Abstract
Plakoglobin and β-catenin are homologous armadillo repeat proteins found in adherens junctions, where they interact with the cytoplasmic domain of classical cadherins and with α-catenin. Plakoglobin, but normally not β-catenin, is also a structural constituent of desmosomes, where it binds to the cytoplasmic domains of the desmosomal cadherins, desmogleins and desmocollins. Here, we report structural, biophysical, and biochemical studies aimed at understanding the molecular basis of selective exclusion of β-catenin and α-catenin from desmosomes. The crystal structure of the plakoglobin armadillo domain bound to phosphorylated E-cadherin shows virtually identical interactions to those observed between β-catenin and E-cadherin. Trypsin sensitivity experiments indicate that the plakoglobin arm domain by itself is more flexible than that of β-catenin. Binding of plakoglobin and β-catenin to the intracellular regions of E-cadherin, desmoglein1, and desmocollin1 was measured by isothermal titration calorimetry. Plakoglobin and β-catenin bind strongly and with similar thermodynamic parameters to E-cadherin. In contrast, β-catenin binds to desmoglein-1 more weakly than does plakoglobin. β-Catenin and plakoglobin bind with similar weak affinities to desmocollin-1. Full affinity binding of desmoglein-1 requires sequences C-terminal to the region homologous to the catenin-binding domain of classical cadherins. Although pulldown assays suggest that the presence of N- and C-terminal β-catenin “tails” that flank the armadillo repeat region reduces the affinity for desmosomal cadherins, calorimetric measurements show no significant effects of the tails on binding to the cadherins. Using purified proteins, we show that desmosomal cadherins and α-catenin compete directly for binding to plakoglobin, consistent with the absence of α-catenin in desmosomes.
Introduction
Adherens junctions and desmosomes are two kinds of intercellular junctions that mediate strong attachments between adjacent cells and are essential for embryonic development and epithelial tissue differentiation (1–5). These junctions share a parallel architecture in which the extracellular regions of transmembrane cadherin cell adhesion proteins mediate cell-cell contacts, whereas the intracellular regions of cadherins associate with the underlying cytoskeleton (3, 4). Adherens junctions contain the classical cadherins, which are functionally linked to the actin-based cytoskeleton (1, 6). Desmosomal cadherins, in contrast, are linked to the intermediate filament system (2).
In adherens junctions, the intracellular region of classical cadherins binds to the protein β-catenin or plakoglobin. β-Catenin binds strongly to the cytoplasmic domain of E-cadherin (Ecyto)2 with a dissociation constant KD of 36 nm or 52 pm, depending on the phosphorylation state of Ecyto (7). β-Catenin and plakoglobin bind to the F-actin-binding protein α-catenin (6). α-Catenin may have a role in linking the cadherin-β-catenin complex to actin, but binding to β-catenin weakens the affinity of α-catenin for F-actin; it appears that α-catenin also serves to regulate actin dynamics and organization (8, 9).
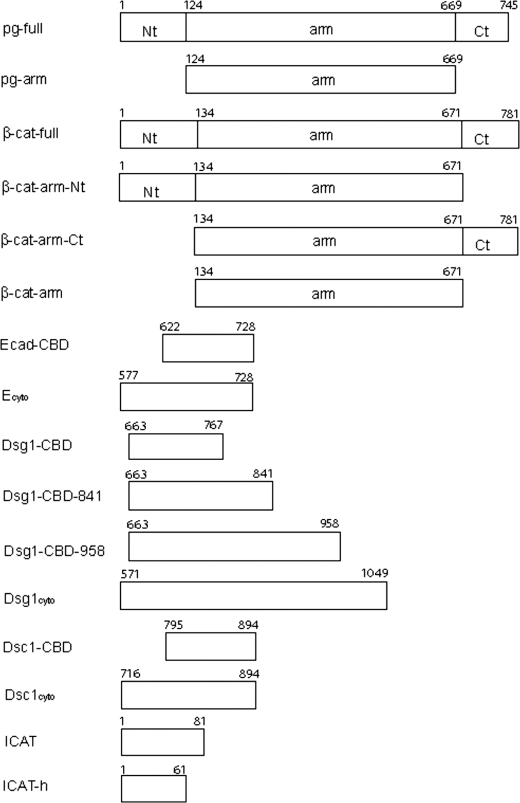
The primary structures of plakoglobin and β-catenin consist of a central armadillo repeat (arm) domain, flanked by the N- and C-terminal “tails” (Fig. 1). The tails appear to be unstructured, as they are highly sensitive to proteolysis both in β-catenin (10) and plakoglobin (see below), and circular dichroism analysis of the plakoglobin N-terminal 79 amino acids reveals no regular secondary structure (data not shown). Moreover, the sequences of these regions are predicted to be intrinsically disordered using the DisEMBL server (11). The sequences of the arm domains of these two proteins are 79% identical, whereas the N- and C-terminal tails display lower identities of 45 and 27%, respectively. The arm domain of β-catenin (βcat-arm) is an elongated structure of 12 arm repeats, each of which consists of three α-helices. Successive arm repeats pack together to form a superhelical structure that features a 95-Å long positively charged groove (10). The groove proves to be the binding site of many partners, including cadherins, Tcf family transcription factors, the adenomatous polyposis coli tumor suppressor Axin, and the transcriptional inhibitor ICAT (see Ref. 7 and references therein). No structural information has been available for plakoglobin, but its high sequence identity with β-catenin arm domains suggests that the two structures are very similar, including their binding sites for cadherins and other ligands.
FIGURE 1.
Constructs used in ITC experiments and pulldown assays. The N- and C-terminal tails and the central armadillo domains of plakoglobin and β-catenin are represented as Nt, Ct, and arm, respectively. The numbers shown on top of each construct represent the beginning and ending residue numbers.
The desmosomal cadherins include four desmoglein isoforms (Dsg1–4) and three desmocollins (Dsc1–3) (5, 12). Analogous to the arrangement in adherens junctions, the intracellular portions of these proteins bind to plakoglobin. Dsgs have large cytoplasmic tails of about 330–490 amino acids that contain an ∼100-amino acid-long plakoglobin-binding domain that is homologous to the β-catenin/plakoglobin binding region of the classical cadherin tails (13–15). There are two splice variants of each Dsc, a longer “a” form and a shorter “b” form. The Dsc a form interacts with plakoglobin, but the Dsc b variants do not because the catenin-binding domain (CBD) is partly deleted (16). The desmosomal cadherin-plakoglobin complex binds to the N-terminal domain of desmoplakin (17), whose C-terminal region associates with intermediate filaments (18, 19).
Deletion studies have shown that plakoglobin arm repeats 1–4 are required for Dsg binding, whereas Dsc binding requires both ends of the arm domain (20, 21). β-Catenin is not normally found in desmosomes, but it is observed in desmosomes of plakoglobin null cells (22, 23) or when desmosomal cadherins are overexpressed (24). Because the arm domain of β-catenin is highly homologous to that of plakoglobin, it was proposed that it can interact with desmosomal cadherins even though it is not normally found in desmosomes. Indeed, the β-catenin arm domain has been found to be associated with Dsg2 in cells that do not express other desmosomal components, suggesting that the arm domain alone cannot distinguish classical cadherins from desmosomal cadherins (25). However, expression of chimeric proteins in which the N- and C-terminal tails of plakoglobin were replaced with those of β-catenin resulted in reduced affinity to Dsg2 as assessed by co-immunoprecipitation, suggesting an inhibitory role of the tail domains of β-catenin (25).
The presence of plakoglobin in desmosomes as well as in adherens junctions suggests an important role in regulating cross-talk between these two kinds of junctions. The interaction between plakoglobin and the classical E-cadherin may be required to initiate the assembly of desmosomes (26), which appear to form after adherens junctions (27), and plakoglobin appears later than β-catenin in developing cell-cell contacts (28). Although plakoglobin can interact with desmosomal cadherins as well as classical cadherins, desmosomal cadherin-bound plakoglobin is not incorporated into adherens junctions. The specific localization of plakoglobin complexes likely depends on its ability to bind α-catenin. It appears that the desmosomal cadherin-plakoglobin is excluded from adherens junctions because it cannot associate with α-catenin, due to the overlap of desmosomal cadherin and α-catenin-binding sites (21, 29, 30). However, this has never been examined directly with purified proteins.
