega). After protein induction for 5 h at 37 °C, the cells were harvested by centrifugation at 6,000 × g for 10 min. Cell pellets were stored at −80 °C until used. For protein purification, pellets were thawed and resuspended in lysis buffer (8 m urea, 10 mm Tris-HCl, pH 8.0, and 100 mm NaH2PO4) containing 1 mm phenylmethylsulfonyl fluoride (Sigma). rLG proteins were purified using a Ni2+-nitrilotriacetic acid-agarose column (Qiagen, Valencia, CA) following the manufacturer's recommendations. Purified recombinant His6-tagged LG proteins were dialyzed sequentially against a solution containing 10 mm Tris-HCl, pH 8.0, 100 mm NaH2PO4, 1 mm phenylmethylsulfonyl fluoride, and 3, 2, 1, or 0.5 m urea, pH 3.0. Finally, the proteins were dialyzed with PBS, pH 3.0, containing 1 mm phenylmethylsulfonyl fluoride. The dialyzed rLG proteins were concentrated at 1 μg/μl with a Centricon YM-10 filter device (Millipore, Bedford, MA) and stored at −80 °C until use. The protein concentration was determined using a protein assay kit (Bio-Rad).
Circular Dichroism Spectroscopy
rLG proteins at 0.2 mg/ml were prepared in PBS. CD spectra were recorded on a Jasco spectropolarimeter (model J-715; Jasco International Co., Japan). Protein samples were analyzed at 23 °C from 180 to 300 nm with a 2-mm path length cell. Three repetitive scans were averaged and smoothed by binomial curve smoothing. The molar ellipticity (in degrees cm2 dmol−1) was calculated on the basis of protein concentration and molar mass for each rLG protein.
Cell Adhesion and Spreading Assays
Cell adhesion assay was performed as described previously (20). Briefly, 24-well plates were coated with 5 μg/ml human placental laminin (Sigma) and various amounts of rLG proteins for 12 h at 4 °C. Synthetic peptides were also coated onto plates by drying them for 12 h at room temperature. The substrate-coated plates were blocked with 1% heat-inactivated BSA in PBS for 1 h at 37 °C and washed twice with PBS. PC12 cells were detached, resuspended in culture media, added to each plate (2 × 105 cells/500 μl), and incubated for 1 h at 37 °C. After incubation, unattached cells were removed by rinsing twice with PBS. The attached cells were fixed with 10% formalin in PBS for 15 min, rinsed twice with PBS, and stained with 0.5% crystal violet for 1 h. Plates were gently rinsed with double-distilled water three times and lysed with 2% SDS for 5 min. Absorbance was measured at 570 nm in a model 550 microplate reader (Bio-Rad). Cell spreading was measured using the photographs that were taken from the cell adhesion assay. To ensure a representative count, each sample was divided into quarters, and one field per quarter was photographed. The area of each spreading cell was determined using a computer equipped with Image-Pro plus software (Media Cybernetics, Silver Spring, MD). At least 200 cells were examined on each occasion.
Adhesion Inhibition Assay
Cells (2 × 105 cells/500 μl) were preincubated with 5 mm EDTA, 10 μg/ml of function-blocking antibody against the integrin β1 subunit (BD Biosciences), Gö6976, rottlerin (Calbiochem), calphostin C (Sigma), synthetic peptides (100 μg/ml), or 100 μg/ml of various glycosaminoglycans, such as heparin, heparan sulfate, dermatan sulfate (chondroitin sulfate B), hyaluronic acid, chondroitin sulfate A, chondroitin sulfate C, and de-N-sulfated heparin (Sigma), for the indicated times. The preincubated cells were then seeded on plates coated with either rLG proteins (25 μg/ml) or Ln2-P3 (22 μg/cm2) and further incubated for 1 h at 37 °C. Attached cells were quantified by either cell counting or absorbance measurement at 570 nm in a model 550 microplate reader (Bio-Rad), as described elsewhere.
Solid-phase Heparin Binding Assay
rLG proteins were coated onto 96-well plates for 12 h at 4 °C, and synthetic peptides were also coated onto the plates by drying them for 12 h at room temperature. The substrate-coated plates were washed twice with PBS containing 0.05% Tween 20 and blocked with 3% heat-inactivated BSA in PBS containing 0.05% Tween 20 for 2 h at 37 °C. The wells were washed twice with PBS containing 0.05% Tween 20, and 10 ng of biotinylated heparin (Calbiochem) was added. After incubation for 1 h at 37 °C, the supernatant was removed, and the wells were washed three times with PBS containing 0.05% Tween 20. To detect the bound biotinylated heparin, 10 ng of streptavidin-conjugated horseradish peroxidase (Calbiochem) was added and further incubated for 1 h at 37 °C. After washing three times with PBS containing 0.05% Tween 20, 0.4 mg/ml of O-phenylenediamine (Sigma) was added and incubated for 10 min at room temperature. After adding 3 m H2SO4, the absorbance was measured at 490 nm in a microplate reader (Bio-Rad).
Treatment of Cells with Enzymes
Cells (6 × 105 cells/200 μl) were pretreated with 2 μg/ml cycloheximide for 2 h and further incubated with 0.1, 0.5, or 1 unit/ml heparitinase I or chondroitinase ABC (Sigma) in PBS containing 2 μg/ml cycloheximide, 2 mm CaCl2, and 0.1% BSA for 90 min at 37 °C. Cells were suspended in serum-free RPMI 1640 medium and then used for the cell adhesion assay, as described elsewhere.
Immunodot Blotting
Cells were washed with PBS and lysed with a buffer (50 mm Tris-HCl, pH 7.4, 0.5 mm EDTA, and 1% Triton X-100) containing a protease inhibitor mixture. Lysates were preincubated with 50 μl of immobilized protein A/G beads (Pierce) for 30 min at 4 °C and then centrifuged. One ml of the supernatants was incubated with 30 μg of His6 or His6-rLG1 proteins for 2 h at 4 °C. The bound materials were immunoprecipitated with 100 μl of immobilized protein A/G beads carrying 1 μg of anti-His6 antibody (Roche Applied Science) for 12 h at 4 °C. After centrifugation, beads carrying the immune complexes were washed four times with a buffer (25 mm Tris-HCl, pH 5.0, and 150 mm NaCl). To elute the immune complexes from the beads, 100 μl of elution buffer (Pierce) were added and centrifuged at 2,500 × g for 1 min. The supernatants were collected, and 10 μl of neutralization buffer (1 m Tris-HCl) was added. The supernatants were dot-blotted onto a nitrocellulose membrane and probed with primary antibody. All blots were detected using horseradish peroxidase-conjugated secondary antibody (Cell Signaling Technology, Beverly, MA) and developed with ECL reagents (iNtRON Biotechnology, Korea).
Immunofluorescence
Cells grown on glass slide chambers precoated with rLG1 (25 μg/ml) or Ln2-P3 (22 μg/cm2) for 30 min were washed in cold PBS, fixed with 3.7% formalin in PBS for 20 min, and permeabilized with 0.5% Triton X-100 in PBS for 5 min at room temperature. After blocking with 1% BSA in PBS for 60 min, cells were incubated with primary antibodies for 12 h at 4 °C. Cells were rinsed with PBS and incubated with secondary antibodies for 1 h at room temperature. Samples were observed and recorded using a fluorescence microscope (Olympus FV300, Japan) equipped with a CDD camera.
Immunoprecipitation
Cells were starved for 12 h by replacing RPMI 1640 medium with 0.1% fetal bovine serum. Cells were trypsinized, washed, and resuspended in serum-free medium containing 0.1% BSA. Cells (4 × 106) were seeded on 100-mm dishes coated with rLG1 (25 μg/ml) or Ln2-P3 (22 μg/cm2) for 30 min and washed with ice-cold PBS. Cells were lysed in RIPA buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1% Nonidet P-40, 1 mm β-glycerophosphate, 2 mm Na3VO4, 1 mm phenylmethylsulfonyl fluoride, and 1× protease inhibitor mixture) for 30 min on ice. Lysates were then spun at 10,000 × g for 15 min at 4 °C, and the supernatants were precleared by adding 20 μl of protein G PLUS-agarose (Upstate Biotechnology, Lake Placid, NY). Precleared lysates were incubated with the indicated antibodies for 2 h at 4 °C, then further incubated with 80 μl of protein G PLUS-agarose for 12 h at 4 °C, and spun at 10,000 × g for 15 min at 4 °C. After washing the beads four times with RIPA buffer, precipitated proteins were resolved by 10% SDS-PAGE and immunoblotted.
