Abstract
MicroRNAs contribute to cancer development by acting as oncogenes or tumor suppressor genes. However, only a few microRNA target genes were determined. We identified a nearly perfect complementarity between miR-206 and the 3′-untranslated regions of both mouse and human notch3. Expression of miR-206 decreased the luciferase activity dose-dependently when cotransfected with the mouse or human notch3 3′-untranslated region-luciferase reporter containing the miR-206 target site in HeLa cells. This suppression was relieved by deletion and mutation of the miR-206-binding site and was partially recovered by expression of notch3 or by a specific inhibitor of miR-206. Interestingly, overexpression of miR-206 decreased the levels of both Notch3 protein and mRNA. Expression of miR-206 markedly induced apoptotic cell death and blocked the anti-apoptotic activity of Notch3. In addition, ectopic expression of miR-206 inhibited HeLa cell migration and focus formation. Therefore, we identified miR-206 as a pro-apoptotic activator of cell death, which was associated with its inhibition of notch3 signaling and tumor formation. The inhibition of cancer cell migration and focus formation by miR-206 strongly suggests that miR-206 may function as a novel tumor suppressor.
Introduction
MicroRNAs (miRNAs)2 regulate gene expression by binding to the 3′-untranslated regions (3′-UTRs) of specific mRNAs (1) and play important roles in development, proliferation, and differentiation (2–4). Recent studies revealed the involvement of miRNAs in apoptosis signaling (5). For instance, miR-15 and miR-16 have been shown to induce apoptosis by targeting Bcl-2 (6). miR-34a can be regulated by p53, which promotes apoptosis (7, 8). Despite the growing evidence for their importance in carcinogenesis (9), limited information is available about their function in human cancers. The future challenges are to identify the biological targets of miRNAs and the signaling pathways they regulate during oncogenesis.
Early studies identified miR-206 as a skeletal muscle- specific miRNA involved in muscle development (10–12). Recent studies showed that miR-206 targets the estrogen receptor (13) and is down-regulated in estrogen receptor-positive breast cancer (14). It is thus postulated that miR-206 may be associated with breast cancer metastasis (15). In addition to its high expression in muscle, miR-206 was also reported to be expressed in brain associated with schizophrenia (16) and in brown adipocytes (17). The level of miR-206 is increased in bone marrow-derived DC19+ WM cells associated with Waldenström macroglobulinemia (18). Despite a broader tissue-specific expression than originally anticipated, the physiological function of miR-206 in cancer progression remains largely unknown.
In this study, we report several novel findings. We identify notch3 (Notch homolog 3) as a novel miR-206 target. We reveal miR-206 as a potent apoptotic cell death inducer via cross-talk with notch3. We further show that miR-206 inhibits tumor cell migration and focus formation. Our study suggests miR-206 as a critical regulator of tumorigenesis.
EXPERIMENTAL PROCEDURES
Real-time Reverse Transcription-PCR Quantification of miRNAs
Real-time reverse transcription-PCR quantification of miRNA expression was carried out using a TaqMan® microRNA assay kit (Applied Biosystems, Foster City, CA) according to the manufacturer's protocol. Small nucleolar RNA 202 was used as an internal control to normalize RNA input in the real-time reverse transcription-PCR assay. The detailed method was described in our recent study (19).
Targeted Gene Prediction of miR-206
The full-length mRNAs of human and mouse notch3 (NM_008716 and NM_000435) were obtained from the NCBI Database. The 3′-UTRs of human and mouse notch3 were extracted and converted to FASTA format. The miRNA mature sequence data base (miRBase) was obtained from the Sanger Institute. The miRanda algorithm was used for finding potential miR-206 target sites in the human and mouse notch3 3′-UTRs. Score threshold was set as −20, and other parameters were the default. Two other algorithms, TargetScan and PicTar, also predicted notch3 as a putative target gene of miR-206.
Expression Construct of miR-206
Mouse genomic regions containing the Mus musculus miR-206 hairpins and ∼150 nucleotides of 5′- and 3′-flanking regions were cloned into the pRNAT-CMV3.2 vector between the XhoI and BamHI sites (GenScript, Piscataway, NJ). The specific miR-206 hairpin inhibitor (IH-310462-07) and miRIDIAN miRNA inhibitor negative control 1 (IN-001000-01) were purchased from Dharmacon (Lafayette CO). This miR-206 inhibitor is designed to specifically target miR-206 by blocking the cleavage of precursor miR-206 to mature miR-206. The full-length notch3 expression vector was obtained from Dr. Michael Wang (University of Michigan).