Few biochemical and no structural data exist for plakoglobin relative to the extensively characterized β-catenin. It is not known, for example, whether sequence differences between these two proteins lead to intrinsic differences in their ability to bind classical and desmosomal cadherins or whether the selective exclusion of β-catenin from desmosomes is due to other factors. Here, we present structural, biophysical, and biochemical analyses with highly purified proteins aimed at assessing the similarities and differences between plakoglobin and β-catenin that allow plakoglobin to serve as a structural component in both adherens junctions and desmosomes. We present the crystal structure of a plakoglobin-E-cadherin complex, and we accurately determine the binding affinity of plakoglobin and β-catenin for various cadherins by calorimetry. Binding assays with purified, intact proteins are used to map the binding sites of desmosomal cadherins on plakoglobin. The effect of the β-catenin tails that flank the arm domain on binding to desmosomal cadherins is assessed quantitatively, and direct competition between desmosomal cadherins and α-catenin is also demonstrated.
EXPERIMENTAL PROCEDURES
Construct Design and Overexpression in E. coli
Two different human plakoglobin constructs, encoding full-length (pg-full residues 1–745) and arm domain (pg-arm residues 124–669), were used for GST pulldown experiments, and full-length plakoglobin was used for ITC experiments. Four human Dsg1 constructs, covering full cytoplasmic domain (Dsg1cyto residues 572–1049), the catenin-binding domain (Dsg1-CBD residues 663–767), and catenin-binding domain with some C-terminal extensions (Dsg1-CBD-841 residues 663–841 and Dsg1-CBD-958 residues 663–958), and two human Dsc1a constructs, encoding the full cytoplasmic domain (Dsc1cyto residues 715–894) and the catenin-binding domain (Dsc1-CBD residues 795–894), were used for ITC experiments and pulldown assays. All these constructs were made using a pGEX-TEV vector to produce glutathione S-transferase (GST) fusion proteins with an N-terminal TEV protease-cleavable GST tag. Murine β-catenin constructs, including βcat-full-(1–781), β-catenin N-tail + arm domain-(1–671), β-catenin arm domain + C-tail-(134–781), and βcat-arm-(134–671), cytoplasmic domain of E-cadherin (Ecyto residues 577–728), E-cadherin CBD (Ecad CBD, residues 622–728), full-length ICAT (ICAT residues 1–81), and helical domain of ICAT (ICAT-h residues 1–61) were described previously (7). These constructs were used for pulldown experiments, and βcat-full, βcat-arm, Ecyto, ICAT, and ICAT-h were also used for ITC experiments. Full-length mouse α-catenin was expressed with a C-terminal His6 tag as described previously (31) and used for competition assays.
The resulting constructs (Fig. 1) were transformed into the Escherichia coli BL21 strain, and each protein was overexpressed as either a GST-fused form or a His6-tagged form. Cells were grown at 37 °C to an A600 of 0.6–0.8 and were then induced with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside. Cells were harvested by centrifugation after additional growth of cells for 3–4 h at 30 °C, and the cell paste was stored at −80 °C.
Purification of GST Fusion Proteins for GST Pulldown Assays
For GST pulldown assays, GST-βcat-full, GST-βcat-arm, GST-pg-arm, GST-Ecyto, GST-Dsg1-CBD, and GST-Dsc1-CBD were purified as follows. Protease inhibitor mixture (CompleteTM, Roche Applied Science) was added into thawed cell paste, and the cells were lysed by French press. After centrifugation of lysed cells at 40,000 rpm for 30 min, cleared lysates were removed and incubated with glutathione-agarose beads (G-agarose; Sigma) for 1 h at 4 °C. Beads were washed first with phosphate-buffered saline buffer containing 1 m NaCl, 0.005% Tween 20, and 5 mm dithiothreitol (DTT) and then with washing buffer consisting of 25 mm Tris-Cl (pH 8.5), 100 mm NaCl. GST fusion proteins were eluted from G-agarose with a 25 mm Tris-Cl (pH 8.5), 25 mm reduced glutathione, and 100 mm NaCl buffer. Each eluted GST fusion protein was further purified by Mono Q column (GE Healthcare), using Tris-Cl (pH 8.0) buffer for GST-Ecyto, GST-Dsg1-CBD and GST-Dsc1-CBD, or Tris-Cl (pH 8.5) buffer for GST-βcat-full, GST-βcat-arm, GST-pg-full, and GST-pg-arm. Each protein was eluted by linear gradient from 0 to 0.5 m NaCl. Peak fractions were pooled and loaded onto a Superdex S200 gel filtration column (GE Healthcare) equilibrated with assay buffer (25 mm Tris-Cl (pH 8.5), 0.1 m NaCl, 2 mm DTT). Purified GST fusion proteins were concentrated to 1 mg/ml and reloaded onto G-agarose beads for pulldown assays.
Purification of Recombinant Proteins for Pulldown Assays and ITC Measurements
For pulldown assays and ITC experiments, pg-full, βcat-full, βcat-arm, Ecyto, Dsg1cyto, Dsg1-CBD-841, Dsg1-CBD-958, Dsg1-CBD, Dsc1cyto, Dsc1-CBD, Ecad CBD, and ICAT were purified as their GST-fused forms. Each GST fusion protein was purified on a G-agarose affinity column as described above, but instead of eluting GST fusion proteins, thrombin (for βcat-arm, Ecyto, and ICAT) or TEV protease (for all other constructs) was added for the cleavage of each GST fusion protein. The cleavage reaction was carried out at 4 °C for 2 h for thrombin and overnight for TEV protease. The cleaved protein, except for βcat-arm, was collected from flow-through and loaded onto a Mono Q column equilibrated with 25 mm Tris-Cl (pH 8.0), 2 mm DTT, 0.5 mm EDTA, and 20 mm NaCl. In the case of βcat-arm, 25 mm ethanolamine (pH 9.5), 2 mm DTT, 0.5 mm EDTA was used for the Mono Q column. Protein was eluted from the Mono Q column with a linear NaCl gradient and then applied to a Superdex S200 gel filtration column equilibrated with buffer H (25 mm HEPES-NaOH (pH 7.5), 75 mm NaCl, and 0.5 mm EDTA) for ITC experiments with plakoglobin or full-length β-catenin, buffer T (25 mm Tris-Cl (pH 8.8), 2 mm DTT, and 0.1 m NaCl) for ITC experiments with βcat-arm, or assay buffer for pulldown assays.
Purification of ICAT-helical domain (ICAT-h) with an N-terminal His6 tag was performed as described previously (32). After elution of His6-tagged proteins from Ni2+-nitrilotriacetic acid-agarose using a linear gradient of imidazole from 20 to 300 mm, the N-terminal His6 tag of ICAT-h was removed by TEV protease by incubation of cleavage reaction mixture at 4 °C overnight. To remove uncleaved ICAT-h and TEV protease, cleavage reaction mixture was reloaded onto Ni2+-nitrilotriacetic acid-agarose beads after dialysis. Cleaved protein was collected from the flow-through and was further purified by gel filtration chromatography and equilibrated with buffer H or with assay buffer. Full-length α-catenin with a C-terminal His6 tag was purified as described previously (31). After gel filtration chromatography, only monomer peak fractions of α-catenin were pooled, concentrated and used for competition assays.
Isothermal Titration Calorimetry (ITC)
ITC measurements were performed at 30 °C using a VP-ITC instrument (MicroCal, Inc.). All purified ligands, pg-full and full-length β-catenin, were concentrated to 200–350 μm and 8–25 μm, respectively, in H buffer. T buffer was used for ITC experiments with βcat-arm because of the insolubility of βcat-arm at pH 7.5. Each titration experiment was initiated by a 2–4-μl injection, followed by 25–35 7–10-μl injections. Blank titrations, which were carried out by injecting ligand into H or T buffer depending on the particular experiment, were subtracted from each data set. Thermodynamic parameters were calculated by using Origin software package (MicroCal, Inc.) and simple thermodynamic equations (7).
GST Pulldown Assays
Pulldown assays using GST-fused cadherins were performed in assay buffer. Competition binding assays using ICAT were performed by GST pulldown assays of GST-pg-arm with Ecyto, Dsg1cyto, Dsg-CBD, Dsc1cyto, and Dsc-CBD. After washing, each reaction mixture was divided in half. Purified ICAT was added into one half and buffer was into the other half as a control. After a 1-h incubation at 4 °C, followed by washing, bound bands in the presence and in the absence of ICAT were compared by Coomassie-stained SDS-PAGE.