Cell Protein Fractionation
The fractionation of membrane and cytosolic proteins was prepared using plasma membrane protein extraction kit according to the instructions of the manufacturer (BioVision, Mountain View, CA). Briefly, cells were starved for 12 h prior to the experiment, detached, and resuspended in serum-free medium containing 0.1% BSA. Cells were seeded (4 × 106/100-mm dish) on rLG1- or Ln2-P3-coated dishes and allowed to attach for the indicated time periods at 37 °C. Cells were lysed in 200 μl of the Homogenize buffer (BioVision), and the cytosol fractions were collected after centrifugation at 10,000 × g for 30 min at 4 °C. The pellets were solubilized in RIPA buffer, and the membrane fractions were recovered after centrifugation at 14,000 × g for 15 min at 4 °C. The protein concentration was determined using the Bradford reagent (Bio-Rad) and analyzed by immunoblotting, as described elsewhere.
Flow Cytometry
Flow cytometric analysis of the cell surface receptor expression level was performed. Cells were detached by gentle treatment with 0.05% trypsin and 0.53 mm EDTA in PBS, washed, and incubated with anti-syndecan or anti-glypican monoclonal antibodies for 45 min at 4 °C. After washing, cells were incubated with fluorescein isothiocyanate-labeled secondary antibodies for 45 min at 4 °C. Finally, cells were analyzed on a FACSCalibur flow cytometer (BD Biosciences).
RT-PCR
Syndecan-1 and -4 and glypican-1 mRNA levels were determined by RT-PCR. Total RNA was reverse-transcribed to cDNA, which allowed for PCR amplification of syndecan-1 (33 cycles) and -4 (28 cycles), glypican-1 (28 cycles), and glyceraldehyde-3-phosphate dehydrogenase (33 cycles, denaturation at 95 °C for 20 s, annealing at 58 °C for 10 s, and extension at 70 °C for 10 s). Syndecan-1 and -4 and glypican-1 primers were designed as follows: forward for syndecan-1, 5′-CCCCACAACCCCTGCCACTCACTA-3′, and reverse for syndecan-1, 5′-AAGGTCTGCTGGGGCTCTAAAACA-3′; forward for syndecan-4, 5′-CATGGGGAGAGGAGTTGAGGATTG-3′, and reverse for syndecan-4, 5′-TGCCCCGAGATACACCACAGAGAC-3′; forward for glypican-1, 5′-GTGGCGCCTACGGTGGAAATGATG-3, and reverse for glypican-1, 5′-TGCCCCTGAGGTCCCTGAAAAACA-3′. Glyceraldehyde-3-phosphate dehydrogenase primers for normalization of the expression level were also prepared, 5′-TGTCAGCAATGCATCCTGTACCACC-3′ and 5′-AAAGTCCGTGGAGACCACCTGGTCC-3′. The PCR products were resolved on a 1% agarose gel, stained with ethidium bromide, and visualized using a UV illuminator.
Transfection
Small interfering RNAs (siRNAs) against rat syndecan-1 and -4, glypican-1, and a nonspecific control siRNA (Invitrogen) were used for gene silencing. PC12 cells were plated at a density of 5 × 105 cells/60-mm dish and transfected with 100 nm syndecan-1 siRNA, 100 nm syndecan-4 siRNA, glypican-1 siRNA, or 100 nm control siRNA using Lipofectamine RNAiMAX transfection reagents and 7 μl of Lipofectamine RNAiMAX (Invitrogen) for 5 h. After 48 h, transfected cells were harvested and analyzed by RT-PCR and flow cytometry, as described elsewhere.
RESULTS
Expression, Purification, and Characterization of Recombinant Laminin α2 LG Domains
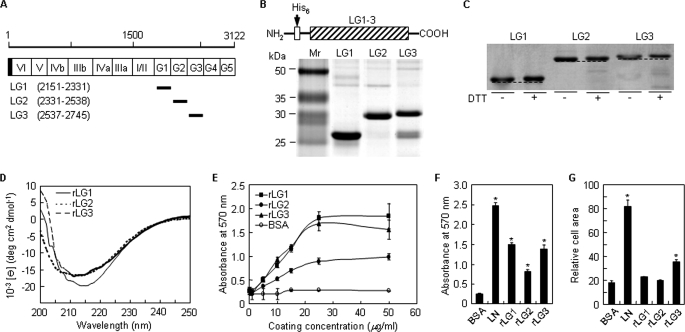
Human laminin α2 chain cDNA was synthesized using mRNAs isolated from human keratinocytes and fibroblasts, and the nucleotide sequences were confirmed by sequence analysis. In this study, we identified a cDNA that corresponded to the laminin α2 chain variant from the human brain (GenBankTM accession number AB208922). Amino acid substitution (V2587A) was detected in the LG3 domain compared with the laminin α2 chain sequence from human placenta (GenBankTM accession number Z26653). Three human laminin α2 C-terminal LG domains (LG1 to LG3) were individually expressed as monomers in E. coli. The corresponding amino acid positions of the rLG proteins in the entire laminin α2 chain are shown in Fig. 1A. The molecular masses of the expected rLG1, rLG2, and rLG3 proteins were 24, 27, and 27 kDa, respectively, and each purified rLG protein showed the predicted molecular mass (Fig. 1B). The rLG3 protein showed several bands, with the largest corresponding to the predicted molecular size, although the smaller bands corresponded to partially degraded products (Fig. 1B). Because laminin α2 LG3 domain of mice and humans is known to contain a cleavage site that is proteolytically processed by a furin-like convertase (13, 21) and the recombinant mouse laminin α2 LG3 protein could not be obtained in mammalian cells (22), we expressed the rLG3 in E. coli to abolish the protease activity and obtained the intact rLG3 with a high degree of efficiency (∼10 mg per liter of bacterial culture).
FIGURE 1.
Analyses of purified rLG proteins of human laminin α2 chain by SDS-PAGE and circular dichroism and their cell adhesion activities. A, schematic diagram of the human laminin α2 LG domains and its recombinant proteins. The amino acid scale is shown on the top, and the domain structures of the laminin α2 chain are indicated with open column. The shaded portion and the closed bars represent the signal peptide and the positions of the recombinant proteins, respectively. Numbers in parentheses correspond to the amino acid positions of the recombinant proteins in the entire laminin α2 chain. B, schematic diagram and SDS-PAGE analysis of the rLG proteins in the human laminin α2 chain. The three rLG proteins were expressed as His6-tagged fusion proteins. The rLG proteins were subjected to SDS-PAGE analysis (13% polyacrylamide gels, reducing) and visualized by Coomassie staining. C, gel mobilities of purified rLG proteins treated with dithiothreitol (DTT) were compared with nontreated rLG proteins under 10% SDS-PAGE conditions. Reduction of rLG proteins prior to electrophoresis resulted in reproducible decrease in gel mobility. D, CD analyses of rLG proteins in PBS, pH 3.0, at 23 °C. The CD spectra of all three rLG proteins showed ellipticity minima at around 210–220 nm, indicating that the purified rLG proteins consist predominantly of β-structure. E, rLG proteins support adhesion of PC12 cells seeded on rLG protein-coated plates for 1 h in serum-free medium in a dose-dependent manner. F, adhesion of PC12 cells seeded on plates coated with laminin (LN; 5 μg/ml) and rLG proteins (25 μg/ml) for 1 h in serum-free medium. G, cell spreading to laminin and rLG proteins. PC12 cells were seeded on laminin (LN; 5 μg/ml)- or rLG protein (25 μg/ml)-coated plates for 3 h in serum-free medium. Cell area was measured using Image-Pro Plus software. Values are expressed as the mean ± S.D. (n = 4). *, p < 0.01.
To determine whether or not an intramolecular disulfide bond formed in the rLG proteins, we subjected the purified recombinant proteins to SDS-PAGE under reducing or nonreducing conditions and looked for mobility differences. Treatment of rLG proteins with 100 mm dithiothreitol prior to SDS-PAGE caused a small but reproducible reduction in gel mobility, suggesting that intramolecular disulfide bonds are present in all three recombinant proteins (Fig. 1C). Next, the secondary structure of rLG proteins was assessed by CD spectroscopy. CD spectra of recombinant His6-LG1, recombinant His6-LG2, and recombinant His6-LG3 showed ellipticity minima at 215, 214, and 211 nm, respectively (Fig. 1D), the characteristic of proteins rich in β-structure. Our findings are in good agreement with reported previously CD values for rLG proteins of laminin-5 (23) and are consistent with the crystal structures of laminin α2 LG5 domain and LG4–5 domain, which reveal a 14-stranded β-sandwich structure (12, 24). Taken together, these results suggest that the bacterially expressed LG domains from laminin-2 are sufficiently well folded to support potential functional activity.
