Abstract
Bacilysin is a non-ribosomally synthesized dipeptide antibiotic that is active against a wide range of bacteria and some fungi. Synthesis of bacilysin (l-alanine-[2,3-epoxycyclohexano-4]-l-alanine) is achieved by proteins in the bac operon, also referred to as the bacABCDE (ywfBCDEF) gene cluster in B. subtilis. Extensive genetic analysis from several strains of B. subtilis suggests that the bacABC gene cluster encodes all the proteins that synthesize the epoxyhexanone ring of l-anticapsin. These data, however, were not consistent with the putative functional annotation for these proteins whereby BacA, a prephenate dehydratase along with a potential isomerase/guanylyl transferase, BacB and an oxidoreductase, BacC, could synthesize l-anticapsin. Here we demonstrate that BacA is a decarboxylase that acts on prephenate. Further, based on the biochemical characterization and the crystal structure of BacB, we show that BacB is an oxidase that catalyzes the synthesis of 2-oxo-3-(4-oxocyclohexa-2,5-dienyl)propanoic acid, a precursor to l-anticapsin. This protein is a bi-cupin, with two putative active sites each containing a bound metal ion. Additional electron density at the active site of the C-terminal domain of BacB could be interpreted as a bound phenylpyruvic acid. A significant decrease in the catalytic activity of a point variant of BacB with a mutation at the N-terminal domain suggests that the N-terminal cupin domain is involved in catalysis.
Introduction
Bacilysin is a non-ribosomally synthesized di-peptide antibiotic that consists of an l-alanine residue at the N terminus attached to a non-proteinogenic amino acid, l-anticapsin. (Fig. 1a) (1). The antimicrobial activity of bacilysin is initiated by the proteolysis of this di-peptide by a peptidase to release l-anticapsin. l-Anticapsin is a competitive inhibitor of glucosamine synthase. The irreversible inhibition of glucosamine synthase results in the lysis of bacterial or fungal cells. Extensive genetic analysis suggests that the non-ribosomal production of bacilysin is achieved by the stepwise activity of enzymes in the bac operon (2). Synthesis of bacilysin in B. subtilis depends on growth conditions such as supplements to media or temperature (3). Apart from the bac operon, a transposon mutagenesis study revealed the involvement of thyA (thymidylate synthase), ybgG (homocysteine methyl transferase), and the oligopeptide permease OppA in bacilysin synthesis (4, 5). The involvement of OppA in bacilysin synthesis suggests that the production of this antibiotic is likely to be regulated by a quorum-sensing pathway that is responsible for sporulation, competence development and surfactant synthesis (4). Bacilysin could be synthesized in vitro using cell-free extracts of B. subtilis A14 with l-alanine and l-anticapsin as the substrates. This process which required ATP and Mg2+ led to the synthesis of both bacilysin as well as the dipeptide l-Ala-l-Ala (3).
FIGURE 1.
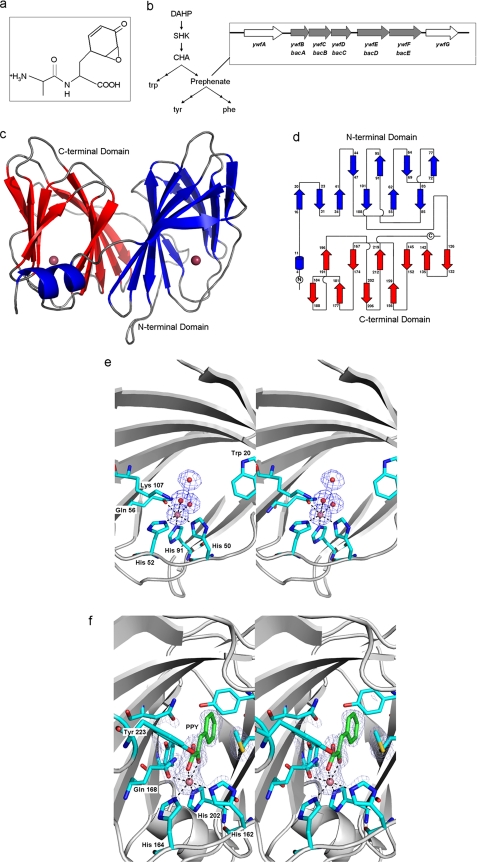
BacB in the context of the bac operon of B. subtilis. a, di-peptide antibiotic bacilysin has an N-terminal alanine and anticapsin at the C terminus. b, biosynthetic route for bacilysin production branches off the aromatic amino acid biosynthesis pathway at prephenate. c, BacB is a bicupin with two putative active sites, each containing a bound metal ion. d, topology diagram of BacB (23). e, electron density map of the active site of the N-terminal domain. The metal ion along with two water molecules is shown along with residue Lys-107 (at the entrance of the β barrel, shown to be crucial for catalytic activity), and residues His-50, His-52, His-91, and Gln-56 (coordinating the metal ion). f, (Fo − Fc) electron density map at the C-terminal domain could be interpreted as a bound PPY. The metal ion along with the coordinating residues His-162, His-164, His-202, and Gln-168 are shown.