Competition between α-catenin and desmosomal cadherins was monitored by pulldown assays using GST-Ecyto, GST-Dsg1cyto, and GST-Dsc1cyto. Each purified 7 μm GST-fused cadherin was incubated with 10 μm pg-full. After washing G-agarose beads, 20 μm α-catenin was added into each reaction. As a control, the same amount of α-catenin was incubated with each 7 μm GST-Ecyto, GST-Dsg1cyto, and GST-Dsc1cyto in the absence of pg-full. For concentration-dependent competition assays of α-catenin, 5, 10, 20, 30, 40, and 50 μm α-catenin was added into each GST-Dsg1cyto/pg-full and GST-Ecyto/pg-full reaction.
Limited Trypsin Digestion
15 μm of purified βcat-arm and pg-arm were incubated with 0.02, 0.04, or 0.08 μg of trypsin in reaction buffer (20 mm Tris-Cl (pH 8.5), 0.1 m NaCl, 2 mm CaCl2) at room temperature. After 15 and 45 min of incubation, each reaction was stopped by adding SDS gel loading buffer and boiling for 5 min. Each reaction was analyzed by SDS-PAGE. For N-terminal sequence analysis, purified 65-kDa fragment of plakoglobin was incubated with 1.4 μg/ml trypsin for the indicated time period, and the reaction was stopped as above. After SDS-PAGE, three fragments, corresponding to 14, 22, and 29 kDa apparent molecular mass, were transferred into polyvinylidene difluoride membrane, and each slice containing each fragment was cut and subjected to N-terminal sequence analysis.
Purification of the Plakoglobin Arm Domain and Crystallization
The arm domain and the C-terminal helix of plakoglobin (pg-armC; residues 124–676) was purified as above by G-agarose affinity chromatography, followed by cleavage of the GST tag and purification on MonoQ, except that the MonoQ column was run in 25 mm ethanolamine (pH 10.0), 2 mm DTT, 0.5 mm EDTA. The catenin-binding domain of E-cadherin was purified as above and phosphorylated in vitro as described previously (33). The purified phosphorylated catenin-binding domain of E-cadherin (pE-CBD) and pg-armC were mixed in a 1.1:1.0 molar ratio, and their complex was further purified by size exclusion chromatography on an S200 column (GE Healthcare) in a buffer containing 25 mm Tris (pH 8.8), 0.1 m NaCl, 2 mm DTT, 0.5 mm EDTA.
The purified pg-armC-pE-CBD complex was crystallized by hanging-drop vapor diffusion. Protein at 8 mg/ml was mixed with mother liquor consisting of 100 mm Tris-Cl (pH 8.0), 17% PEG3350, and 100 mm ammonium sulfate. Crystals appeared after 3–4 days of incubation at 16.5 °C and were flash-frozen into liquid nitrogen after transfer to reservoir solution containing 25% ethylene glycol.
Data Collection, Data Processing, and Structure Determination
Crystals of the pg-armC-pE-CBD complex are in space group P21, with two copies of the complex in the asymmetric unit. A complete 2.8 Å data set was obtained from a single crystal on the 23ID-B microfocus beamline at the Advanced Photon Source, by translating the crystal after every 15–20 1° images. Diffraction data were integrated and scaled with the HKL2000 program (34) (Table 1).
TABLE 1.
Crystallographic data collection and refinement statistics for the plakoglobin-pE-CBD complex
| Space group | P21 |
| Unit cell parameters | a = 76.0 Å, b = 76.2 Å, c = 122.9 Å, β = 97.2° |
| Resolution | 50 to 2.8 Å (2.9 to 2.8 Å) |
| % complete | 99.2 (99.1) |
| Rsym | 0.09 (0.40) |
| 〈I/σI〉 | 13.0 (2.3) |
| Average multiplicity | 3.3 (3.0) |
| Rwork | 20.2% |
| Rfree | 25.9% |
| No. of residues | 1267 |
| No. of sulfate groups | 2 |
| Root mean square from ideal | |
| Bond lengths | 0.002 Å |
| Bond angles | 0.53° |
| Ramachandran plot | |
| Most favored | 92.3% |
| Additionally allowed | 7.7% |
| Generously allowed | 0.0% |
| Disallowed | 0.0% |
The program PHASER (35) was used to solve the pg-armC-pE-CBD structure by molecular replacement. The arm domain of β-catenin (Protein Data Bank code 1I7W) was used as the search model; the cadherin structure was left out of the search model to avoid potential model bias. Initial rigid body refinement gave R and Rfree values of 0.445 and 0.452, respectively. The electron density map calculated from these phases revealed extra density for pE-CBD. Model building was performed with COOT. Initial refinement indicated that the C-terminal region of plakoglobin was slightly different in the two crystallographically independent copies, so no noncrystallographic symmetry restraints were applied during refinement. Multiple rounds of manual rebuilding, positional refinement, grouped temperature factor refinement, and TLS refinement were performed using the PHENIX package. The later stages of refinement used three TLS groups for pga and three TLS group for pE-CBD, determined by TLS Motion Determination (TLSMD) server. Refinement statistics are shown in Table 1.
RESULTS
Structure of the Plakoglobin-E-cadherin Complex
To visualize directly the interaction of E-cadherin with plakoglobin and compare it with β-catenin, we co-crystallized the minimal E-cadherin CBD with human plakoglobin. Because the arm repeat domain of plakoglobin behaves poorly, it was necessary to try several different plakoglobin fragments. Recently, the structures of full-length zebrafish β-catenin and a fragment of human β-catenin containing the arm domain and the N-terminal region of the C-terminal tail were reported (36). The sequence of the C-terminal tail that immediately follows the arm domain forms a single α-helix that is linked by a loop to the last arm repeat. The helix forms hydrophobic contacts with the last helix of arm repeat 12. Based on these observations, we generated a plakoglobin construct, residues 124–676, that contains both the arm repeat domain and the equivalent C-terminal helix. Although we were unable to form crystals of this (or any other) plakoglobin construct alone or bound to the bacterially expressed E-cadherin CBD, we obtained crystals of this construct bound to the E-cadherin CBD that had been phosphorylated by casein kinase 2. We have shown previously that casein kinase 2-mediated phosphorylation of cadherin changes its affinity for β-catenin from 36 nm to 52 pm and results in the structuring of 10 residues that interact with arm repeats 2–5 of β-catenin (7, 33). ITC measurements (data not shown) indicate that plakoglobin binds to the phosphorylated E-cadherin CBD with an affinity of <1 nm. Accurate measurement of this picomolar range affinity by ITC would require the displacement method (7), but the amounts of plakoglobin required for this experiment were prohibitive.
The overall structure of the arm domain of plakoglobin is very similar to that of β-catenin, as expected from their high sequence identity. It consists of 12 arm repeats, with each repeat composed of three α-helices, H1, H2, and H3, except that H1 is absent in arm repeats 1 and 7 (Fig. 2). The C-α atoms of the plakoglobin and β-catenin arm domains superimpose with an root mean square deviation of 1.2 Å (509 residues) (Fig. 2). The pE-CBD structure is virtually identical to that observed bound to β-catenin (33), forming the same interactions with residues conserved in the two armadillo proteins. A noteworthy difference, however, is that apart from five residues (697–701) that are disordered, the entire cadherin CBD sequence is visible in both copies of the pg-arm complex, whereas different regions of the cadherin primary structure were disordered in the various crystallographically independent copies of the Ecyto-β-catenin complexes (33).
FIGURE 2.
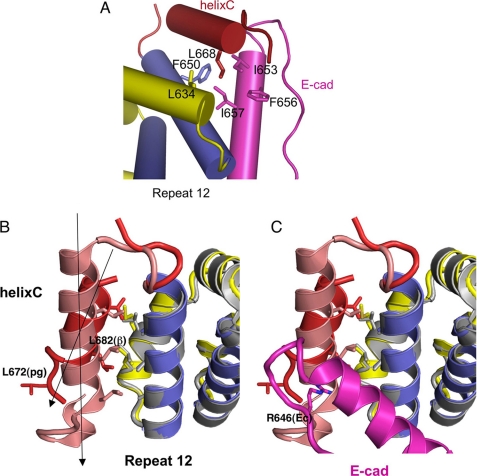
Structure of the plakoglobin arm domain and C-terminal helix bound to phosphorylated E-cadherin. A, overall structure of the plakoglobin-pE-CBD complex. Plakoglobin arm repeat helices are shown in yellow (H1 and H2) and blue (H3), and helix C is shown in red. The pE-CBD is shown in magenta. B, superposition of the plakoglobin arm domain (yellow H1 and H2 and blue H3) with the β-catenin arm domain (gray) as seen in the β-cat-arm-pEcyto complex (33). The orientation is that same as in A. H1 and H2 helices are shown in gray and H3 in blue. The pEcyto structure is shown in cyan. C, superposition of the pE-cadherin CBD bound to plakoglobin (magenta) and β-catenin (cyan). The arm repeat domains of β-catenin and plakoglobin were superimposed, and the transformation was used to compare the bound cadherins.