Laminin and Laminin α2 LG Domains Promote Cell Adhesion
Because mouse laminin α2 LG domains are known to contain five cell binding sequences (14), recombinant human laminin α2 LG proteins were examined for cell adhesion activity to identify the cell binding domains. PC12 cells adhered to rLG1 and rLG3 in a dose-dependent manner but weakly attached to rLG2 (Fig. 1E). Cell adhesion activity of rLG1, rLG2, and rLG3 reached the maximum level at ∼25 μg/ml in PC12 cells (Fig. 1E). The levels of cell adhesion to rLG1 and rLG3 were found to have ∼42% reduction compared with human placental laminin (Fig. 1F). Laminin and rLG3 induced cell spreading by 5- and 2-fold compared with BSA control, respectively, whereas rLG1 and rLG2 had no effect on cell spreading (Fig. 1G). These results suggest that human laminin α2 LG1 and LG3 domains have a cell adhesion site(s).
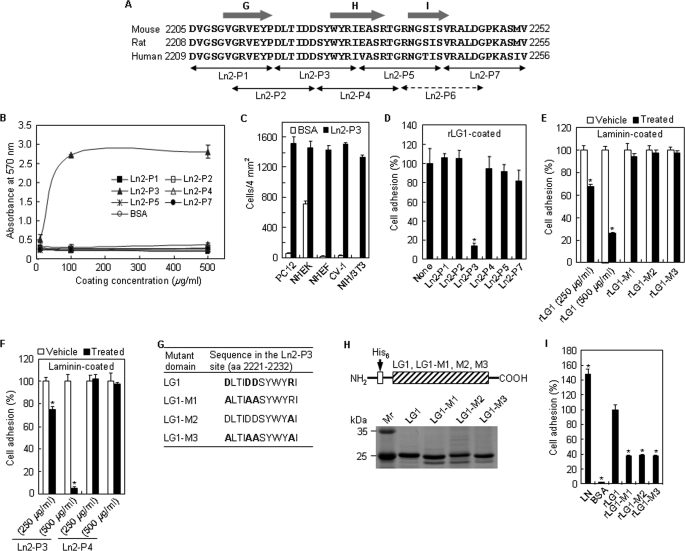
DLTIDDSYWYRI Motif within the Human Laminin α2 LG1 Domain Promotes Cell Adhesion
After establishing that rLG1 mediates adhesion of PC12 cells, this study turned to identifying the essential cell binding sequences conferring cell adhesion activity of the human laminin α2 LG1 domain. Accordingly, six overlapping 12-mer peptides covering amino acids 2209–2256 derived from the LG1 domain were synthesized (Fig. 2A), and we tested their cell adhesion activities. Because we could not obtain enough amounts of Ln2-P6 peptide (amino acids 2239–2250) after purification, we ruled out this peptide in this study. As shown in Fig. 2B, Ln2-P3 promoted adhesion of PC12 cells; the peptide displayed a strong cell adhesion activity in a dose-dependent manner. The cell adhesion activity of Ln2-P3 reached the maximum level at ∼100 μg/ml in PC12 cells. On the other hand, other peptides displayed no cell adhesion activity, even at high coating concentrations (Fig. 2B). Because Ln2-P3 displayed cell adhesion activity in PC12 cells, it was further investigated whether Ln2-P3 would also mediate adhesion of other types of cells. Like PC12 cells, normal human epidermal keratinocytes (passage 2), normal human dermal fibroblasts (passage 4), CV-1, and NIH/3T3 displayed a strong cell adhesion activity to Ln2-P3 at 100 μg/ml (Fig. 2C), demonstrating that Ln2-P3 has a broad cell-type specificity of adhesion. This result is supported by a previous report (25), which explains that peptide F-9 (RYVVLPRPVCFEKGMNYTVR, amino acids 641–660) within the β1 chain of laminin induces the adhesion of various cell types, including murine melanoma, murine fibrosarcoma, rat glioma, rat pheochromocytoma, and bovine aortic endothelial cells. Next, the ability of Ln2-P3 to compete for cell adhesion to rLG1 was examined to verify the role of Ln2-P3 in the cell adhesion activity of intact LG1 domain (Fig. 2D). Ln2-P3 inhibited cell adhesion to rLG1 by ∼86%, whereas none of the other peptides showed reduced adhesion activity in PC12 cells pretreated with 100 μg/ml peptides. To further investigate whether the Ln2-P3 motif within the LG1 domain plays a crucial role in cell adhesion to intact laminin, we tested the ability of the rLG1 protein and Ln2-P3 to inhibit cell adhesion to laminin. rLG1 showed weak inhibition of cell adhesion to laminin by ∼33% in PC12 cells pretreated with 250 μg/ml protein. However, rLG1 inhibited cell adhesion to laminin by ∼75% in PC12 cells pretreated with 500 μg/ml protein (Fig. 2E). This suggests that other cell surface receptors induce cell adhesion to laminin and that integrins and α-dystroglycan bind laminin α2 chain LG domains, agreeing with previous reports (3, 13). In contrast, three mutant LG1 proteins did not inhibit cell adhesion to laminin (Fig. 2E), suggesting that charged amino acid residues in the Ln2-P3 motif are important for cell adhesion to intact laminin. Similarly, Ln2-P3 showed weak inhibition of cell adhesion to laminin by ∼25% in PC12 cells pretreated with 250 μg/ml peptide (Fig. 2F). The peptide, however, inhibited cell adhesion to laminin by ∼95% in PC12 cells pretreated with 500 μg/ml peptide, whereas the other peptide did not inhibit cell adhesion to laminin in PC12 cells, even at high pretreatment concentration (Fig. 2F). This suggests that the Ln2-P3 displays inhibition activity to laminin similar to rLG1, its parent molecule. These results are consistent with previous reports (26–28) and indicate that synthetic peptides derived from laminin α chain inhibit cell adhesion to laminin at high concentrations (400–1,000 μg/ml). Taken together, these results suggest that the Ln2-P3 motif within the LG1 domain is important for cell adhesion to laminin.
FIGURE 2.
Novel motif DLTIDDSYWYRI (Ln2-P3) within the LG1 domain promotes cell adhesion. A, amino acid sequence alignment of part of the laminin α2 LG1 domain from humans, rats, and mice. The arrows indicate the locations of the synthetic peptides. The predicted β-strand structures are represented by the gray arrows. B, dose-dependent cell adhesion to immobilized synthetic peptides. The synthetic peptides were coated onto plates by drying them for 12 h at room temperature, and PC12 cells were allowed to adhere for 1 h in serum-free medium. Values are expressed as the mean ± S.D. (n = 4). C, cell adhesion to Ln2-P3 in normal human epidermal keratinocytes (passage 2), normal human dermal fibroblasts (passage 4), CV-1, and NIH/3T3. Ln2-P3 (100 μg/ml) was coated onto plates by drying them for 12 h at room temperature, and cells were allowed to adhere for 30 min in serum-free medium. Values are expressed as the mean ± S.D. (n = 4). D, inhibition of cell adhesion to rLG1 by Ln2-P3. PC12 cells were pretreated with the various peptides (100 μg/ml) or without the peptide (None) for 10 min at room temperature and then seeded on plates precoated with rLG1 (25 μg/ml) for 1 h in serum-free medium. The number of adherent cells was quantified by cell counting. Values are expressed as a percentage of the value for cells pretreated without the peptide (mean ± S.D., n = 4). *, p < 0.01. E, inhibition of cell adhesion to laminin by rLG1. PC12 cells were pretreated with rLG1 (250 and 500 μg/ml) and mutant rLG1 proteins (250 μg/ml) or without the proteins (Vehicle) for 30 min at 37 °C and then seeded on plates precoated with laminin (5 μg/ml) for 30 min in serum-free medium. Cell counting and expression of values were the same as in D. *, p < 0.01. F, inhibition of cell adhesion to laminin by Ln2-P3. PC12 cells were pretreated with the peptides (250 and 500 μg/ml) or without the peptides (Vehicle) for 15 min at 37 °C and then seeded on plates precoated with laminin (5 μg/ml) for 30 min in serum-free medium. Cell counting and expression of values were the same as in D. *, p < 0.01. G, site-directed mutant domains of acidic and/or basic residues in the Ln2-P3 site of laminin α2 LG1 domain. The acidic and basic residues Asp and Arg in the Ln2-P3 site of laminin α2 LG1 domain, indicated by boldface type, were substituted to Ala. H, schematic diagram and SDS-PAGE analysis of the mutant rLG1 proteins in the human laminin α2 chain. The three mutant rLG1 proteins were expressed as His6-tagged fusion proteins. I, adhesion of PC12 cells seeded on plates coated with laminin (LN; 5 μg/ml), rLG1 (25 μg/ml), and three mutant rLG1 proteins (25 μg/ml) for 30 min in serum-free medium. The number of adherent cells was quantified by cell counting. Values are expressed as a percentage from the value of rLG1-treated cells (mean ± S.D., n = 4). *, p < 0.01.