Several aspects of the stepwise synthesis of bacilysin have been elucidated by the use of deletion mutants in different strains of B. subtilis. These studies revealed that the deletion of BacD in the bacABCDE operon results in the accumulation of anticapsin, the potent antibiotic component of bacilysin. BacD is a ligase, whereas BacE confers self-protection to bacilysin (5, 6). Biosynthesis of anticapsin branches off the prephenate metabolic pathway and the subsequent synthesis of this antibiotic proceeds by a non-ribosomal mechanism (Fig. 1b) (7). Functional roles for BacA, BacB, and BacC in the bac operon could only be inferred based on the sequence similarity of these proteins to other enzymes (4). BacA was thus proposed to function as a prephenate dehydratase, whereas BacC was annotated as a putative dioxygenase, although no experimental evidence for these activities has been demonstrated in vitro for these proteins (5). BacB was inferred to be similar to isomerase/guanylyl transferases, although the poor sequence similarity between BacB and these proteins suggested that this functional assignment was not likely to be robust. Indeed, these functional annotations were not consistent with genetic data that suggested that the conversion of prephenate to l-anticapsin could be achieved within two to three enzymatic steps (5). Experimental evidence for this hypothesis came from the observation that proteins of the bacABC cluster were sufficient to produce anticapsin and disruption of any of these genes inhibited anticapsin synthesis. An intriguing feature of this synthesis mechanism is the variation across different strains- no recombinant bacilysin is produced upon the transformation of the bac operon in B. coagulans, B. licheniformis, or B. megaterium. On the other hand, introduction of the bacA and bacB genes alone were sufficient to support bacilysin production in B. amyloliquefaciens GSB272 (5). Whereas it is yet possible that certain undetermined factors in the genetic background of B. amyloliquefaciens support ligation and self-protection functions, these data suggest that BacA and BacB constitute a minimal subset of the non-ribosomal machinery for l-anticapsin synthesis. In the absence of unambiguous bioinformatics or in vitro enzymatic data, we proposed to characterize the BacA and BacB enzymes in vitro. The identification of the enzymatic activities of these two enzymes along with the fortuitous identification of a bound phenylpyruvate (PPY)2 at the active site of BacB provides a biochemical rationale for the role of these enzymes in l-anticapsin production as well as a potential route to the regulation of bacilysin synthesis.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification of BacA and BacB
The cloning, expression, and purification of recombinant BacB has been reported earlier (8). Briefly, Escherichia coli BL21(DE3)PLysS cells expressing recombinant BacB were grown in Luria Broth. Recombinant BacB, with a polyhistidine tag at the C terminus, was partially purified using cobalt-NTA resin (Talon, Clontech Inc.). The protein was further purified by size exclusion chromatography using a Sephacryl S-200 column (Amersham Biosciences). The purity of BacB was examined using SDS-PAGE as well as mass spectrometry (MALDI-TOF, Bruker Daltonics, Inc.). The bacA gene, which precedes bacB in the bac operon, was cloned into the pET28a expression vector. Similar to BacB, recombinant BacA was transformed into E. coli BL21 (DE3) cells and purified using cobalt-NTA affinity chromatography.
Crystallization and Structure Determination of BacB
Initial screening for crystallization conditions of BacB was performed using crystallization kits from Hampton Research Inc. Needle-shaped crystals could be obtained in more than one crystallization condition; however a range of polyethylene glycol (PEG 1, 3.3, 4, and 8 K) and different ratios of 2-methane-4-pentane-diol (MPD) had to be examined to obtain diffraction quality crystals. Modification of the crystallization protocol was the key to obtaining crystals of a different morphology from the poor-quality plate-like or needle-like forms to three-dimensional crystals, which belonged to either monoclinic or tetragonal space groups. A single crystal of BacB was mounted on a cryo-loop using 20% PEG400 as the cryo-protectant. The monoclinic crystal form diffracted up to 1.9 Å whereas the tetragonal form crystals of BacB diffracted up to 2.2 Å at the home source. Diffraction data were processed using MOSFLM (9) and were scaled using SCALA (10). Initial molecular replacement trials using different cupin domain models were unsuccessful. Subsequently, efforts were made to solve this structure using isomorphous derivatives. Purified recombinant BacB was purple-green in color, which suggested a bound metal ion in the purified protein. The change in color of the cobalt-NTA affinity beads used in the first step of purification suggested that the metal ion bound with BacB could be Co2+. This was subsequently noted to be the metal cofactor that was most suited for the catalytic activity of BacB (supplemental Fig. S3). The presence of the bound Co2+ ion could also be confirmed by x-ray fluorescence (data not shown). The high redundancy (∼22) and completeness of the data in the tetragonal form of BacB provided an excellent dataset for structure determination using single wavelength anomalous dispersion (SAD). The positions of the metal ions were located using PHENIX (11), followed by iterative model building and refinement using Coot (12), Arp/Warp (13), and Refmac5 (10). The data collection, phasing, and refinement statistics are compiled in Table 1.
TABLE 1.
Data collection, phasing, and refinement statistics
Initial phase information was obtained from SAD using the data of tetragonal form of BacB. Phase information for the structure in the monoclinic space group was obtained by molecular replacement using the structure from the tetragonal form.
| Tetragonal form | Monoclinic form | ||
|---|---|---|---|
| I. Data Collection | |||
| Wavelength | 1.5418 Å | 1.5418 Å | |
| Resolution (Å) | 35.27-2.21 (2.33-2.21)a | 26.21-1.86 (1.96-1.86)a | |
| Unit cell parameters | a = b = 68.7 Å, c = 211.5 Å | a = 45.9 Å, b = 118.5 Å, c = 46.9 Å, β = 97.2° | |
| Space group | P41212 | P21 | |
| Total number of reflections | 573250 (76162) | 226869 (26730) | |
| Number of unique reflections | 25906 (3264) | 39949 (4989) | |
| Completeness (%) | 98.1 (87.0) | 95.5 (81.9) | |
| Multiplicity | 22.1 (23.3) | 5.7 (5.4) | |
| Rmergeb (%) | 10.2 (38.1) | 6.7 (59.8) | |
| <I>/σ (I) | 31.3 (10.1) | 15.3 (2.7) | |
| II. Phasing | |||
| Co2+ (SAD) | Molecular Replacement | ||
| Resolution bin (Å) | FOM | Model: A chain of Tetragonal form | |
| 35.0-8.43 | 0.31 | Phaser Statistics | |
| 8.43-5.29 | 0.36 | Z-Score46.24 | |
| 5.29-4.12 | 0.34 | LLG Final 1491.35 | |
| 4.12-3.49 | 0.32 | ||
| 3.49-3.08 | 0.28 | ||
| 3.08-2.78 | 0.23 | ||
| 2.78-2.56 | 0.18 | ||
| 2.56-2.38 | 0.14 | ||
| Mean | 0.25 | ||
| III. Refinement | |||
| Rcrystc (%) | 0.22 | 0.19 | |
| Rfreed (%) | 0.28 | 0.23 | |
| Rmsbond (Å) | 0.01 | 0.01 | |
| Rmsangle (degree) | 1.52 | 1.48 | |
a Values for outer shells are given in parentheses.
b Rmerge = Σj|<I> − Ij|/Σ<I> where Ij is the intensity of the jth reflection and <I> is the average intensity.
c Rcryst = Σhkl|Fo − Fc|/Σhkl|Fo|.
d Rfree was calculated as for Rcryst but on 5% of the data excluded from the refinement calculation.