In addition to the arm domain, the C-terminal helix, residues 661–668, is visible in the plakoglobin-pE-cadherin complex (Fig. 3). This helix, designated helix C, is connected to H3 of arm repeat 12 by a flexible and partly disordered loop and packs against a hydrophobic patch on repeat 12. This arrangement is similar to that observed in uncomplexed β-catenin structures (36), where it was suggested that this is a common structural feature of the β-catenin family that functions as a cap to prevent the energetically unfavorable exposure of the repeat 12 surface.
FIGURE 3.
Helix C in plakoglobin and β-catenin. A, close up of the hydrophobic interaction between plakoglobin (helix C in red and repeat 12 in blue and yellow) and E-cadherin (magenta). B, comparison of the hydrophobic interactions between helix C (plakoglobin, red; β-catenin, pink) and arm repeat 12 (plakoglobin, blue and yellow; β-catenin, gray). The β-catenin structure is from Ref. 36, Protein Data Bank code 2Z6H. C, same as B but with the E-cadherin CBD (magenta) bound to plakoglobin.
The relative positions of helix C and repeat 12 H3 are quite distinct in plakoglobin and β-catenin; in plakoglobin, helix C is tilted by about 25° relative to repeat 12 H3, whereas these two helices are almost parallel in β-catenin (Fig. 3). Despite this difference, key hydrophobic interactions between helix C and repeat 12 are preserved. In plakoglobin, Phe650, Leu649, and Ala646 of repeat 12 H3 pack against Val664 and Leu668 of helix C. On the other hand, helix C of β-catenin is one turn longer than that of plakoglobin, and a leucine in this extra turn (Leu682) contacts Ala652 of repeat 12. Plakoglobin residue Leu672, equivalent to β-catenin Leu682, cannot be part of the α-helix in the complex, because it would clash with Arg646 of E-cadherin. It is not clear whether this difference is intrinsic to the proteins or whether the presence of cadherin in the plakoglobin complex changes the relative positions of the helices, as the tilted orientation of helix C avoids steric clashes with E-cadherin (Fig. 3). In any case, the fact that the relative orientation and length of helix C are different in the two structures suggests some plasticity of this helix that allows it to accommodate interactions of β-catenin family members with a variety of ligands, while maintaining hydrophobic contacts with repeat 12. For example, the helical domain of the transcriptional inhibitor ICAT binds to the β-catenin arm domain through a hydrophobic helix-helix packing with H3 of repeat 12, which might displace helix C completely. Indeed, the affinity of ICAT for β-catenin is the same whether helix C is present (7).
Thermodynamics of Plakoglobin-Cadherin Interactions
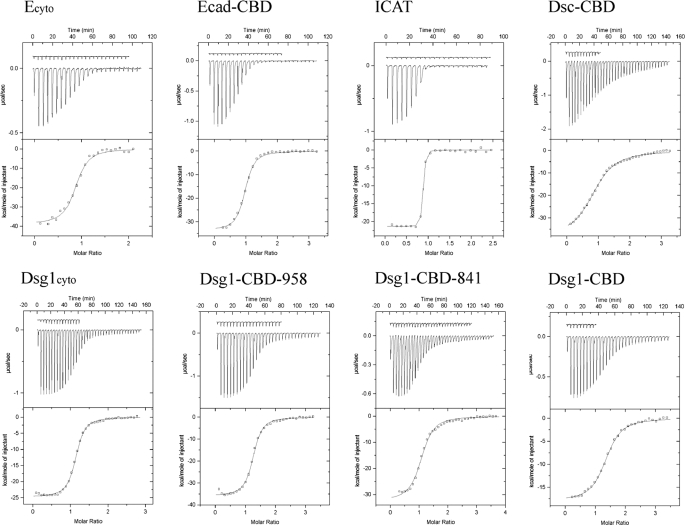
ITC was used to determine accurately the binding parameters of purified plakoglobin and the cytoplasmic domains of selected classical and desmosomal cadherins. Fig. 1 shows the protein constructs used in these experiments, and Fig. 4 shows representative ITC experiments, including titration data and binding curves calculated using the best fit parameters. Table 2 summarizes the thermodynamic parameters. All ligands bind to plakoglobin in a 1:1 ratio. Plakoglobin binds to the full 152-amino acid cytoplasmic domain of E-cadherin (Ecyto) with a dissociation constant KD of 85 nm. A fragment of the E-cadherin domain slightly longer than that visible in the crystal structure of Ecyto bound to β-catenin (33), residues 622–728, binds similarly, thereby defining a minimal CBD.
FIGURE 4.
Ligand binding to full-length plakoglobin determined by ITC. Representative experiments are shown. All isothermal calorimetric experiments were performed at 30 °C in H buffer. The raw heat signals obtained by a series of injections of each ligand into a solution of full-length plakoglobin are shown on top, and the binding curve calculated using the best fit parameters obtained by a nonlinear least squares fit is shown on the bottom. The base-line heats that are subtracted from the raw data, obtained by injecting the ligand into H buffer, are shown vertically offset from the binding data.
TABLE 2.
ITC data for plakoglobin ligand binding
All measurements were performed in H buffer.
| Ligand | KD | ΔH | TΔS | ΔG |
|---|---|---|---|---|
| nm | kcal/mol | kcal/mol | kcal/mol | |
| E-cadherin-CBD | 116 ± 31 | −31.3 ± 0.4 | −21.9 | −9.4 |
| Ecyto | 85 ± 22 | −38.2 ± 0.9 | −28.4 | −9.8 |
| Dsg1-CBD (residues 667–767) | 510 ± 36 | −16.9 ± 0.2 | −8.2 | −8.7 |
| Dsg1-CBD-841 | 496 ± 28 | −32.2 ± 0.5 | −23.5 | −8.7 |
| Dsg1-CBD-958 | 189 ± 18 | −36.0 ± 0.3 | −26.7 | −9.3 |
| Dsg1cyto | 200 ± 14 | −21.7 ± 0.1 | −12.4 | −9.3 |
| Dsc1-CBD | 2900 ± 200 | −37.9 ± 0.6 | −30.2 | −7.7 |
| Dsc1cyto | 2200 ± 300 | −31.3 ± 0.6 | −23.5 | −7.8 |
| ICAT-h | 11.1 ± 2.6 | −9.2 ± 0.1 | 1.9 | −11.0 |
| ICAT | 3.7 ± 1.0 | −21.3 ± 0.1 | −9.5 | −11.8 |
The Dsg1 cytoplasmic domain (Dsg1cyto) is 479 amino acids in length and that of Dsc1 is 180 amino acids in length. However, only about 100 amino acids of the desmosomal cadherin cytoplasmic regions are homologous to the E-cadherin CBD. Dsg1cyto binds to plakoglobin more weakly than does Ecyto, with a KD of 200 nm (Table 2). This contrasts with the apparently stronger binding of Dsg1 measured by less direct methods (37). Dsg1 residues 663–767 (Dsg1-CBD) bear significant sequence homology to the E-cadherin CBD. Dsg1-CBD binds with a slightly reduced affinity of 510 nm relative to the full domain. As shown in Table 2, extending the construct to residue 958, which includes most of the desmoglein-specific cytoplasmic region (38), restores full binding affinity. In contrast, a similar analysis of the desmocollin-1 cytoplasmic domain shows that the region of this protein homologous to the E-cadherin CBD binds to plakoglobin with the same affinity as the slightly larger full cytoplasmic domain. The interaction of Dsc1 with plakoglobin is much weaker than that of Dsg1 or E-cadherin, however, with a KD of 2.9 μm.