We further verified the role of Ln2-P3 in the cell adhesion activity of the intact LG1 domain by mutant rLG1 with mutations in the Ln2-P3 site. Because charged amino acid residues are important for the interaction between the LG domains of some laminin α chains and integrins or heparin-like cell surface receptors, mutant rLG1 proteins with mutations in the Ln2-P3 site were prepared by substituting the acidic and/or basic amino acids in the Ln2-P3 site with Ala (Fig. 2G). The molecular weights of the expected rLG1-M1, rLG1-M2, and rLG1-M3 proteins were ∼24 kDa, and each purified mutant protein showed the predicted molecular mass (Fig. 2H). As shown in Fig. 2I, all mutant proteins showed reduced adhesion activity in PC12 cells; the levels of cell adhesion to three mutant proteins were reduced by ∼62–63% from that of rLG1. These results indicate that the DLTIDDSYWYRI motif (amino acids 2221–2232) functions as a cell-binding site in the human laminin α2 LG1 domain and that charged amino acid residues in the motif are essential for cell adhesion activity of the intact LG1 domain.
LG1 Domain and Ln2-P3 Motif Mediate Cell Adhesion via Cell Surface Heparan Sulfate and Dermatan Sulfate
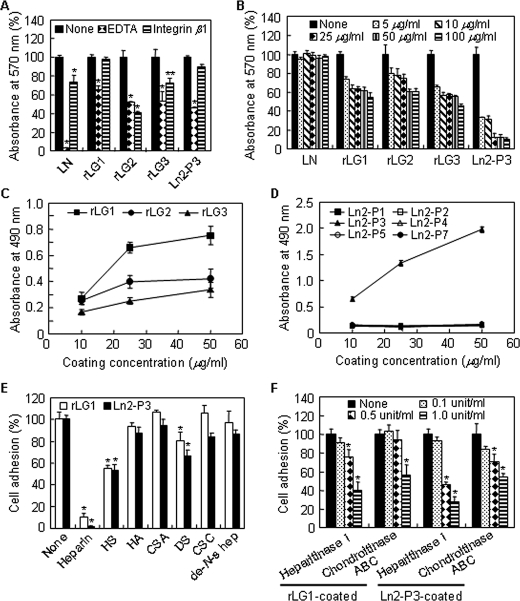
Binding of integrins and α-dystroglycan to their ligands is known to require a divalent cation, such as Ca2+ (23). Therefore, the effect of EDTA, a metal-chelating reagent, on cell adhesion to laminin, LG domains, and the Ln2-P3 motif was investigated to determine the role of divalent cations. Cell adhesion to laminin was completely inhibited in PC12 cells cultured in the presence of 5 mm EDTA compared with control cells, but cell adhesion to rLG1, rLG2, rLG3, or Ln2-P3 was partially inhibited by EDTA treatment (Fig. 3A). To better understand the interaction of laminin, LG domains, and Ln2-P3 motif with integrins, we examined the effect of monoclonal function-blocking antibody against the integrin β1 subunit as a competitor for cell adhesion. Integrin β1 antibody partially inhibited cell adhesion to laminin, rLG2, and rLG3 relative to vehicle-treated control, but rLG1 and Ln2-P3 did not (Fig. 3A). We next examined the effect of heparin on cell adhesion to laminin, LG domains, and Ln2-P3 motif. Cell adhesion to LG domains and Ln2-P3, but not laminin, was significantly inhibited by heparin in a dose-dependent manner; the inhibition activity of heparin reached the maximum level at ∼100 μg/ml in PC12 cells (Fig. 3B). Based on these data, 100 μg/ml heparin was used for the rest of the experiments. We further evaluated the binding interactions between heparin and LG domains or synthetic peptides via the solid-phase binding assay using immobilized biotinylated heparin. rLG1 and Ln2-P3 showed strong heparin binding activity in a dose-dependent manner, but rLG2, rLG3, and other peptides did not (Fig. 3, C and D). These results suggest that LG1 and Ln2-P3 are critical for cell adhesion and heparin binding.
FIGURE 3.
Cell adhesion activity of rLG1 is mediated by heparin binding activity. A, cell adhesion to laminin, rLG proteins, and Ln2-P3 in PC12 cells pretreated with EDTA or integrin β1 antibody. PC12 cells were pretreated with 5 mm EDTA or 10 μg/ml of monoclonal function-blocking antibody against the integrin β1 subunit for 15 min at 37 °C and then seeded on plates precoated with laminin (LN; 5 μg/ml), rLG proteins (25 μg/ml), or Ln2-P3 (22 μg/cm2) for 1 h in serum-free medium. B, cell adhesion to laminin, rLG proteins, and Ln2-P3 in PC12 cells pretreated with heparin for 10 min at room temperature. The assay conditions were the same as described in A. C and D, adhesion of heparin to rLG1 and Ln2-P3. Enzyme-linked immunosorbent assay plates were coated with rLG proteins or synthetic peptides for 12 h at 4 °C, blocked with 3% BSA for 2 h at 37 °C, and then incubated with 10 ng/well of biotinylated heparin for 1 h at 37 °C. The biotinylated heparin bound to rLG proteins or synthetic peptides was detected using streptavidin-conjugated horseradish peroxidase. E, inhibition of cell adhesion to rLG1 and Ln2-P3 in cells pretreated with heparan sulfate and dermatan sulfate. PC12 cells were pretreated with 100 μg/ml of heparin, heparan sulfate (HS), hyaluronic acid (HA), chondroitin sulfate A (CSA), dermatan sulfate (DS), chondroitin sulfate C (CSC), or de-N-sulfated heparin (de-N-s hep) for 10 min at room temperature and seeded on plates precoated with rLG1(25 μg/ml) or Ln2-P3 (22 μg/cm2) for 1 h in serum-free medium. The number of adherent cells was quantified by cell counting. F, inhibition of cell adhesion to rLG1 and Ln2-P3 in cells pretreated with heparitinase I or chondroitinase ABC for 90 min at 37 °C. The assay conditions were the same as described in E. Values are expressed as a percentage of the value for cells pretreated without EDTA or integrin β1 antibody, heparin, glycosaminoglycans, or enzymes (None) in A, B, E, and F (mean ± S.D., n = 3). Values are expressed as the mean ± S.D. (n = 4) in C and D. *, p < 0.01; **, p < 0.05.
Because LG1 and Ln2-P3 contain a heparin binding region, we next examined the specificity of heparin binding of rLG1 and Ln2-P3 via adhesion inhibition assays using various glycosaminoglycans as competitors. Cell adhesion to rLG1 and Ln2-P3 in cells pretreated with heparin was completely inhibited, whereas the adhesion failed to inhibit in cells pretreated with hyaluronic acid, chondroitin sulfate A, chondroitin sulfate C, and de-N-sulfated heparin (Fig. 3E). On the other hand, heparan sulfate (∼44% for rLG1 and 46% for Ln2-P3) and dermatan sulfate (∼19% for rLG1 and 33% for Ln2-P3) partially inhibited cell adhesion to both rLG1 and Ln2-P3 (Fig. 3E). To confirm the involvement of heparan sulfate and dermatan sulfate in cell adhesion, changes in cell adhesion levels to rLG1 and Ln2-P3 were evaluated after enzyme digestion of PC12 cells with heparitinase I or chondroitinase ABC. As expected, preincubation of PC12 cells with heparitinase I and chondroitinase ABC significantly inhibited cell adhesion to rLG1 and Ln2-P3, respectively, in conjunction with increasing enzyme activities (Fig. 3F). Changes of cell adhesion levels were higher in the heparitinase I-treated cells than in the chondroitinase ABC-treated cells (Fig. 3F). Overall, these results suggest that LG1 and Ln2-P3 mediate cell adhesion via cell surface heparan sulfate and dermatan sulfate and that the affinities of LG1 and Ln2-P3 might be higher to cell surface heparan sulfate than to dermatan sulfate.