2,4-Dinitrophenylhydrazine (DNPH) Assay
The presence of PPY in BacB was determined using previously published experimental procedures (14). A stock solution of DNPH (0.125 g/liter) was dissolved in 2 m HCl. Increasing concentrations of the BacB protein extract was incubated with 50 μl of DNPH stock solution in 200 μl of the reaction mixture. The reaction mixture was incubated at 37 °C for 10 min to enable the formation of the DNPH-PPY adduct. The reaction was stopped by the addition of 50 μl of 0.6 m NaOH. A UV/visible spectrum was recorded with a wavelength scan between 600 and 350 nm. The supernatant used for the PPY assay was also subjected to ESI analysis in both positive and negative ionization modes.
Characterization of the Reaction Products of BacA and BacB
The reaction products of BacA and BacB were characterized using high pressure liquid chromatography (HPLC) using a Shimadzu SPD-20A instrument. The reaction products were separated on a C-18 Zorbax analytical column (Agilent, Inc) at a flow rate of 0.5 ml/min. with an acetonitrile gradient from 0 to 20%. Samples for HPLC analysis were prepared by incubating 100 μm prephenate with 1 μm BacA and 0.5 μm BacB enzymes at 37 °C for various time intervals (1, 5, 10, and 60 min). The chromatogram was recorded at two wavelengths, 220 and 280 nm.
UV Spectrophotometric Analysis
The UV absorbance spectra were recorded on a Varian-Cary 100 Bio-UV/visible spectrophotometer. In the case of the activity assays for BacA and BacB, the time course spectra between 340 and 220 nm were obtained with the sample in a 100-μl cuvette.
NMR Spectroscopy
All NMR spectra were acquired on a Bruker Avance 700 MHz spectrometer equipped with a 5-mm triple resonance cryoprobe with single (z axis) pulsed field gradient accessory. Spectra were acquired at 303 K. NMR samples were prepared in buffers made in 90% H2O/10% D2O. One-dimensional 1H NMR spectra were acquired using excitation sculpting with gradients for water suppression (15). NMR data were processed on an INTEL PC Work station running on Suse Linux 10.1 using NMRPipe/NMRDraw processing software. The directly detected time domain data were processed by applying a 90° phase-shifted squared sine bell filter. Data sets were zero-filled prior to Fourier transformation. All chemical shifts were referenced to external DSS.
The procedure for the time course measurements of BacA and BacB reactions was performed in two steps. First, the reaction mixture was incubated with 4 μm BacA for 15 min. Subsequently, BacB was added to a final concentration of 5 μm. The reaction samples were prepared in 5 mm Tris-HCl buffer, pH-8.1. The product formation was monitored based on the new peaks in the 1H NMR spectra that were absent in the spectrum of prephenate. The chemical structures of the products were deduced based on the consistency between the observed 1H NMR spectra as well as mass spectrometric data.
Enzyme Assays for BacA and BacB
Barium prephenate (Sigma-Aldrich, Inc.) was dissolved in 50 mm Tris-HCl buffer (pH 8.1). In the case of BacA, the enzyme assay was started by adding 0.5 μm BacA to the reaction mixture (110 μl) at room temperature. The reaction mixture was incubated for 20 min prior to the detection of the product. The substrate for the reaction catalyzed by BacB is the product of BacA. Thus 0.2 μm BacB was added to the reaction mixture and immediately the product formation was monitored for 10 min. Varying concentrations of prephenate viz., 10, 30, 60, 100, 200, 400, 700, and 1000 μm were examined. Appropriate controls to account for the absorbance of BacA, BacB, and the auto-oxidation of the product of BacA were incorporated into the procedure. All assays were performed in triplicate.
RESULTS
The Crystal Structure of BacB
Cupin proteins, in general, share poor sequence similarity although their structural features are remarkably conserved. This feature has been alluded to as evidence that proteins with the cupin fold exploit reaction space to acquire diverse functional roles (16). The only sequence feature common to these proteins are the so-called cupin motifs: GX5HXHX3,4EX6G and GX5PXGX2HX3N separated by a loop that can vary in length from 15–50 residues (17). An intriguing feature of this motif is that although the cupin domain is duplicated in bi-cupins, the sequence motif(s) are not necessarily duplicated (18). Put together, the low sequence but moderate structural conservation among cupin proteins implies that predicting the functional roles can be difficult by conventional bioinformatics or knowledge-based strategies. B. subtilis BacB is a classic illustration of this problem. Although BacB was identified as a bicupin based on the presence of the cupin sequence motifs, the putative functional assignment as an isomerase/guanylyl transferase could not be reconciled with its role in anticapsin synthesis. Indeed, initial efforts to solve the structure of BacB by molecular replacement using different cupin domain models were unsuccessful. Eventually, the structure was solved by SAD with the anomalous scattering from the bound cobalt ion at the active site.
BacB is a bi-cupin protein with two cupin (double-stranded β helix) domains fused in a tightly packed arrangement. The crystal structure of the BacB monomer, the most compact bicupin structure known to date, consists of 225 residues (Fig. 1, c and d). BacB comprises two ten-stranded β barrels, with a small α-helical stretch forming the base of the C-terminal barrel. Each cupin domain is characterized by a sandwich arrangement of a six-stranded and a four-stranded antiparallel β-sheet. Each of the 10 β-strands in the barrel is approximately perpendicular to the barrel axis. The two cupin domains of BacB are related by an approximate 2-fold axis (involving a rotation of 173.3°) and superpose fairly well with a root mean-squared deviation (R.M.S.D.) of 0.95 Å. The N-terminal domain comprises residues 16–109, whereas the cupin domain at the C terminus comprises residues 126–219. Residues 5–15 adopt an α-helical conformation, whereas residues 16–20 from the N-terminal cupin domain are involved in subunit contacts in the asymmetric unit. The two cupin domains are linked by a largely flexible stretch comprising residues 110–125. There are two molecules of BacB in the asymmetric unit of the crystal. BacB, however, is a monomer in solution as seen by size exclusion chromatography studies (data not shown).