We previously demonstrated, using a variety of methods, that the E-cadherin cytoplasmic domain is an intrinsically unstructured protein (39). This correlates with the highly unfavorable entropy of binding to β-catenin, presumably because of the loss of configurational entropy over roughly 100 residues (7). Similarly unfavorable binding entropies are observed for the interaction of the homologous desmosomal cadherins with plakoglobin (Table 2), suggesting that the equivalent regions of Dsg and Dsc are also intrinsically unstructured. Moreover, the desmoglein-specific cytoplasmic region required for full affinity is also an intrinsically unstructured protein (38). These observations are consistent with the abnormally large apparent molecular masses of these proteins on sizing columns (data not shown).
β-Catenin Binds to Desmosomal Cadherins
Plakoglobin and β-catenin display 79% sequence identity in their armadillo repeat domains, and the crystal structures of E-cadherin bound to these proteins reveal identical interactions. Several regions of E-cadherin critical for the interaction with β-catenin, in particular regions III and V (33), are conserved in desmogleins and desmocollins. These observations are consistent with the observation of β-catenin-desmosomal cadherin complexes under some circumstances (22–24).
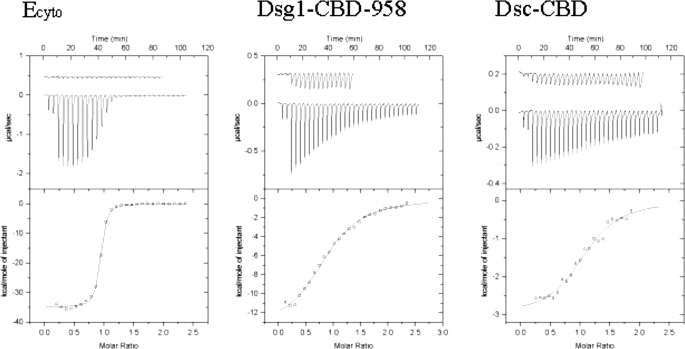
ITC measurements show that β-catenin binds to the same regions of Dsg1 and Dsc1 that display full affinity for plakoglobin. Surprisingly, β-catenin binds to Dsg1 ∼10-fold more weakly than does plakoglobin, whereas both proteins bind with similarly weak affinity to Dsc1 (Fig. 5; Table 3). The thermodynamics of Dsc1 binding differ markedly between plakoglobin and β-catenin, however. Plakoglobin binds to Dsc1 with a large, favorable binding enthalpy that is compensated by a large, unfavorable entropy. In contrast, β-catenin binds to Dsc1 with a small binding enthalpy and a slightly favorable entropy term. This could reflect intrinsic differences in the β-catenin and plakoglobin arm repeats such as flexibility, different modes of binding, or a combination of both.
FIGURE 5.
Cadherin binding to full-length β-catenin determined by ITC. Representative experiments are shown. All isothermal calorimetric experiments were performed at 30 °C in H buffer. The raw heat signals obtained by a series of injections of each ligand into a solution of full-length β-catenin are shown on top, and the binding curve calculated using the best fit parameters obtained by a nonlinear least squares fit is shown on the bottom. The base-line heats that are subtracted from the raw data, obtained by injecting the ligand into H buffer, are shown vertically offset from the binding data.
TABLE 3.
ITC data for full-length β-catenin-full-cadherin binding
All measurements were performed in H buffer.
| Ligand | KD | ΔH | TΔS | ΔG |
|---|---|---|---|---|
| nm | kcal/mol | kcal/mol | kcal/mol | |
| Ecyto | 41 ± 3.6 | −35.2 ± 0.2 | −24.9 | −10.3 |
| Dsg1-CBD-958 | 2200 ± 200 | −13.7 ± 0.4 | −5.9 | −7.8 |
| Dsc1-CBD | 2700 ± 400 | −3.0 ± 0.1 | 4.7 | −7.7 |
Plakoglobin Is More Flexible than β-Catenin
The difference in binding affinity of plakoglobin and β-catenin to Dsg1 indicates that the arm domains of plakoglobin and β-catenin are not completely interchangeable, despite the high level of sequence identity. Indeed, it has been noted that β-catenin cannot restore plakoglobin adhesive function in plakoglobin knock-out cells (40). The proteolytic sensitivity of plakoglobin was tested to probe for differences in flexibility between it and β-catenin. When either full-length or the armadillo repeat domain of β-catenin is digested with trypsin, fragments with apparent molecular masses of 40 and 14 kDa on SDS-PAGE result from cleavage at Arg550 in repeat 10. These fragments remain associated on anion exchange and gel filtration columns, indicating that trypsin nicks a flexible loop in repeat 10 but does not otherwise digest this domain (10). Surprisingly, trypsin digestion of the plakoglobin arm domain under the same conditions produced multiple fragments, showing that pg-arm is much more vulnerable to trypsin (Fig. 6a); a 14-kDa fragment is produced, but the larger 40-kDa fragment was not observed. Instead, a 21-kDa fragment and multiple bands around 30 kDa were detected.
FIGURE 6.
Proteolytic sensitivity of β-catenin and plakoglobin. a, pg-arm or βcat-arm was incubated with the indicated amount of trypsin for 15 and 45 min at room temperature. After incubation, each reaction was added by SDS sample buffer and was boiled. Untreated pg-arm and βcat-arm were shown as controls. b, 66-kDa plakoglobin fragment was incubated with trypsin for 0, 5, 10, 15, and 30 min and 1, 2, and 3 h. The asterisks indicate degradation products used for N-terminal sequence analysis. c, schematic drawings of primary structures of β-catenin and plakoglobin showing the location of trypsin cleavage sites. Numbered boxes represent the 12 armadillo repeats, and NH2 and COOH represent the N- and C-terminal tails. Numbered arrows show the location of the cleavage sites. The sequence around the cleaved Arg or Lys residues (shown in boldface) is indicated.
N-terminal sequencing was used to characterize the fragments that result from limited trypsinolysis of plakoglobin (Fig. 6, b and c). A purified 65-kDa plakoglobin fragment that includes the arm domain and full C-terminal tail was used in this analysis (residues 82–745; the N-terminal cleavage results from thrombin digestion to remove a GST affinity tag). The digestion produced three stable bands with apparent molecular mass of 29 (doublet), 21, and 14 kDa on a 15% SDS-polyacrylamide gel (Fig. 6b). The 14-kDa band is a mixture of two fragments, one starting at Ser125, which is the N-terminal boundary of the arm domain, and the other at His541, which corresponds to Arg550 in β-catenin. Although the precise molecular weights of these fragments were not determined, the location of Arg and Lys residues makes it likely that the fragment starting at Ser125 spans amino acids 125–261 or 125–265, and the fragment starting at His541 corresponds to residues 541–661 or 541–674. Because Arg661 corresponds to Arg671 of β-catenin, which is the C-terminal boundary of the arm domain, Arg661 is the likely C-terminal boundary of this fragment. Similar analysis of the 21-kDa tryptic fragment suggests that this fragment spans residues 478–661. N-terminal sequence analysis of the 29-kDa doublet revealed cleavage sites at Lys124, Lys394, and Lys426; the resulting fragments would be 125–394, 395–661, and 427–661, whose calculated molecular masses are 30, 29, and 26 kDa, respectively.
As summarized in Fig. 6c, of the five tryptic cleavage sites within the plakoglobin arm domain, four are conserved in βcat-arm, but only two of these, Lys124 and Arg540, are cleaved in β-catenin (Lys133 and Arg550). The other two sites, which correspond to Lys435 and Arg486 of β-catenin, are located in the H1 helices of repeats 8 and 9, respectively. The additional cleavage site, Lys394, corresponds to Gly403 of β-catenin, which is located in H2 helix of repeat 7. Finally, we see no stable fragments for the region spanning repeats 4–6. The greater proteolytic sensitivity of the plakoglobin arm domain suggests that it is significantly more flexible or conformationally heterogeneous than that of β-catenin.
Binding Sites of Desmosomal Cadherins on Plakoglobin
Even though the catenin-binding domains of E-cadherin and desmosomal cadherins have high sequence similarity, previous studies suggest that the interaction of desmosomal cadherins with plakoglobin is somewhat different from that of E-cadherin. E-cadherin interacts with all 12 arm repeats (Fig. 2). In contrast, the Dsg-binding site was mapped to the first four arm repeats of plakoglobin, and Dsc binding appears to require both N- and C-terminal ends of the arm domain (21). These mapping experiments were performed with mutants of plakoglobin in which various arm repeats were deleted. However, the crystal structures of plakoglobin and β-catenin indicate that the structure of any repeat depends upon interactions with its neighbors, and it is likely that deletion of repeats would destabilize the structure near the sites of such deletions (10). We attempted to bacterially express a plakoglobin mutant in which arm repeats 6–10 were deleted, connecting repeat 5 directly to repeat 12; helix C was included in this construct to prevent exposure of hydrophobic residues of repeat 12 (see above). However, this deletion mutant was not soluble and was found exclusively in inclusion bodies (data not shown).