Syndecan-1 Mediates LG1- and Ln2-P3-induced Cell Adhesion
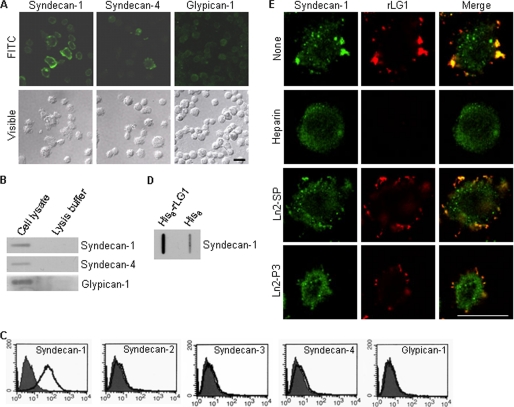
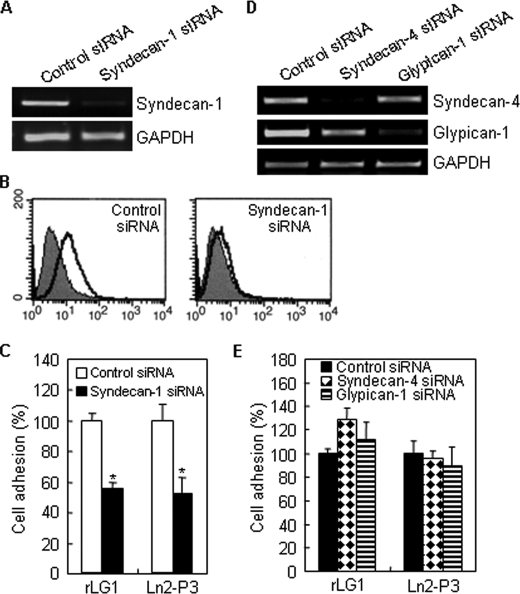
Currently, the identities of the receptors for the human laminin α2 LG1 domain and synthetic peptides derived from LG1 domain are unclear. Therefore, we identified what specific adhesion receptor for LG1 and Ln2-P3 mediates adhesion of PC12 cells. Because rLG1 and Ln2-P3 were shown to bind to both heparan sulfate and dermatan sulfate, we set out to identify the particular cell surface proteoglycan receptor(s) through which LG1 and Ln2-P3 mediate cell adhesion. Furthermore, numerous studies show that LG domains and synthetic peptides derived from the laminin α chain bind to syndecans in several cells and cell types (16, 24, 29–31). Our first step was to identify the syndecans family that is expressed on the surface of PC12 cells. PC12 cells expressed syndecan-1 and -4 (Fig. 4, A and B) but not syndecan-2 and -3 (data not shown). Glypicans are also cell surface heparin sulfate proteoglycans. Immunostaining (Fig. 4A) and immunodot blot analysis (Fig. 4B) showed that glypican-1 is expressed in PC12 cells. These results are consistent with a previous report (32) showing that glypican-1 is expressed in PC12 cells. We further analyzed the levels of expression of syndecans and glypican-1 by flow cytometry. PC12 cells only expressed syndecan-1 at the cell surface, but syndecan-2, -3, and -4, and glypican-1 were not found (Fig. 4C). The results suggest that syndecan-1 is available as a cell surface receptor for the LG1 domain and Ln2-P3 motif in PC12 cells. Next, to identify a cell surface receptor for LG1 domain, we used immunodot blot analysis to show that syndecan-1 markedly bound to rLG1 (Fig. 4D), demonstrating that LG1 binds to PC12 cells through the syndecan-1. Thus, to confirm that syndecan-1 is a receptor for the LG1 and Ln2-P3, we investigated whether rLG1 and syndecan-1 exist as associated complex in PC12 cells. Immunofluorescence analysis showed the colocalization of rLG1 with syndecan-1 (Fig. 4E). The binding of rLG1 to syndecan-1 was completely inhibited by heparin and less effectively inhibited by Ln2-P3 compared with vehicle or scrambled peptide (Fig. 4E). These results indicate that the LG1 domain interacts with syndecan-1 on the surface of PC12 cells and that Ln2-P3 may also interact with syndecan-1 in the cells. Nevertheless, we did not eliminate the possibility that other glypican isoforms except glypican-1 could function as a cell surface receptor for LG1 domain and Ln2-P3 motif in PC12 cells.
FIGURE 4.
Syndecan-1 binds to LG1 domain and in part to Ln2-P3 motif. A, expression of syndecan-1 and -4 and glypican-1 in PC12 cells. Cells were seeded on glass slide chambers for 2 days and immunostained with relevant antibodies (green). FITC, fluorescein isothiocyanate. B, immunodot blot analysis of the syndecan-1 and -4 and glypican-1 in PC12 cells. C, fluorescence-activated cell sorter analysis of PC12 cells assessing expression of syndecan isoforms and glypican-1 by using relevant antibodies (white area) compared with IgG-control (gray area). D, lysates were incubated with beads containing His6 alone or His6-rLG1, and the bound proteins were subjected to immunodot blotting and analyzed for syndecan-1 expression. Results are representative of three independent experiments. E, colocalization of rLG1 with syndecan-1. PC12 cells were seeded on glass slide chambers for 2 days, fixed, treated with heparin, scrambled peptide (Ln2-SP), Ln2-P3, or without the molecule (None) for 2 h at room temperature and further incubated with 25 μg/ml of rLG1 for 12 h at 4 °C. The cells were immunostained with anti-syndecan-1 antibody (green) and anti-His6 antibody (red). Scale bar, 20 μm.
Suppression of Syndecan-1 Inhibits Cell Adhesion to LG1 Domain and Ln2-P3 Motif
A direct causal role for syndecan-1 and -4 and glypican-1 in adhesion of PC12 cells to LG1 and Ln2-P3 was further addressed by examining the consequence of suppressing syndecan-1 and -4 and glypican-1 with RNA interference. Thus, we attenuated syndecan-1 and -4 and glypican-1 expression by transfecting cells with the corresponding siRNAs, to examine the effect on the adhesion of PC12 cells. As expected, the expression of syndecan-1 and -4 and glypican-1 in transfected cells was significantly reduced compared with control siRNA-transfected cells (Fig. 5, A, B, and D). Suppression of syndecan-1 expression significantly inhibited cell adhesion to rLG1 and Ln2-P3 (Fig. 5C), but the suppression of syndecan-4 and glypican-1 did not inhibit cell adhesion (Fig. 5E). Overall, these data confirm that syndecan-1, but not syndecan-4 and glypican-1, functions as a receptor for adhesion of PC12 cells to LG1 and Ln2-P3.
FIGURE 5.
PC12 cells attach to rLG1 and Ln2-P3 through syndecan-1. PC12 cells were transfected with control siRNA, syndecan-1 siRNA, syndecan-4 siRNA, or glypican-1. A, RT-PCR analysis of the expression of syndecan-1. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. B, fluorescence-activated cell sorter analysis of syndecan-1 siRNA-transfected cells assessing syndecan-1 expression by using an antibody against syndecan-1 (white area) compared with IgG control (gray area). C, inhibition of cell adhesion to rLG1 and Ln2-P3 in syndecan-1 siRNA-transfected cells. siRNA-transfected cells were seeded on rLG1- or Ln2-P3-coated plates for 1 h in serum-free medium. The number of adherent cells was quantified by cell counting. Results are expressed as a percentage from the value of control siRNA-transfected cells (mean ± S.D., n = 3). *, p < 0.01. D, RT-PCR analysis of the expression of syndecan-4 and glypican-1. E, adhesion of syndecan-4 siRNA- and glypican-1 siRNA-transfected cells to rLG1 and Ln2-P3 showing no difference to the control siRNA-transfected cells. The assay conditions were the same as described in C.