Mouse and Human notch3 3′-UTR-Luciferase Vector Construction and Luciferase Assay
To generate the luciferase reporter vectors, notch3 3′-UTR segments were amplified by PCR from the mouse and human cDNAs and inserted into the pMIR-REPORT vector between HindIII and SacI sites (Ambion, Austin, TX). PCR with appropriate primers also generated inserts with deleted miR-206 complementary sites. Wild-type and mutant vectors with deleted miR-206 complementary sites were confirmed by sequencing. Primer sequences are listed in supplemental Table S1.
Twenty-four h before transfection, 5 × 104 HeLa cells were plated per well in a 24-well plate. Thirty ng of notch3 3′-UTR-luciferase construct, different concentrations of the miR-206 expression construct, and 30 ng of pSV-β-galactosidase control vector (Promega, Madison, WI) were transfected using FuGENE HD (Roche Applied Science). Thirty-six h after transfection, luciferase and β-galactosidase assays were performed using the luciferase assay system and Beta-Glo® assay system (Promega). Luciferase activities were normalized to galactosidase activities for each transfected well. For each experiment, wells were transfected in triplicate, and each well was assayed in triplicate.
Western Blotting
Cells grown in 6-cm culture dishes were washed three times with ice-cold phosphate-buffered saline and collected in radioimmune precipitation assay lysis buffer with protease inhibitors. After a 30-min incubation on ice, cell lysates were sonicated and centrifuged, and the clear supernatants were saved as whole-cell lysate. Cell lysates were resolved by SDS-PAGE and transferred to nitrocellulose membranes according to standard procedures. Membranes were washed with Tris-buffered saline containing 0.05% Tween 20 (TBST), blocked for 1 h with TBST containing 5% nonfat milk, and then incubated with primary antibodies (ab23426, Abcam) at 1:000 dilution in TBST containing 5% nonfat milk overnight at 4 °C. Membranes were washed with TBST before incubation with horseradish peroxidase-conjugated secondary antibody (3 μg/liter in TBST containing 5% nonfat milk) for 1 h at room temperature. Membranes were washed four times with TBST before antibody binding was visualized by SuperSignal West Femto maximum sensitivity substrate (catalog no. 34095, Thermo Fisher Scientific) according to the manufacturer's protocol.
Apoptosis Assay
Apoptosis was assessed and quantified by staining for annexin V-phycoerythrin and 7-aminoactinomycin (Pharmingen) according to the manufacturer's protocols. In brief, HeLa cells (1 × 106) were transfected with the expression constructs of miR-206 (6 μg) and/or notch3 (6 μg) for 36 h. Cells were washed with cold phosphate-buffered saline buffer at pH 7.4. After the cells were centrifuged at 2000 × g for 5 min, they were resuspended in 100 μl of binding buffer. Cells (1 × 105) were then stained with annexin V-phycoerythrin (5 μl) and 7-aminoactinomycin (5 μl) by incubation for 15 min at room temperature in the dark. Following this, results were acquired by flow cytometry (FACSCalibur, BD Biosciences) and analyzed by CellQuest software (BD Biosciences).
Colorimetric Assay of Caspase Activities
Measurements were performed using a kit for caspase 3 (catalog no. K106–100, BioVision, Inc., Mountain View, CA) in accordance with the manufacturer's instructions. The kits detect specific colorimetric substrates that, when cleaved, increase light absorption at 405 nm. Briefly, HeLa cells were cultured overnight in 60-mm culture dishes and then transfected with the various vectors as indicated. Cells were resuspended in the cell lysis buffer provided with the kits and incubated on ice for 10 min, and cell lysates were centrifuged at 10,000 × g for 1 min at 4 °C to collect the supernatants. Protein concentrations were determined using the DC protein assay (catalog no. 500-0112, Bio-Rad). Proteins (200 μg for each assay) were incubated with reaction buffer and substrate at 37 °C for 2 h and then measured in a microtiter plate reader.
Wound Healing Assay
HeLa cells seeded at 1 × 106 in 5 ml of Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum were cultured overnight at 37 °C in 6-cm plates. The next day, miR-206 expression vector (6 μg) and/or notch3 plasmid (6 μg) was transfected into HeLa cell using Lipofectamine 2000 (Invitrogen). After 36 h, transfected cells were grown to confluence and wounded by dragging a 1-ml pipette tip through the monolayer. Cells were washed using prewarmed phosphate-buffered saline to remove cellular debris and allowed to migrate for 16 h. Wound closure or cell migration images were photographed when the scrape wound was introduced (0 h) and at a designated time (16 h) after wounding using a DMI6000 inverted microscope. The relative surface traveled by the leading edge was assessed using LAS AF6000 Version 1.8.0 software. The individual gaps were measured at each time point, and the speed of migration was acquired by dividing the length of the gap by the culture time. The residual gap between the migrating cells from the opposing wound edge was expressed as a percentage of the initial scraped area. Three replicates each of two independent experiments were performed.