To map the binding sites of desmosomal cadherins using soluble, stable, well folded proteins, we designed a competition binding assay that exploits the transcriptional inhibitor ICAT (41). ICAT prevents binding of Tcf/Lef transcription factors to β-catenin by occupying a groove formed by repeats 5–9 that interacts with an extended peptide common to most β-catenin ligands (32, 42). High affinity binding of ICAT is due to a compact helical domain, designated ICAT-h, which interacts with repeats 11 and 12 of β-catenin (7, 32, 42). Residues in β-catenin repeat 12 that interact with ICAT-h, and other residues in the β-catenin arm repeat 5–9 groove that interact with extended region of ICAT, are conserved in plakoglobin. Also, as noted above, the residues that interact directly with E-cadherin are identical in β-catenin and plakoglobin. Plakoglobin (Table 2) and β-catenin (7) bind to ICAT and ICAT-h with similar affinities and thermodynamics, strongly suggesting that the same interactions are present in the plakoglobin-ICAT complex. Thus, ICAT can be used to inhibit binding of ligands requiring repeats 5–12 of plakoglobin, and ICAT-h will selectively compete with ligands binding to repeats 11–12.
ICAT was added to complexes of cadherins bound to a GST fusion of the plakoglobin arm repeat domain (GST-pg-arm; Fig. 1) immobilized on glutathione-agarose beads. After incubation and washing, proteins bound to plakoglobin were analyzed by SDS-PAGE and visualized by Coomassie staining. As shown in Fig. 7a, Ecyto and the Dsc1-CBD were displaced by ICAT, whereas significant amounts of Dsg1-CBD remained bound. This cannot be due to a stronger affinity as E-cadherin, which binds more strongly to plakoglobin than Dsg1 (Table 2), is displaced by ICAT. Instead, the Dsg1-CBD and ICAT appeared to form a ternary complex with GST-pg-arm. To rule out the possibility that the presence of both ICAT and Dsg1-CBD on the beads arises from mixtures of two binary plakoglobin complexes (i.e. ICAT or Dsg1-CBD), the pulldown format was reversed by using a GST fusion of the Dsg1-CBD to which purified full-length plakoglobin was added to form the binary plakoglobin-Dsg1-CBD complex. ICAT, ICAT-h, Ecyto, or Dsc1cyto was then added to the immobilized binary complex. ICAT and ICAT-h formed ternary complexes with the GST-Dsg1-CBD-plakoglobin complex (Fig. 7b). Note that the ITC data indicate that extending the Dsg1-CBD construct C-terminally to residue 958 provides a 2.5× enhancement of affinity (Table 2). It is possible that these extra residues interact with the more C-terminal armadillo repeats. However, the 2.5× affinity difference corresponds to a very small difference in binding energy, and we did not observe binding of the extended region (i.e. residues 768–958) in trans (data not shown). Assuming that ICAT interacts with repeats 5–12 of plakoglobin as observed in the crystal structure of β-cat-arm-ICAT complex, these results indicate that the majority of the binding energy of Dsg1 is provided by interaction with repeats 1–4 of plakoglobin, confirming earlier deletion mutagenesis data (21).
FIGURE 7.
Mapping of the desmosomal cadherin-binding sites on plakoglobin. a, GST-pg-arm at a concentration of 10 μm was incubated with 10 μm catenin-binding domain of Dsg1 or full cytoplasmic domains of E-cadherin or Dsc1 for 1 h. After isolation of protein complexes with glutathione-agarose, 10 μm ICAT was added into each reaction, and each reaction mixture was incubated for another 1 h at 4 °C. Protein complexes were purified with glutathione-agarose, and each purified protein complex in the absence or presence of ICAT was analyzed by SDS-PAGE and Coomassie staining. Molecular markers are shown on the right. b, GST-Dsg1-CBD at a concentration of 10 μm was incubated with 10 μm pg-full. After protein complexes were co-precipitated on glutathione-agarose, ICAT, ICAT-h, Dsc1cyto, or Ecyto each at a concentration of 10 μm was added into each reaction, and the beads were centrifuged. Supernatants (S) and pelleted beads (P) were analyzed by SDS-PAGE and visualized with Coomassie Blue stain. ICAT and ICAT-h stain very poorly relative to plakoglobin, and their bands in the pellet with plakoglobin have been marked with white boxes. c, similar binding assays were performed using GST-Dsc1cyto, instead of GST-Dsg1-CBD. After isolation of protein complex of GST-Dsc1cyto and pg-full, ICAT, ICAT-h, or Dsg1-CBD was added into each reaction. Although ICAT and ICAT-h stain poorly, it is clear that neither protein co-sediments with plakoglobin-bound Dsc1.
Similar experiments between ICAT and Dsc1 indicate that, unlike Dsg1, the binding site for Dsc1 at least partly overlaps that of ICAT (Fig. 7c). To more finely map the region of overlap, ICAT-h and Dsg1, whose binding sites were mapped to repeats 11–12 and repeats 1–4, respectively, were used for competition assays. Plakoglobin binding to Dsc1 was reduced in the presence of either of these ligands (Fig. 7c), suggesting that Dsc1 interacts with the arm repeats 1–4 and 11–12. The data do not rule out, however, that the Dsc1-binding site also includes the central region of the arm domain. Indeed, the strong conservation of the extended peptide-binding site (cadherin region III (33)) among classical and desmosomal cadherins suggests that this site is also involved in both Dsc and Dsg binding, even if it is not absolutely required for detectable binding.
Do the β-Catenin Tails Affect Cadherin Binding?
It has been suggested that the N- and C-terminal tails of β-catenin that flank the arm domains, which are less homologous to their plakoglobin counterparts, weaken the affinity for desmosomal cadherins such that β-catenin cannot bind to these proteins. The arm domains of plakoglobin and β-catenin can bind to Dsg2, whereas a chimeric protein comprising the plakoglobin arm domain flanked by the β-catenin tails binds Dsg2 less well than wild-type plakoglobin (25). Dsg1 binds to β-catenin 10 times less well than to plakoglobin (Tables 2 and 3). We therefore asked whether the tails of β-catenin are responsible for this difference by measuring the affinity of the β-catenin arm domain for various cadherins using purified proteins. We reported previously that the β-catenin tails have no effect on the binding of E-cadherin (7). As shown in Tables 3 and 4, the β-catenin arm domain binds to the three tested cadherin cytoplasmic regions with affinities very similar to that of the full-length protein. Note that because the arm domain tends to aggregate below pH 8.0, these experiments were performed at pH 8.8. However, we showed earlier that the binding to E-cadherin has little or no dependence on pH (7). Therefore, the tails of β-catenin do not influence the affinity of this protein for classical or desmosomal cadherins.
TABLE 4.
ITC measurements of β-cat-arm-cadherin interactions
Measurements were in T buffer.
| Ligand | KD | ΔH | TΔS | ΔG |
|---|---|---|---|---|
| nm | kcal/mol | kcal/mol | kcal/mol | |
| Ecyto | 8.2 ± 5.7 | −32.3 ± 0.7 | −22.4 | −9.8 |
| Dsg1-CBD-958 | 2200 ± 200 | −17.0 ± 0.6 | −9.2 | −7.8 |
| Dsc1-CBD | 3900 ± 400 | −11.4 ± 0.6 | −3.9 | −7.5 |
The quantitative data obtained by ITC contrast with results of pulldown experiments of cadherin interaction with full-length β-catenin or constructs lacking one or both tails, where the tails appear to diminish binding (data not shown). We speculate that the unstructured acidic tails of β-catenin and plakoglobin produce artifacts in co-precipitation assays, perhaps because of nonspecific electrostatic effects that give rise to different background levels of binding, or that deletion of these sequences produces changes in solubility. This might also explain the effects of the tails in electrophoretic mobility shift assays when studying the interaction of Tcf transcription factors with β-catenin and plakoglobin (43), despite the observation that Lef-1 shows no difference in affinity to full-length or tail-deleted β-catenin by ITC (7).
It has been reported that the tails of plakoglobin can influence binding of cadherins and other ligands (44). Unfortunately, the purified plakoglobin arm domain tends to aggregate at all pH values tested, making it impossible to test rigorously the effect of the plakoglobin tails by precise ITC measurements.