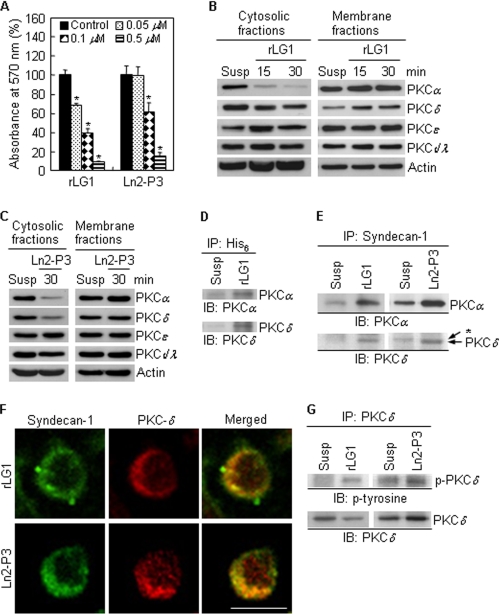
Stimulation of LG1 Domain and Ln2-P3 Motif via the Syndecan-1 Results in Translocation of PKCδ from the Cytosol to the Syndecan-1 and Induces Phosphorylation of PKCδ
Syndecans play an important role in cellular functions, such as cell proliferation, cell-matrix and cell-cell adhesion, cell migration, and focal adhesion formation (33, 34). Specifically, syndecan-4 has been known to regulate the localization of PKCα from the cytosol to the plasma membrane and the stability of PKCα (35, 36); however, the downstream signaling pathways mediated by other syndecan isoforms are obscure. First of all, we investigated whether PKC is involved in LG1- and Ln2-P3-mediated cell adhesion. Cell adhesion activities to rLG1 and Ln2-P3 were significantly inhibited in PC12 cells pretreated with calphostin C, an inhibitor of PKC, compared with cells without pretreatment in a dose-dependent manner (Fig. 6A). In addition, calphostin C did not affect cell growth at the indicated concentrations and times (data not shown), suggesting that the inhibition of cell adhesion to rLG1 and Ln2-P3 is not caused by cellular cytotoxicity and that PKC is required for adhesion of PC12 cells. Next, to test which PKC isoforms are involved in the signaling pathways mediated by either LG1 or Ln2-P3, we determined the levels of PKC isoforms (α, δ, ϵ, λ, and ι) in the cytosolic and membrane fractions of PC12 cells seeded on rLG1- and Ln2-P3-coated dishes. rLG1 induced translocation of the PKCα and PKCδ from the cytosol to the membrane fraction (Fig. 6B), whereas Ln2-P3 showed only a slight translocation (Fig. 6C). On the other hand, the levels of PKCϵ and -ι/λ in the cytosolic and membrane fractions of PC12 cells were not affected by cell adhesion to rLG1 and Ln2-P3 (Fig. 6, B and C). Overall, these data suggest that interaction of syndecan-1 with either LG1 domain or Ln2-P3 motif regulates localization of PKCα and -δ.
FIGURE 6.
Localization and tyrosine phosphorylation of PKCδ are regulated by LG1 and Ln2-P3. A, inhibition of cell adhesion to rLG1 and Ln2-P3 by calphostin C treatment. PC12 cells were pretreated with calphostin C, an inhibitor for PKC, for 15 min at 37 °C and then seeded on plates precoated with rLG1 and Ln2-P3 for 1 h in serum-free medium. Values are expressed as a percentage of the value for cells pretreated without calphostin C (mean ± S.D., n = 3). *, p < 0.01. B and C, immunoblots of PKC isoforms from cytosolic and membrane fractions of cells cultured on rLG1- or Ln2-P3-coated dishes. PC12 cells were suspended for 15 min and then seeded on rLG1- or Ln2-P3-coated dishes for the indicated times. Susp, suspension of PC12 cells for 45 min. D and E, interactions of His6 and syndecan-1 with PKCα and -δ, respectively, in PC12 cells seeded on rLG1- or Ln2-P3-coated dishes for 30 min. Lysates were subjected to immunoprecipitation and analyzed for PKCα and -δ expression. *, nonspecific signal. IP, immunoprecipitation; IB, immunoblotting. F, colocalization of PKCδ with syndecan-1. PC12 cells were seeded on glass slide chambers precoated with rLG1 or Ln2-P3 for 30 min and immunostained with anti-syndecan-1 antibody (green) and anti-PKCδ antibody (red). Scale bar, 10 μm. G, interaction of PKCδ with phosphotyrosine in PC12 cells seeded on rLG1- or Ln2-P3-coated dishes for 30 min.
Because PKCα and -δ are translocated from the cytosol to the plasma membrane by LG1- and Ln2-P3-mediated cell adhesion, it is important to identify the targeting molecule(s) that is bound to PKCα and -δ. Therefore, we examined whether the PKCα and -δ would be translocated to the cell surface receptor syndecan-1. We investigated whether PKCα or PKCδ and syndecan-1 exist as associated complexes in cells cultured on rLG1- and Ln2-P3-coated dishes. Our first step was to identify whether His6 or syndecan-1 and PKCα and -δ exist as associated complexes in PC12 cells cultured on rLG1- and Ln2-P3-coated dishes. In anti-His6 immunoprecipitates, PKCα and -δ were observed in cells cultured on rLG1-coated dishes, indicating that rLG1 associates with PKCα and -δ (Fig. 6D). Similarly, in syndecan-1 immunoprecipitates, PKCα and -δ were observed in cells cultured on rLG1- and Ln2-P3-coated dishes (Fig. 6E). However, the association of syndecan-1 and PKCα, but not that of syndecan-1 and PKCδ, was also found in suspended cells (Fig. 6E), suggesting that the association of syndecan-1 and PKCα rises in a cell adhesion-independent manner. Immunofluorescence analysis also showed the colocalization of PKCδ with syndecan-1 (Fig. 6F) and that of PKCα with syndecan-1 (supplemental Fig. 1) in cells cultured on rLG1- and Ln2-P3-coated glass slide chambers. Growth factors, such as epidermal growth factor and platelet-derived growth factor, are known to induce the tyrosine phosphorylation of PKCδ (37). Therefore, we next examined whether the tyrosine phosphorylation of PKCδ would be affected by LG1 and Ln2-P3 in PC12 cells. We determined protein levels of p-PKCδ-Tyr in cells cultured on rLG1- and Ln2-P3-coated dishes. Adhesion of PC12 cells to rLG1 (Fig. 6G, left panel) or Ln2-P3 (Fig. 6G, right panel) induced tyrosine phosphorylation of PKCδ. This suggests that p-PKCδ-Tyr affects cell adhesion when phosphorylated by stimulation of LG1 domain and Ln2-P3 motif via the syndecan-1.
Taken together, these results indicate that PKCδ is translocated from the cytosol to the cell surface receptor syndecan-1 by LG1- and Ln2-P3-mediated cell adhesion and that Ln2-P3 motif and LG1 domain, containing the motif, within human laminin α2 chain promote cell adhesion through the syndecan-1 and the PKCδ signaling pathway.
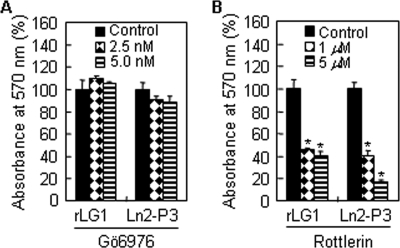
Functional Role of PKCδ in LG1- and Ln2-P3-induced Cell Adhesion
Finally, we focused on examining the functional roles of PKCα and -δ in cell adhesion. Thus, we performed adhesion assay in the presence of the PKCα/β inhibitor Gö6976 (IC50 = 2.3 nm) or the PKCδ inhibitor rottlerin (IC50 = 3–6 μm). Gö6976 treatment did not impair adhesion of PC12 cells to rLG1 or Ln2-P3 (Fig. 7A); however, rottlerin treatment significantly reduced adhesion of PC12 cells to rLG1 and Ln2-P3 (Fig. 7B). In addition, Gö6976 and rottlerin did not affect cell viability at the indicated concentrations and times (data not shown). These results clearly indicate that the PKCδ signaling is associated with LG1- and Ln2-P3-mediated cell adhesion.
FIGURE 7.
Role of PKCα and -δ in LG1- and Ln2-P3-induced cell adhesion. A and B, Gö6976, a PKCα inhibitor, did not inhibit adhesion of PC12 cells seeded on rLG1- or Ln2-P3-coated plates, but rottlerin, a PKCδ inhibitor, significantly inhibited cell adhesion. PC12 cells were pretreated with Gö6976 or rottlerin for 15 min at 37 °C and seeded on rLG1- or Ln2-P3-coated plates for 1 h in serum-free medium. Values are expressed as a percentage of the value for cells pretreated without Gö6976 or rottlerin (mean ± S.D., n = 3). *, p < 0.01.
DISCUSSION
Numerous reports indicate that mouse laminin α2 LG domains have been shown to have several biological functions (11, 12, 15) and that several active sites for cell adhesion and heparin binding have been identified within the mouse laminin α2 LG4 domain (14, 17). However, cell binding sequences within LG1-LG3 domains of the human laminin α2 chain and their receptors have not been identified. Therefore, we individually expressed the three human laminin α2 LG domains (LG1, LG2, and LG3) as monomeric, soluble fusion proteins, and we examined their proper folding and biological function. CD spectroscopy, a sensitive method for determining protein secondary structures, was used to assess the folding of purified rLG proteins. CD spectra of recombinant His6-tagged LG proteins are characteristic of proteins rich in β-structure, suggesting that rLG proteins have proper folding. We demonstrated that the human laminin α2 LG1 domain has cell adhesion activity and binds to syndecan-1. More significantly, we identified a novel motif (DLTIDDSYWYRI, amino acids 2221–2232) that is crucial for cell adhesion and syndecan-1 binding within the human laminin α2 LG1 domain to investigate the possibility for biomedical usefulness. The motif had a broad cell-type specificity of adhesion and also induced neurite outgrowth in PC12 cells (supplemental Fig. 2). The present finding agrees with a previous report (15) that synthetic peptides derived from laminin α1 and α2 chains promote neurite outgrowth in various neuronal cells.