The Metal Ion Binding Site(s) of BacB
Two metal ions could be clearly modeled into the electron density at the putative active sites of BacB in both domains. The location of this metal ion in both active sites is such that the metal cofactor binding site remains partially exposed to the solvent. The two metal ion sites within a BacB monomer are separated by ∼27 Å. These are coordinated by a distorted bi-pyramidal (octahedral) geometry in both the active sites of BacB (Fig. 1, e and f). The residues coordinating the metal ion in the N-terminal domain are His-50, His-52, His-91, and Gln-56, whereas those in the C-terminal domain include His-162, His-164, His-202, and Gln-168. In the N-terminal domain, the remaining coordination sphere is completed by two water molecules, whereas the C-terminal domain revealed additional density for a bound ligand (discussed in the next section). The metal ion coordination distances are in the range of 2.1–2.3 Å. The residues surrounding the active site(s) are predominantly hydrophobic. The N-terminal domain is rich in valine and leucine residues (Val-24, Val-41, Val-47, Val-69, Val-101, and Leu-37, Leu-65), whereas the C-terminal domain is dominated by methionine and tyrosine residues (Met-135, Met-148, Met-159, Met-179, Met-188, and Tyr-177, Tyr-194, Tyr-223). The residues Lys-107 and Trp-20 at the entrance of the N-terminal cupin domain appear poised for substrate recognition. This functional role was subsequently confirmed by mutational analysis. Asn-224 and Ile-32 adopt a similar position in the C-terminal cupin domain. The base of C-terminal domain is formed by Tyr-11 and Phe-12, a position occupied by Tyr-121 and Phe-122 in the N-terminal cupin domain. There are three salt bridges between Arg-31 and Glu-200, Arg-74 and Asp-124, and Arg-108 and Glu-167 that serve to tether the compact bi-domain arrangement. In addition to these interdomain interactions, a short helix from the N-terminal domain traverses across to the C-terminal barrel. A noteworthy feature of the two cupin domains is that despite sequence variations in the residues that surround the cupin domains, the cavities formed by the two cupin domains are fairly similar (28.1 and 29.5 Å3 for the N- and the C-terminal domains).
Experimental Evidence for the Bound Phenylpyruvate at the C-terminal Domain of BacB
Clear density to accommodate an aromatic ligand was seen in the refined electron density map at 1.9 Å (Fig. 1f). Despite identical metal ion coordination, additional electron density for the bound ligand was seen in the C-terminal cupin domain alone in both crystal forms of BacB. This electron density at the C-terminal domain could not be explained on the basis of chemical entities added at various steps of purification or additives in the crystallization condition. First, the ligand is planar. It has a carboxylic acid group reacting with the metal ion and an aromatic ring which points away from the metal ion. Based on these features, a search in MSDchem resulted in two suitable contenders, PPY and phenylalanine (Phe) (19). The planarity seen in the electron density map clearly suggests PPY over phenylalanine (Fig. 1f). Tyr-223 makes a hydrogen bond with the PPY bound at the active site of the C terminus and could thus be involved in positioning this substrate in the active site. PPY was not added at any point during the purification and crystallization of this protein, as the underlying assumption at that stage was that we were working on a potential mannose isomerase or a guanylyl transferase. It thus appears likely that the ligand bound BacB at some point of protein synthesis in E. coli. Efforts made post-facto to obtain BacB in the absence of PPY were not successful.
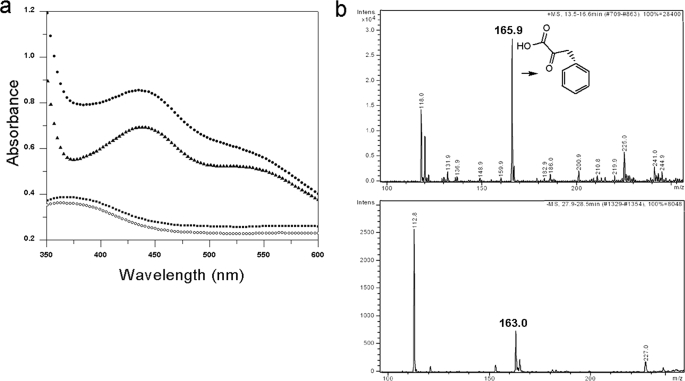
To confirm the presence of PPY at the active site, an assay based on DNPH was carried out. More generally, the DNPH assay is adopted for the estimation of pyruvic acid based on the formation of an adduct of the ketone group of pyruvate with DNPH. Using a slightly modified version of the conventional assay protocol, the supernatant of denatured BacB was checked for the formation of the DNPH adduct by measuring the absorbance at 420 nm and 515 nm (Fig. 2a). The assay was robust, providing clear evidence for PPY over the controls, viz., phenylpyruvic acid-disodium salt taken as a positive control versus phenylalanine as a negative control. The ESI-MS spectrum of PPY extracted from BacB in both positive as well as in negative ion modes is shown in Fig. 2b. The theoretical mass of PPY is 164.16 Da. The mass of PPY was confirmed by the peak at 165.9 Da in positive ionization and by a peak at 163.0 Da in the negative ionization mode.
FIGURE 2.
Experimental evidence for a bound phenylpyruvate at the C-terminal active site of BacB. a, DNPH assay with denatured BacB extract. (○), DNPH alone; (■), phenylalanine treated with DNPH (negative control); (▴), phenylpyruvic acid-sodium salt treated with DNPH (positive control); (●), BacB treated with DNPH. b, ESI-spectrum of BacB extract in positive as well as in negative ion modes shows the presence of phenylpyruvic acid.