Overlap with α-Catenin Site
Because plakoglobin is common to desmosomes and adherens junctions, an important question is why α-catenin is found only in adherens junction. The α-catenin-binding site of β-catenin (residues 118–149) and plakoglobin (residues 109–140) resides in the sequence just N-terminal to the arm domain. Deletion studies have shown that the α-catenin and desmosomal cadherin-binding sites overlap (20), and alanine scanning mutagenesis of plakoglobin combined with co-immunoprecipitation assays showed that residues Ile127, Leu130, Ile131, Leu150, Leu151, and Leu231 are required for the interaction with Dsg, Dsc, and α-catenin but not with Ecyto. The first three residues lie in the α-catenin-binding site, again suggesting that the desmosomal cadherin-binding site overlaps that of α-catenin (30). However, the effects of these mutations on the structure of plakoglobin were not tested; for example, Leu151 and Leu231 lie in the hydrophobic core of the first two arm repeats, so mutation of these residues to alanine likely alters the structure of plakoglobin in this region. Thus, it is important to test whether there is direct competition between α-catenin and desmosomal cadherins for plakoglobin using correctly folded native proteins.
Competition assays were performed using purified α-catenin, plakoglobin, and GST-fused cadherins to test directly whether α-catenin and desmosomal cadherins compete for binding to plakoglobin. Only the GST-Ecyto-pg-full complex interacts with α-catenin to form a ternary complex (Fig. 8). Addition of α-catenin to the GST-Dsc1/CBD-pg mixture releases some of the bound plakoglobin, consistent with competition between GST-Dsc1-CBD and α-catenin for binding to plakoglobin. In this case, the binding affinities of α-catenin and Dsc1 are likely to be on the same order of magnitude. In contrast, α-catenin does compete with Dsg1-CBD for the interaction with pg-full, suggesting that Dsg1 has higher affinity. Addition of increasing concentrations of α-catenin did not result in any binding to the GST-Dsg1-plakoglobin complex over background (Fig. 8). These experiments show directly that desmosomal cadherin-plakoglobin complexes cannot associate with α-catenin, explaining the absence of the latter from desmosomes.
FIGURE 8.
Overlapping binding sites of α-catenin and desmosomal cadherins on plakoglobin. Upper panel, GST-Ecyto, GST-Dsg1cyto, or GST-Dsc1cyto each at a concentration of 7 μm was incubated with 10 μm plakoglobin and increasing amounts of α-catenin as indicated. Protein complexes were isolated with glutathione-agarose and analyzed by SDS-PAGE and Coomassie staining. Lower panel, same binding assays were carried out in the absence of plakoglobin as a control for nonspecific background binding of α-catenin to glutathione-agarose.
DISCUSSION
The data presented here provide a rigorous foundation for understanding the interaction of cadherins with β-catenin and plakoglobin. The crystal structure of the plakoglobin-pE-CBD complex shows that plakoglobin and β-catenin are structurally very similar. We also show that these two armadillo proteins bind to E-cadherin with similar thermodynamic parameters. Sequence alignments indicate that residues critical for the interaction of E-cadherin with plakoglobin and β-catenin arm repeats 5–9 (region III (33)) and the N-terminal repeats (the “cap” or region V (33)) are strongly conserved in all desmoglein and desmocollin isoforms, suggesting that they interact similarly with these regions of plakoglobin. The interaction with the C-terminal repeats is less obviously conserved between desmosomal and classical cadherins, and the basis of why desmocollins but not desmogleins appear to interact with this region of plakoglobin is not clear. There are other strongly conserved regions within Dsg and Dsc isoforms (for example, region IV defined in Ref. 33), but these differ from classical cadherins, so their contribution to binding, if any, cannot be ascertained without detailed structural information.
We have not measured the affinity of other Dsg or Dsc isoforms for plakoglobin, but we have found that the E- and N-cadherin cytoplasmic domains, which share 64% identity, bind with the same affinity to β-catenin.3 The sequence homology between the catenin binding regions of Dsg1 and other Dsg isoforms is in the range ∼50–70%, and similar homologies are present between Dsc1 and other Dscs. These observations suggest that Dsgs and Dscs isoforms bind to plakoglobin with affinities comparable with those found here for Dsg1 and Dsc1.
Despite their similar modes of interaction with E-cadherin, β-catenin and plakoglobin are biologically and physically distinct, the latter evidenced by a significant difference in their proteolytic sensitivities when not bound to a ligand. We have shown that the cytoplasmic portion of the classical E-cadherin binds more strongly to both of these proteins than do the desmosomal proteins Dsg1 or Dsc1. Both β-catenin and plakoglobin are found in adherens junctions; β-catenin is seen in developing junctions, whereas both β-catenin and plakoglobin are associated with more mature junctions (28).
Desmosome formation appears to require prior adherens junction assembly (26, 27, 45). Plakoglobin had been reported to bind to Dsg1 better than to E-cadherin (37), leading to the suggestion that once Dsg is recruited to plakoglobin in the adherens junction, it displaces classical cadherins from plakoglobin to initiate desmosome assembly (26). The ITC data indicate that Dsg1 would not readily replace E-cadherin bound to plakoglobin, although this would depend on the relative concentrations of these two cadherins. It is also possible that post-translational modifications of plakoglobin and/or Dsg modify the strength of their interaction, as has been documented for the phosphorylation of E-cadherin (7, 33). Dsg2 is phosphorylated by the epidermal growth factor receptor (46), and several tyrosine kinases, including epidermal growth factor receptor, Src, and Fer/Fyn, have been reported to phosphorylate plakoglobin (47, 48). It is not known, however, whether these modifications affect the affinity of plakoglobin-cadherin interactions.
α-Catenin and β-catenin are not found in desmosomes (49). The competition data confirm the notion that the exclusion of α-catenin from desmosomes arises from the overlap of its binding site on plakoglobin with desmosomal cadherins. The 10-fold weaker binding of Dsg1 to β-catenin versus plakoglobin is consistent with the absence of β-catenin from desmosomes, although this depends both on the intrinsic affinity and the presently unknown relative cellular concentrations of these two armadillo proteins. However, β-catenin and plakoglobin bind with similar affinity to Dsc1. The ITC data indicate that the tails that flank the β-catenin armadillo domain do not alter the affinity for classical or desmosomal cadherins, again arguing against an autoinhibitory mechanism in which one or both tails compete with ligands for access to the arm repeats (7, 25). It seems more likely that other proteins, post-translational modifications, and/or control of the relative levels of β-catenin and plakoglobin contribute to the exclusion of β-catenin from desmosomes.
Plakoglobin binds to Dsc1 much more weakly than to either E-cadherin or Dsg1. It remains to be seen how this weak affinity relates to the biology of desmosomal cadherins, in particular their precise roles in desmosome formation and function. There are clear differences in the interaction regions of Dsgs and Dscs with plakoglobin, as illustrated by the observation that Dsg1 can form ternary complexes with ICAT, whereas Dsc1 cannot. It is possible that the different modes of interaction allow other partners of either the cadherin or plakoglobin to join the higher order desmosome assembly under different conditions.
Acknowledgments
We thank Kathleen Green for providing cDNAs for the desmosomal cadherins, Pamela Cowin for the plakoglobin cDNA, and Sofiya Fridman for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM56169.
The atomic coordinates and structure factors (code 3IFQ) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
H.-J. Choi, J. C. Gross, S. Pokutta, and W. I. Weis, unpublished observations.
- Ecyto
- E-cadherin cytoplasmic domain
- β-cat-arm
- β-catenin armadillo repeat domain
- CBD
- catenin-binding domain
- pE-CBD
- phosphorylated catenin-binding domain of E-cadherin
- Dsc1cyto
- Dsc1 cytoplasmic domain
- Dsg1cyto
- Dsg1 cytoplasmic domain
- pE-CBD
- phosphorylated E-cadherin catenin-binding domain
- G-agarose
- glutathione-agarose
- ICAT
- inhibitor of catenin and TCF
- ICAT-h
- ICAT helical domain
- ITC
- isothermal titration calorimetry
- pg-arm
- plakoglobin armadillo repeat domain
- GST
- glutathione S-transferase
- DTT
- dithiothreitol
- TEV
- tobacco etch virus
- pg-armC
- pg-arm domain and C-terminal helix.