The conformations of proteins and peptides are usually essential to elucidating their mechanisms of actions. To predict the location of the Ln2-P3 motif in the crystal structure of the laminin α2 LG1 domain, we aligned the amino acid sequences of Ln2-P3 of the laminin α2 LG1 domain among mice, rats, and humans using a structure-based sequence alignment. An analysis of the crystal structure of laminin α2 LG5 domain predicts that all the LG domains in the laminin α chains have a 14-stranded β-sandwich structure (12). Previous reports indicate that peptides AG-73 (RKRLQVQLSIRT, amino acids 2719–2730) and MG-73 (amino acids 2780–2791), which are located within β-strand C of the mouse laminin α1 and α2 chain, respectively, possess heparin binding activity and bind to syndecan-1 (16). F4 peptide that has heparin binding activity is located in the H-strand of the mouse laminin α5 chain (30, 38, 39). Moreover, peptides A3G75aR (NSFMALYLSKGR, amino acids 1412–1423) and A4G82 (TLFLAHGRLVFM, amino acids 1514–1525), which also have heparin binding activity, are located in the connecting loop of the E and F strands of the human laminin α3 chain and mouse laminin α4 chain, respectively. Similarly, Ln2-P3 corresponded to the loop region between the β strands G and H.
Similar studies have been reported previously for mouse laminin α2 LG1 domain with somewhat different results (14), which indicate that the MG-10 peptide (SYWYRIEASRTG, amino acids 2223–2234) of the mouse laminin α2 LG1 domain supports cell spreading and binds to integrins. In this study, Ln2-P4 (SYWYRIVASRTG, amino acids 2227–2238) of the human laminin α2 LG1 domain, a homologous to sequence of MG-10, did not show cell adhesion activity, even at high coating concentrations (Fig. 2, A and B, and supplemental Fig. 3). Therefore, to elucidate real differences between species, we determined cell adhesion activities of related peptides and compared them. Here, we observed that the MG-10 peptide significantly promoted adhesion of PC12 cells (supplemental Fig. 3). Overall, these findings indicate that substitution of the Val residue with the Glu residue dramatically increases cell adhesion activity, suggesting that the Glu residue in MG-10 peptide plays an important role in cell adhesion. This finding is partly supported by a previous report (40), which explains that Glu residue in LALERKDHSG motif of thrombospondin-1 N-terminal domain is required for integrin α6β1 binding.
Currently, the identities of the receptors for the human laminin α2 LG1 domain and synthetic peptides derived from LG1 domain are largely unknown. Here, we found that the LG1 domain and the Ln2-P3 motif bound to the heparan sulfate and demartan sulfate chains of syndecan-1. Moreover, the binding affinity of heparan sulfate is higher than that of demartan sulfate. The difference in binding affinity may be due to fine structural differences (24). Similarly, it has been reported that the heparan sulfate and chondroitin 4-sulfate chains of syndecan-1 bind to the human laminin α3 LG4/5 domain; however, heparan sulfate has a higher affinity than chondroitin 4-sulfate (24). Four different sites of sulfation are found at the N-, 3-O-, and 6-O-positions of glucosamine and at the 2-O-position of hexuronic acid residues (41). The sulfation pattern of heparan sulfate is also tissue-specific (42). These differences in sulfation pattern could change the affinities of a growth factor or extracellular matrix protein. For example, fibroblast growth factor-2 binding requires N-sulfate and 2-O-sulfation of iduronic acid, whereas fibronectin binding to heparin sulfate requires N-sulfation (43, 44). In this study, we showed that de-N-sulfated heparin, a derivative in which N-sulfate groups of the N-sulfated glucosamine residues of heparin was removed, did not inhibit PC12 cell adhesion to LG1 and Ln2-P3, suggesting that N-sulfation might be important for cell binding.
Although a previous report has demonstrated that syndecan-4 directly binds to PKCα through the cytoplasmic domain and increases localization of PKCα to focal adhesions (35), the mechanism by which syndecan-1 promotes cell adhesion had remained largely unknown. Here, we investigated that interaction of syndecan-1 with PKCδ was dependent on cell adhesion to LG1 and Ln2-P3, which was prevented by the PKCδ inhibitor rottlerin. These findings suggest that PKCδ could play an important role for LG1- and Ln2-P3-mediated cell adhesion via the syndecan-1. In this study, we also found that recruitment of PKCα to the syndecan-1 is independent on cell adhesion to LG1 and Ln2-P3. Furthermore, inhibition of PKCα by the specific inhibitor Gö6976 does not prevent cell adhesion to LG1 and Ln2-P3, suggesting that activation of PKCα may be required for regulating the cell-cell contacts of PC12 cells. In fact, syndecan-1 is an important mediator of cell-cell contacts (45), and PKCα and -ϵ are selectively targeted cell-cell contacts of pituitary GH3B6 cells (46).
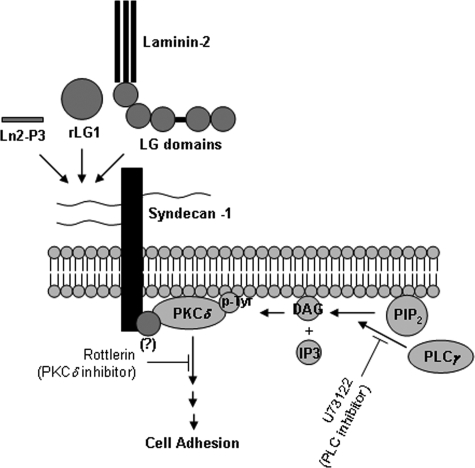
The activation of PKC results from the increase in intracellular diacylglycerol levels, which is mediated by phospholipase C (47), suggesting that phospholipase C may be required for the activation of PKCδ and the syndecan-1-mediated cell adhesion. In this study, we demonstrated that adhesion of PC12 cells to LG1 and Ln2-P3 is mediated by syndecan-1, which induces the tyrosine phosphorylation of PKCδ, and that the phospholipase C inhibitor U73122 significantly inhibits cell adhesion to LG1 and Ln2-P3 (supplemental Fig. 4). These findings are supported by a previous report (48), which explains that U73122 treatment reduces adhesion of PC12 cells to laminin and collagen IV. Furthermore, tyrosine phosphorylation of PKCδ is blocked by U73122 in rat parotid acinar cells (49). To identify the upstream regulators of PKCδ activation and tyrosine phosphorylation, future studies will be needed (Fig. 8). In conclusion, our findings constitute the first report that explains that the DLTIDDSYWYRI motif and the LG1 domain, containing the motif, within the human laminin α2 chain are the major binding sites for heparan sulfate and dermatan sulfate chains of syndecan-1, which recruits PKCδ to the membrane and induces the tyrosine phosphorylation of PKCδ, thus promoting cell adhesion.
FIGURE 8.
Proposed pathway for promotion of cell adhesion by laminin-2, Ln2-P3 motif, and LG1 domain, containing the motif, within the human laminin α2 chain.
Supplementary Material
This work was supported by grants from the Korea Science and Engineering Foundation through the Intellectual Biointerface Engineering Center at Seoul National University (to B.-M. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- LG
- large globular
- PKC
- protein kinase C
- rLG
- recombinant LG
- His6
- histidine 6
- Ln2-P3
- PPFLMLLKGSTR motif
- BSA
- bovine serum albumin
- PBS
- phosphate-buffered saline
- RT
- reverse transcription.