Role of BacA and BacB in Anticapsin Synthesis
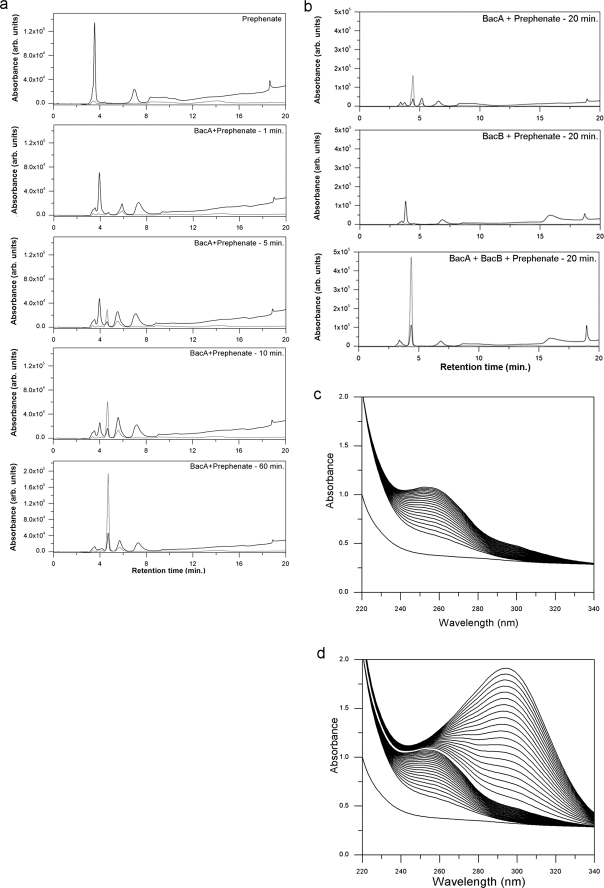
The synthesis of bacilysin branches off the pathway for aromatic amino acid synthesis at prephenate. We therefore examined if prephenic acid was modified by either BacA or BacB or both. This was done by incubating prephenic acid for 20 min at 37 °C with either BacA or BacB. The reaction products were separated using HPLC as described earlier. In the absence of BacA, the prephenate peak at 220 nm appears at a retention time of ∼4 min. Addition of BacA leads to the gradual decrease of this peak with the appearance of two new peaks at 4.5 and 5 min retention. This time course analysis of the product formed upon the incubation of BacA with prephenate is shown in Fig. 3, a and b. The variation in the product profile confirmed that prephenate is a substrate for BacA and not for BacB. Addition of BacB to this reaction mixture (BacA and prephenate) led to a change in the HPLC chromatogram with the disappearance of the 5-min peak, now replaced by one at the 4.5-min retention time. This new compound (profile shown in grey in Fig. 3b) suggested that the product of BacA served as a substrate for BacB. We note that prolonged incubation of prephenate with BacA also leads to the same product (Figs. 3a and 4a).
FIGURE 3.
HPLC and UV spectrophotometric studies of the products of BacA and BacB enzymes. a, HPLC elution profiles of the BacA product with prephenate at different time points (1, 5, 10, and 60 min; 220 nm, 280 nm). b, HPLC elution profile of prephenate incubated with BacA alone, prephenate with BacB alone and prephenate with BacA and BacB. c, UV spectrophotometric time course measurement of the catalytic activity of BacA on prephenate. These spectra were recorded at intervals of 30 s. d, UV spectrophotometric time course measurement of the catalytic activity of BacB. These spectra were recorded at intervals of 30 s.
FIGURE 4.
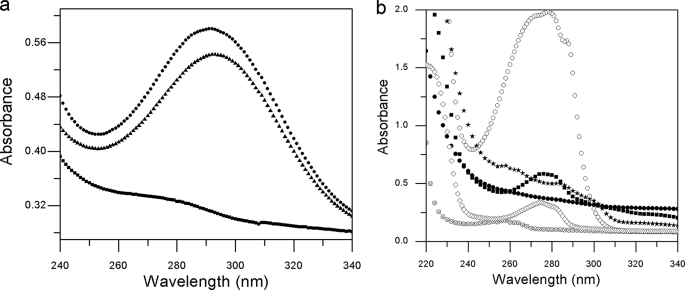
Auto-oxidation of the product of BacA. a, product obtained by the activity of BacA on prephenate is prone to oxidation upon prolonged incubation. The spectroscopic profile of this product resembles that of BacB. Here, (■), prephenate alone incubated for 12 h; (▴), BacA with prephenate incubated for 12 h; (●), BacA and BacB with prephenate incubated for 15 min. b, reference UV spectra of potential substrates and products of the Bac proteins. (■), 4-hydroxy phenylpyruvic acid, (⊗), l-phenylalanine; (○), dl-tryptophan; (●), prephenate; (◊), l-tyrosine, (★), phenylpyruvic acid.
The change in the UV spectra of prephenate upon incubation with BacA is shown in Fig. 3c. These time-course measurements revealed that incubation of BacA with prephenate led to a product with a λmax of ∼260 nm. Subsequent addition of BacB results in a compound with an absorption maximum at 293 nm with a ∼5-fold increase in the absorbance (Fig. 3d). For a comparison, the spectra for all the compounds analyzed in this study (4-hydroxyphenylpyruvic acid, l-phenylalanine, d,l-tryptophan, prephenate, l-tyrosine, phenylpyruvic acid) recorded between 340 and 240 nm is shown in Fig. 4b.