REFERENCES
- 1.Yap A. S., Brieher W. M., Gumbiner B. M. (1997) Annu. Rev. Cell Dev. Biol. 13, 119–146 [DOI] [PubMed] [Google Scholar]
- 2.Kowalczyk A. P., Bornslaeger E. A., Norvell S. M., Palka H. L., Green K. J. (1999) Int. Rev. Cytol. 185, 237–302 [DOI] [PubMed] [Google Scholar]
- 3.Gumbiner B. M. (2005) Nat. Rev. Mol. Cell Biol. 6, 622–634 [DOI] [PubMed] [Google Scholar]
- 4.Perez-Moreno M., Jamora C., Fuchs E. (2003) Cell 112, 535–548 [DOI] [PubMed] [Google Scholar]
- 5.Green K. J., Simpson C. L. (2007) J. Invest. Dermatol. 127, 2499–2515 [DOI] [PubMed] [Google Scholar]
- 6.Pokutta S., Weis W. I. (2007) Annu. Rev. Cell Dev. Biol. 23, 237–261 [DOI] [PubMed] [Google Scholar]
- 7.Choi H. J., Huber A. H., Weis W. I. (2006) J. Biol. Chem. 281, 1027–1038 [DOI] [PubMed] [Google Scholar]
- 8.Drees F., Pokutta S., Yamada S., Nelson W. J., Weis W. I. (2005) Cell 123, 903–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamada S., Pokutta S., Drees F., Weis W. I., Nelson W. J. (2005) Cell 123, 889–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huber A. H., Nelson W. J., Weis W. I. (1997) Cell 90, 871–882 [DOI] [PubMed] [Google Scholar]
- 11.Linding R., Jensen L. J., Diella F., Bork P., Gibson T. J., Russell R. B. (2003) Structure 11, 1453–1459 [DOI] [PubMed] [Google Scholar]
- 12.Garrod D. R., Merritt A. J., Nie Z. (2002) Curr. Opin. Cell Biol. 14, 537–545 [DOI] [PubMed] [Google Scholar]
- 13.Troyanovsky S. M., Eshkind L. G., Troyanovsky R. B., Leube R. E., Franke W. W. (1993) Cell 72, 561–574 [DOI] [PubMed] [Google Scholar]
- 14.Mathur M., Goodwin L., Cowin P. (1994) J. Biol. Chem. 269, 14075–14080 [PubMed] [Google Scholar]
- 15.Troyanovsky S. M., Troyanovsky R. B., Eshkind L. G., Krutovskikh V. A., Leube R. E., Franke W. W. (1994) J. Cell Biol. 127, 151–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Troyanovsky S. M., Troyanovsky R. B., Eshkind L. G., Leube R. E., Franke W. W. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 10790–10794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kowalczyk A. P., Bornslaeger E. A., Borgwardt J. E., Palka H. L., Dhaliwal A. S., Corcoran C. M., Denning M. F., Green K. J. (1997) J. Cell Biol. 139, 773–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kouklis P. D., Hutton E., Fuchs E. (1994) J. Cell Biol. 127, 1049–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stappenbeck T. S., Green K. J. (1992) J. Cell Biol. 116, 1197–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wahl J. K., Sacco P. A., McGranahan-Sadler T. M., Sauppé L. M., Wheelock M. J., Johnson K. R. (1996) J. Cell Sci. 109, 1143–1154 [DOI] [PubMed] [Google Scholar]
- 21.Witcher L. L., Collins R., Puttagunta S., Mechanic S. E., Munson M., Gumbiner B., Cowin P. (1996) J. Biol. Chem. 271, 10904–10909 [DOI] [PubMed] [Google Scholar]
- 22.Bierkamp C., Schwarz H., Huber O., Kemler R. (1999) Development 126, 371–381 [DOI] [PubMed] [Google Scholar]
- 23.Acehan D., Petzold C., Gumper I., Sabatini D. D., Müller E. J., Cowin P., Stokes D. L. (2008) J. Invest. Dermatol. 128, 2665–2675 [DOI] [PubMed] [Google Scholar]
- 24.Norvell S. M., Green K. J. (1998) J. Cell Sci. 111, 1305–1318 [DOI] [PubMed] [Google Scholar]
- 25.Wahl J. K., 3rd, Nieset J. E., Sacco-Bubulya P. A., Sadler T. M., Johnson K. R., Wheelock M. J. (2000) J. Cell Sci. 113, 1737–1745 [DOI] [PubMed] [Google Scholar]
- 26.Lewis J. E., Wahl J. K., 3rd, Sass K. M., Jensen P. J., Johnson K. R., Wheelock M. J. (1997) J. Cell Biol. 136, 919–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vasioukhin V., Bauer C., Yin M., Fuchs E. (2000) Cell 100, 209–219 [DOI] [PubMed] [Google Scholar]
- 28.Adams C. L., Nelson W. J., Smith S. J. (1996) J. Cell Biol. 135, 1899–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Troyanovsky R. B., Chitaev N. A., Troyanovsky S. M. (1996) J. Cell Sci. 109, 3069–3078 [DOI] [PubMed] [Google Scholar]
- 30.Chitaev N. A., Averbakh A. Z., Troyanovsky R. B., Troyanovsky S. M. (1998) J. Cell Sci. 111, 1941–1949 [DOI] [PubMed] [Google Scholar]
- 31.Pokutta S., Weis W. I. (2000) Mol. Cell 5, 533–543 [DOI] [PubMed] [Google Scholar]
- 32.Daniels D. L., Weis W. I. (2002) Mol. Cell 10, 573–584 [DOI] [PubMed] [Google Scholar]
- 33.Huber A. H., Weis W. I. (2001) Cell 105, 391–402 [DOI] [PubMed] [Google Scholar]
- 34.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 35.Storoni L. C., McCoy A. J., Read R. J. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 432–438 [DOI] [PubMed] [Google Scholar]
- 36.Xing Y., Takemaru K., Liu J., Berndt J. D., Zheng J. J., Moon R. T., Xu W. (2008) Structure 16, 478–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chitaev N. A., Leube R. E., Troyanovsky R. B., Eshkind L. G., Franke W. W., Troyanovsky S. M. (1996) J. Cell Biol. 133, 359–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kami K., Chidgey M., Dafforn T., Overduin M. (2009) J. Mol. Biol. 386, 531–543 [DOI] [PubMed] [Google Scholar]
- 39.Huber A. H., Stewart D. B., Laurents D. V., Nelson W. J., Weis W. I. (2001) J. Biol. Chem. 276, 12301–12309 [DOI] [PubMed] [Google Scholar]
- 40.Yin T., Getsios S., Caldelari R., Kowalczyk A. P., Müller E. J., Jones J. C., Green K. J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 5420–5425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tago K., Nakamura T., Nishita M., Hyodo J., Nagai S., Murata Y., Adachi S., Ohwada S., Morishita Y., Shibuya H., Akiyama T. (2000) Genes Dev. 14, 1741–1749 [PMC free article] [PubMed] [Google Scholar]
- 42.Graham T. A., Clements W. K., Kimelman D., Xu W. (2002) Mol. Cell 10, 563–571 [DOI] [PubMed] [Google Scholar]
- 43.Zhurinsky J., Shtutman M., Ben-Ze'ev A. (2000) Mol. Cell. Biol. 20, 4238–4252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Solanas G., Miravet S., Casagolda D., Castaño J., Raurell I., Corrionero A., de Herreros A. G., Duñach M. (2004) J. Biol. Chem. 279, 49849–49856 [DOI] [PubMed] [Google Scholar]
- 45.Jamora C., Fuchs E. (2002) Nat. Cell Biol. 4, E101–E108 [DOI] [PubMed] [Google Scholar]
- 46.Lorch J. H., Klessner J., Park J. K., Getsios S., Wu Y. L., Stack M. S., Green K. J. (2004) J. Biol. Chem. 279, 37191–37200 [DOI] [PubMed] [Google Scholar]
- 47.Gaudry C. A., Palka H. L., Dusek R. L., Huen A. C., Khandekar M. J., Hudson L. G., Green K. J. (2001) J. Biol. Chem. 276, 24871–24880 [DOI] [PubMed] [Google Scholar]
- 48.Miravet S., Piedra J., Castaño J., Raurell I., Francí C., Duñach M., García de Herreros A. (2003) Mol. Cell. Biol. 23, 7391–7402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Plott R. T., Amagai M., Udey M. C., Stanley J. R. (1994) J. Invest. Dermatol. 103, 168–172 [DOI] [PubMed] [Google Scholar]