REFERENCES
- 1.Colognato H., Yurchenco P. D. (2000) Dev. Dyn. 218, 213–234 [DOI] [PubMed] [Google Scholar]
- 2.Ekblom M., Falk M., Salmivirta K., Durbeej M., Ekblom P. (1998) Ann. N.Y. Acad. Sci. 857, 194–211 [DOI] [PubMed] [Google Scholar]
- 3.Suzuki N., Yokoyama F., Nomizu M. (2005) Connect. Tissue Res. 46, 142–152 [DOI] [PubMed] [Google Scholar]
- 4.Leivo I., Engvall E. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 1544–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sewry C. A., D'Alessandro M., Wilson L. A., Sorokin L. M., Naom I., Bruno S., Ferlini A., Dubowitz V., Muntoni F. (1997) Neuropediatrics 28, 217–222 [DOI] [PubMed] [Google Scholar]
- 6.Edwards J. P., Hatton P. A., Wareham A. C. (1998) Brain Res. 788, 262–268 [DOI] [PubMed] [Google Scholar]
- 7.Feltri M. L., Wrabetz L. (2005) J. Peripher. Nerv. Syst. 10, 128–143 [DOI] [PubMed] [Google Scholar]
- 8.Masaki T., Matsumura K., Saito F., Sunada Y., Shimizu T., Yorifuji H., Motoyoshi K., Kamakura K. (2000) Acta Neuropathol. 99, 289–295 [DOI] [PubMed] [Google Scholar]
- 9.Matsumura K., Yamada H., Saito F., Sunada Y., Shimizu T. (1997) Neuromuscul. Disord. 7, 7–12 [DOI] [PubMed] [Google Scholar]
- 10.Vuolteenaho R., Nissinen M., Sainio K., Byers M., Eddy R., Hirvonen H., Shows T. B., Sariola H., Engvall E., Tryggvason K. (1994) J. Cell Biol. 124, 381–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wizemann H., Garbe J. H., Friedrich M. V., Timpl R., Sasaki T., Hohenester E. (2003) J. Mol. Biol. 332, 635–642 [DOI] [PubMed] [Google Scholar]
- 12.Tisi D., Talts J. F., Timpl R., Hohenester E. (2000) EMBO J. 19, 1432–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smirnov S. P., McDearmon E. L., Li S., Ervasti J. M., Tryggvason K., Yurchenco P. D. (2002) J. Biol. Chem. 277, 18928–18937 [DOI] [PubMed] [Google Scholar]
- 14.Nomizu M., Song S. Y., Kuratomi Y., Tanaka M., Kim W. H., Kleinman H. K., Yamada Y. (1996) FEBS Lett. 396, 37–42 [DOI] [PubMed] [Google Scholar]
- 15.Richard B. L., Nomizu M., Yamada Y., Kleinman H. K. (1996) Exp. Cell Res. 228, 98–105 [DOI] [PubMed] [Google Scholar]
- 16.Hoffman M. P., Nomizu M., Roque E., Lee S., Jung D. W., Yamada Y., Kleinman H. K. (1998) J. Biol. Chem. 273, 28633–28641 [DOI] [PubMed] [Google Scholar]
- 17.Suzuki N., Nakatsuka H., Mochizuki M., Nishi N., Kadoya Y., Utani A., Oishi S., Fujii N., Kleinman H. K., Nomizu M. (2003) J. Biol. Chem. 278, 45697–45705 [DOI] [PubMed] [Google Scholar]
- 18.Yeo I. S., Oh J. E., Jeong L., Lee T. S., Lee S. J., Park W. H., Min B. M. (2008) Biomacromolecules 9, 1106–1116 [DOI] [PubMed] [Google Scholar]
- 19.Kim J. M., Park W. H., Min B. M. (2005) Exp. Cell Res. 304, 317–327 [DOI] [PubMed] [Google Scholar]
- 20.Okazaki I., Suzuki N., Nishi N., Utani A., Matsuura H., Shinkai H., Yamashita H., Kitagawa Y., Nomizu M. (2002) J. Biol. Chem. 277, 37070–37078 [DOI] [PubMed] [Google Scholar]
- 21.Talts J. F., Mann K., Yamada Y., Timpl R. (1998) FEBS Lett. 426, 71–76 [DOI] [PubMed] [Google Scholar]
- 22.Talts J. F., Timpl R. (1999) FEBS Lett. 458, 319–323 [DOI] [PubMed] [Google Scholar]
- 23.Hall H., Bozic D., Michel K., Hubbell J. A. (2003) Mol. Cell. Neurosci. 24, 1062–1073 [DOI] [PubMed] [Google Scholar]
- 24.Okamoto O., Bachy S., Odenthal U., Bernaud J., Rigal D., Lortat-Jacob H., Smyth N., Rousselle P. (2003) J. Biol. Chem. 278, 44168–44177 [DOI] [PubMed] [Google Scholar]
- 25.Charonis A. S., Skubitz A. P., Koliakos G. G., Reger L. A., Dege J., Vogel A. M., Wohlhueter R., Furcht L. T. (1988) J. Cell Biol. 107, 1253–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gehlsen K. R., Sriramarao P., Furcht L. T., Skubitz A. P. (1992) J. Cell Biol. 117, 449–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tashiro K., Nagata I., Yamashita N., Okazaki K., Ogomori K., Tashiro N., Anai M. (1994) Biochem. J. 302, 73–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoshida I., Tashiro K., Monji A., Nagata I., Hayashi Y., Mitsuyama Y., Tashiro N. (1999) J. Cell. Physiol. 179, 18–28 [DOI] [PubMed] [Google Scholar]
- 29.Hozumi K., Suzuki N., Nielsen P. K., Nomizu M., Yamada Y. (2006) J. Biol. Chem. 281, 32929–32940 [DOI] [PubMed] [Google Scholar]
- 30.Utani A., Nomizu M., Matsuura H., Kato K., Kobayashi T., Takeda U., Aota S., Nielsen P. K., Shinkai H. (2001) J. Biol. Chem. 276, 28779–28788 [DOI] [PubMed] [Google Scholar]
- 31.Yamashita H., Goto A., Kadowaki T., Kitagawa Y. (2004) Biochem. J. 382, 933–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malavé C., Villegas G. M., Hernández M., Martínez J. C., Castillo C., Suárez de Mata Z., Villegas R. (2003) Brain Res. 983, 74–83 [DOI] [PubMed] [Google Scholar]
- 33.Carey D. J. (1997) Biochem. J. 327, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Couchman J. R., Woods A. (1999) J. Cell Sci. 112, 3415–3420 [DOI] [PubMed] [Google Scholar]
- 35.Lim S. T., Longley R. L., Couchman J. R., Woods A. (2003) J. Biol. Chem. 278, 13795–13802 [DOI] [PubMed] [Google Scholar]
- 36.Keum E., Kim Y., Kim J., Kwon S., Lim Y., Han I., Oh E. S. (2004) Biochem. J. 378, 1007–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steinberg S. F. (2004) Biochem. J. 384, 449–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nielsen P. K., Gho Y. S., Hoffman M. P., Watanabe H., Makino M., Nomizu M., Yamada Y. (2000) J. Biol. Chem. 275, 14517–14523 [DOI] [PubMed] [Google Scholar]
- 39.Yamaguchi H., Yamashita H., Mori H., Okazaki I., Nomizu M., Beck K., Kitagawa Y. (2000) J. Biol. Chem. 275, 29458–29465 [DOI] [PubMed] [Google Scholar]
- 40.Calzada M. J., Sipes J. M., Krutzsch H. C., Yurchenco P. D., Annis D. S., Mosher D. F., Roberts D. D. (2003) J. Biol. Chem. 278, 40679–40687 [DOI] [PubMed] [Google Scholar]
- 41.Salmivirta M., Lidholt K., Lindahl U. (1996) FASEB J. 10, 1270–1279 [DOI] [PubMed] [Google Scholar]
- 42.Maccarana M., Sakura Y., Tawada A., Yoshida K., Lindahl U. (1996) J. Biol. Chem. 271, 17804–17810 [DOI] [PubMed] [Google Scholar]
- 43.Lundin L., Larsson H., Kreuger J., Kanda S., Lindahl U., Salmivirta M., Claesson-Welsh L. (2000) J. Biol. Chem. 275, 24653–24660 [DOI] [PubMed] [Google Scholar]
- 44.Lyon M., Rushton G., Askari J. A., Humphries M. J., Gallagher J. T. (2000) J. Biol. Chem. 275, 4599–4606 [DOI] [PubMed] [Google Scholar]
- 45.Stanley M. J., Liebersbach B. F., Liu W., Anhalt D. J., Sanderson R. D. (1995) J. Biol. Chem. 270, 5077–5083 [DOI] [PubMed] [Google Scholar]
- 46.Quittau-Prévostel C., Delaunay N., Collazos A., Vallentin A., Joubert D. (2004) J. Cell Sci. 117, 63–72 [DOI] [PubMed] [Google Scholar]
- 47.Berridge M. J., Irvine R. F. (1989) Nature 341, 197–205 [DOI] [PubMed] [Google Scholar]
- 48.Vossmeyer D., Hofmann W., Löster K., Reutter W., Danker K. (2002) J. Biol. Chem. 277, 4636–4643 [DOI] [PubMed] [Google Scholar]
- 49.Benes C., Soltoff S. P. (2001) Am. J. Physiol. Cell Physiol. 280, C1498–C1510 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.