NMR Spectroscopy for the Identification of the Reaction Products
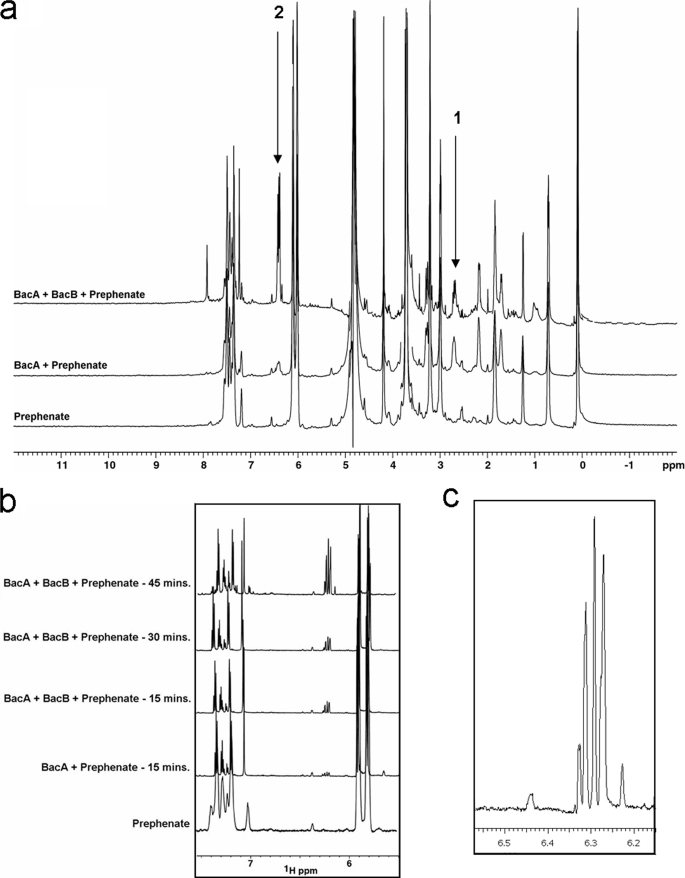
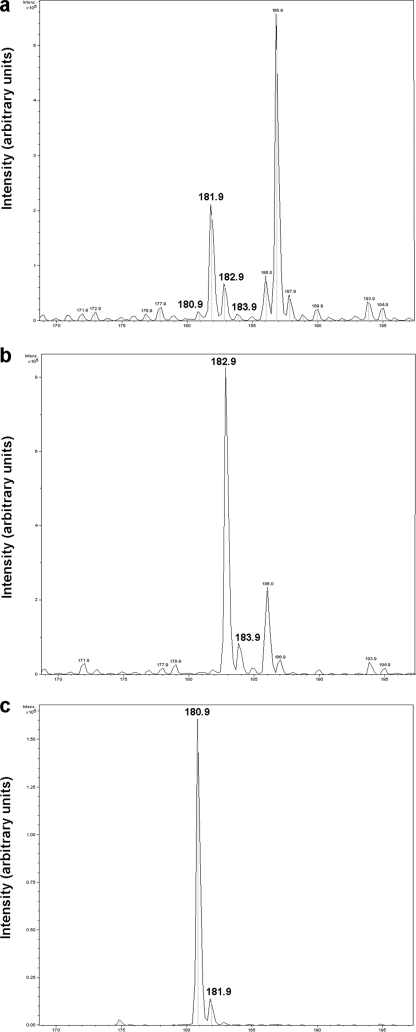
1H NMR spectra of prephenate and the product of the reaction catalyzed by BacA and BacB are shown in Fig. 5. The NMR spectrum shows the conversion of prephenate (s, δ 5.92 and 5.83 ppm) into a product that has unique 1H chemical shifts centered at 6.28, 6.34 ppm in the aromatic region and at 2.78 ppm in the aliphatic region of the spectrum. The chemical shift dispersion of the product suggests its formation to be an outcome of a decarboxylation and an oxidation reaction. These spectra were compared with a simulated 1H NMR spectrum with molecular structures generated using ChemDraw (Cambridge Software, Inc.) (supplemental Fig. S1). Assignment of resonances originating from the product (Fig. 6) agrees well with the simulated NMR spectrum. The chemical shift assignment of prephenate was based on the recorded chemical shift of the pure sample. The identification of the reaction products of BacA and BacB is also consistent with the mass spectra of these compounds (Fig. 7). The mass of barium prephenate is 361.49 Da. The mass obtained by ESI-MS was 226.9 Da in the positive ion mode which corresponds to the mass of prephenate without barium (calculated mass 226.04 Da). Incubation of BacA with prephenate results in products with a mass of 181.9 (182.9 and 183.9 Da with isotopic splitting) in the positive ionization mode (Fig. 7a). Similarly, the incubation of BacA and BacB with prephenate results in products with masses 182.9 (183.9 Da) in the positive ion mode and 180.9 (181.9 Da) in the negative ionization mode with isotopic splitting (Fig. 7, b and c).
FIGURE 5.
1H NMR spectrometric studies. a, overlay of 1H NMR spectra corresponding to prephenate alone, BacA incubated with prephenate, and BacA and BacB incubated with prephenate are shown. These spectra show the product formation through chemical shifts peak 1 (around 3.0 ppm) upon incubating prephenate with BacA and peak 2 (around 6.28–6.34 ppm) upon incubating prephenate with both BacA and BacB. These chemical shifts match the predicted 1H NMR spectra of the products (also see supplemental Fig. S1). The details of sample preparation and data acquisition are discussed under “Experimental Procedures.” Approximately 70% pure barium prephenate (Sigma-Aldrich) was dissolved in 5 mm Tris-HCl buffer, pH 8.1, for the activity assays. b, time course recording of 1H NMR spectra shows an increase in the product formation (6.28–6.34 ppm) upon incubating prephenate with both BacA and BacB. c, magnified 1H NMR spectrum of the product (6.28–6.34 ppm) by the action of BacA and BacB enzymes upon incubation with prephenate for 45 min.
FIGURE 6.
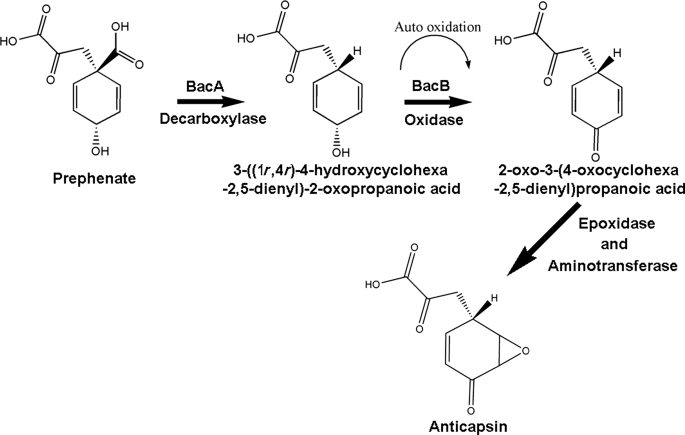
Schematic of the reactions catalyzed by BacA and BacB. Prephenate is converted into an unstable intermediate 3-((1r-4r)-4-hydroxycyclohexa-2,5-dienyl)-2-oxopropanoic acid after a decarboxylation reaction catalyzed by BacA. BacB catalyzes the oxidation of this product to yield 2-oxo-3-(4-oxocyclohexa-2,5-dienyl)propanoic acid. The synthesis of l-anticapsin from 2-oxo-3-(4-oxocyclohexa-2,5-dienyl)propanoic acid would further require a ring epoxidation and a transamination reaction in the ketone group.
FIGURE 7.
Mass spectra of the reaction products of BacA and BacB. a, ESI spectra of BacA incubated with prephenate in the positive ionization mode. b, ESI spectra of the product formed by the incubation of BacA and BacB with prephenate in positive ionization mode. c, ESI spectra of the product formed by the incubation of BacA and BacB with prephenate in and in negative ionization mode. The mass of barium prephenate is 361.49 Da. The deconvoluted mass obtained by ESI-MS is 226.9 Da in the positive ion mode, which corresponds to the mass of prephenate without barium (calculated mass 226.04 Da). Incubation of BacA with prephenate results in products with masses of 181.9, 182.9, and 183.9 Da in the positive ionization mode (isotopic splitting). The difference in mass between the substrate and the product (45 Da) confirms the decarboxylation reaction catalyzed by BacA. The incubation of BacA and BacB with prephenate results in products with masses 182.9 (183.9 Da) in the positive ion mode and 180.9 (181.9 Da) in the negative ionization mode (with isotopic splitting).
The N-terminal Domain of BacB Participates in Catalysis
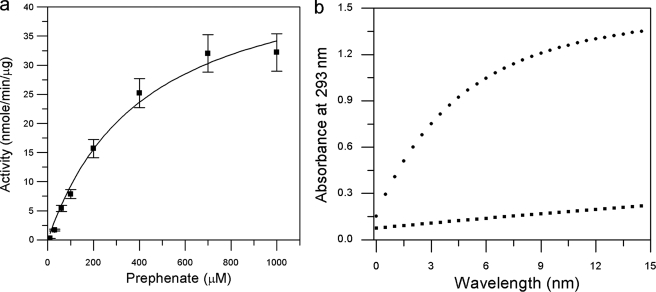
BacB catalyzes the oxidation of the product of BacA to 2-oxo-3-(4-oxocyclohexa-2,5-dienyl)propanoic acid. The enzyme kinetics of BacB appears to follow a quasi steady state approximation with a single binding site with an apparent Michaelis-Menten Km value being 423 ± 81 μm and Vmax value of 53.49 ± 4.56 nmol/min/μg. The kcat value is 1471 min−1 (Fig. 8a). As seen from the crystal structure (and later verified by a biochemical assay), the C-terminal domain binds PPY with very high affinity. In an effort to further examine the roles of these two domains, critical residues identified in the crystal structure were examined. Thus mutations were carried out both N- and C-terminal domains. One point variant of BacB, the K107A mutant, showed a significant decrease in catalytic activity when compared with the native enzyme (Fig. 8b, supplemental Fig. S2). Lys-107 is located at the entrance of the N-terminal cupin domain (Fig. 1e). The oxidation catalyzed by BacB is thus appears to be localized to the N-terminal cupin domain.
FIGURE 8.
The catalytic activity of BacB and K107A variant. a, Michaelis-Menten plot for the activity of the native BacB. The kinetic parameters are as noted in the text. b, N-terminal domain of BacB participates in catalysis. (●), product formed by the incubation of BacA and BacB with prephenate for 15 min (λmax, 293 nm); (■), incubation of BacA and the K107A mutant of BacB with prephenate for 15 min.
DISCUSSION
The dipeptide antibiotic bacilysin (l-alanine-l-anticapsin) is a potent anti-microbial agent and has been noted to be effective against multi-drug resistant Staphylococcus aureus. Although extensive genetic studies have revealed the proteins responsible for the synthesis of this antibiotic in vivo, details of the enzymatic steps in the synthesis of this antibiotic remained elusive. The main drawback to understanding the synthesis of this simple, yet effective antibiotic was the mismatch between bioinformatics predictions on the role of the Bac operon proteins, especially BacA and BacB and genetic data that suggested that these two genes alone were sufficient for l-anticapsin production. A simplistic interpretation of genetic data implied that there could be three enzymatic reactions involved in the production of l-anticapsin in B. subtilis 168 spp. However, in the case of B. subtilis pumulis the bacAB genes alone were sufficient to produce l-anticapsin. Functional annotations for these gene products based on sequence similarity were confusing- BacA shares some sequence similarity with prephenate dehydratase whereas BacB is similar to an isomerase/guanylyl transferase. Another intriguing aspect of l-anticapsin production was the inhibition of its synthesis by both alanine as well as phenylalanine. Surprisingly, the latter observation was easier to rationalize based on the crystal structure. The presence of PPY at the active site of the C-terminal domain suggested a potential role of this domain in the regulation of l-anticapsin, and hence bacilysin synthesis. Nevertheless, the high resolution crystal structure by itself was insufficient to unambiguously assign its function, given that none of the predicted enzymatic activities could be demonstrated in vitro. Thus whereas the bound PPY at the C-terminal active site of BacB suggested a structural rationale for inhibition, the function of BacB remained elusive. The role of BacB was thus examined in the context of the activity of BacA, the enzyme preceding bacB in the bacABC gene cluster.
Most non-ribosomally synthesized peptides and polyketides are products of non-ribosomal peptide synthetase (NRPS) and polyketide synthase (PKS) protein complexes that work in an assembly line fashion (20, 21). It was therefore imperative to characterize the function of BacA to understand the function of BacB. To evaluate if BacA was either a prephenate dehydrogenase or a prephenate dehydratase, experiments were carried out on BacA with prephenic acid. The spectroscopic characterization of the reaction of BacA with prephenate as a substrate revealed that BacA is a decarboxylase that catalyzes the synthesis of 3-((1r-4r)-4-hydroxycyclohexa-2,5-dienyl)-2-oxopropanoic acid. The experimentally observed activity of BacA is thus different from either of the predicted functions viz., prephenate dehydrogenase or dehydratase. The conclusion that BacA is a decarboxylase is further ratified by the coupling between the reactions catalyzed by BacA and BacB. If BacA were to be a prephenate dehydrogenase it would produce 4-hydroxy phenylpyruvic acid whereas a prephenate dehydratase would yield phenylpyruvic acid. BacB was inactive with both these substrates.
The decarboxylation of prephenate catalyzed by BacA involves the replacement of the carboxylic group by hydrogen in the ortho position of the 6-membered ring. The appearance of a peak around 3 ppm (Fig. 5a) shows that the -COOH group is eliminated from prephenate by BacA. The subsequent reaction catalyzed by BacB yields a new peak in the 1H NMR spectrum centered at 6.2 ppm. This can be explained by an oxidation of the -OH group to =O in the para position of the ring. This interpretation is consistent with absorption changes in the UV spectrum with a shift in the absorption maximum from 260 to 293 nm. In this context of bioinformatics-based annotation vis-à-vis experimentally observed functions, the result of a structure-based phylogenetic analysis is worth noting (22). In this study using a three-dimensional structure-based phylogenetic analysis, BacB clusters more closely with quercetinases than any other cupin enzyme suggesting that the gross functional class of BacB could be closer to an oxidoreductase rather than an isomerase or a transferase as classified earlier (5). The catalytic activity of BacA and BacB where prephenate is converted into an intermediate followed by the formation of 2-oxo-3-(4-oxocyclohexa-2,5-dienyl) propanoic acid by BacB is schematically represented in Fig. 6. The biochemical characterization of BacA and BacB thus suggest that the production of l-anticapsin can be carried out from 2-oxo-3-(4-oxocyclohexa-2,5-dienyl)propanoic acid by a ring epoxidation and a transamination reaction in the ketone group. The genetic data that BacA and BacB alone are sufficient for l-anticapsin synthesis thus implies that other enzymes, apart from the bac operon members, could play a role in l-anticapsin synthesis. To conclude, the characterization of the reaction products of BacA and BacB as well as the crystal structure of BacB, suggests that B. subtilis BacB catalyzes the synthesis of a stable intermediate en route to l-anticapsin.
Supplementary Material
This work was supported in part by grants from the Department of Science and Technology, Department of Biotechnology, and the Council for Scientific and Industrial Research, India.
The atomic coordinates and structure factors (codes 3H7J and 3H7Y) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- PPY
- phenylpyruvate
- DNPH
- 2,4-dinitrophenylhydrazine
- ESI-MS
- electron spray ionization mass spectrometry
- PEG
- polyethylene glycol
- R.M.S.D.
- root mean square deviation
- NTA
- nitrilotriacetic acid
- SAD
- single wavelength anomalous dispersion.
REFERENCES
- 1.Kenig M., Abraham E. P. (1976) J. Gen. Microbiol. 94, 37–45 [DOI] [PubMed] [Google Scholar]
- 2.Finking R., Marahiel M. A. (2004) Annu. Rev. Microbiol. 58, 453–488 [DOI] [PubMed] [Google Scholar]
- 3.Sakajoh M., Solomon N. A., Demain A. L. (1987) J. Ind. Microbiol. 2, 201–208 [Google Scholar]
- 4.Inaoka T., Takahashi K., Ohnishi-Kameyama M., Yoshida M., Ochi K. (2003) J. Biol. Chem. 278, 2169–2176 [DOI] [PubMed] [Google Scholar]
- 5.Steinborn G., Hajirezaei M. R., Hofemeister J. (2005) Arch. Microbiol. 183, 71–79 [DOI] [PubMed] [Google Scholar]
- 6.Tabata K., Ikeda H., Hashimoto S. (2005) J. Bacteriol. 187, 5195–5202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roscoe J., Abraham E. P. (1966) Biochem. J. 99, 793–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rajavel M., Gopal B. (2006) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 62, 1259–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leslie A. G. W. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 1696–1702 [DOI] [PubMed] [Google Scholar]
- 10.Collaborative Computational Project Number 4. (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 11.Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 12.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 13.Lamzin V. S., Wilson K. S. (1993) Acta Crystallogr. D Biol. Crystallogr. 49, 129–147 [DOI] [PubMed] [Google Scholar]
- 14.Anthon G. E., Barrett M. D. (2003) J. Sci. Food Agric. 83, 1210–1213 [Google Scholar]
- 15.Hwang T. L., Shaka A. J. (1995) J. Magn. Reson. 112, 275–279 [Google Scholar]
- 16.Anantharaman V., Aravind L., Koonin E. V. (2003) Curr. Opin. Chem. Biol. 7, 12–20 [DOI] [PubMed] [Google Scholar]
- 17.Dunwell J. M., Purvis A., Khuri S. (2004) Phytochemistry 65, 7–17 [DOI] [PubMed] [Google Scholar]
- 18.Gopal B., Madan L. L., Betz S. F., Kossiakoff A. A. (2005) Biochemistry 44, 193–201 [DOI] [PubMed] [Google Scholar]
- 19.Dimitropoulos D., Ionides J., Henrick K. (2006) Current Protocols in Bioinformatics, pp. 14.13.11–14.13.13, John Wiley and Sons, Hoboken, NJ: [DOI] [PubMed] [Google Scholar]
- 20.Walsh C. T. (2004) Science 303, 1805–1810 [DOI] [PubMed] [Google Scholar]
- 21.Marahiel M. A., Stachelhaus T., Mootz H. D. (1997) Chem. Rev. 97, 2651–2674 [DOI] [PubMed] [Google Scholar]
- 22.Agarwal G., Rajavel M., Gopal B., Srinivasan N. (2009) PLoS One 4, e5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams A., Gilbert D. R., Westhead D. R. (2003) Protein Eng. 16, 913–923 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.