Focus Formation Assay
Transfections were performed using Lipofectamine 2000 according to the manufacturer's instructions. Cells (0.5 × 106) in 35-mm plastic dishes were transfected with 3 μg of ptRNA-miR-206 vector. Two days after transfection, transfected cells were suspended with 8 ml of 0.4% top agar (Sigma) and 2× Dulbecco's modified Eagle's medium supplemented with 20% fetal bovine serum before being poured onto 6-cm tissue culture dishes coated with 3.5 ml of 0.7% bottom agar. The plates were prepared in triplicate. Fourteen days later, three areas/plate were chosen randomly, the number of visible colonies was counted, and the size of the colonies was measured. Green fluorescence was used as a marker to determine whether the cells were transfected with a GFP-control or GFP-miR-206 vector. Only foci formed from GFP-control or GFP-miR-206 vector-transfected cells were used for statistical analysis. For a detailed method, see Ref. 20.
Statistical Analysis
All experiments were repeated at least three times, and error bars represent S.E. Statistical analyses were carried out using Student's unpaired t test; p < 0.01 was considered statistically significant.
RESULTS
miR-206 Targets notch3
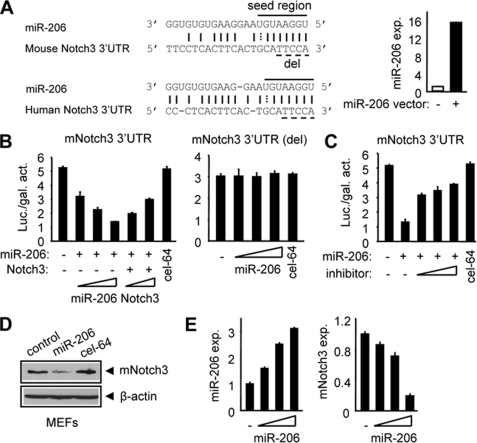
In search for miR-206 target genes, we identified a nearly perfect complementarity between miR-206 and the 3′-UTR of both mouse and human notch3 (including the seed region) using the miRanda algorithm (Fig. 1A, left), which was confirmed by TargetScan and PicTar. The conservation of the seed match region between miR-206 and the notch3 3′-UTR was also observed in other species (supplemental Fig. S1). To elucidate the role of miR-206 in regulating notch3 expression and function, we constructed a miR-206 expression plasmid. When the miR-206 expression vector was transfected into human cervical cancer HeLa cells, the expression of miR-206 was highly induced as determined by real-time PCR analysis (Fig. 1A, right). HeLa cells had minimal endogenous miR-206 expression, which provided a good in vitro cell model system to determine a real interaction between exogenously expressed miR-206 and notch3 and should exclude effects from endogenous miR-206.
FIGURE 1.
miR-206 targets and regulates mouse notch3. A: left, sequence alignment between miR-206 and the 3′-UTR of notch3 in mouse and human. Solid line, seed match region; dashed line, seed-deleted region. Right, real-time PCR analysis of miR-206 expression (exp.) in HeLa cells transfected with a control (−) or miR-206 (+; 400 ng) expression vector. B: HeLa cells cotransfected with a luciferase reporter containing the mouse notch3 3′-UTR (left) or the miR-206-binding site-deleted (del) mouse notch3 3′-UTR (right), a miR-206 expression plasmid (50, 100, and 200 ng), or a full-length notch3 expression plasmid (50 and 100 ng). cel-miR-64 (200 ng) is a non-related miRNA that did not target notch3 and was used as a negative control. Luciferase activities were measured 24 h post-transfection and normalized to the corresponding vector control. Each transfection was done in triplicate, and the experiment was repeated three times. A representative experiment is shown (mean ± S.E.). Luc./gal. activity, luciferase/galactosidase activity. C: luciferase activities determined in HeLa cells transfected with a mouse notch3 3′-UTR-reporter in the presence or absence of the miR-206 expression plasmid (200 ng) or a miR-206 inhibitor (final concentrations of 10, 20, and 40 pmol). Data are represented as the mean ± S.E. D: immunoblot showing that miR-206 (4 μg, 6-cm plate) induced a decrease in endogenous mouse Notch3 protein in mouse MEFs. β-Actin was used as a loading control. cel-miR-64 (4 μg), a non-related miRNA that did not target notch3, was used as a negative control. E: real-time PCR analysis of miR-206 and mouse notch3 expression in SHP+/+ MEFs overexpressed with a miR-206 expression plasmid (50, 100, and 200 ng). Data are represented as the mean ± S.E.
To determine whether miR-206 can decrease the expression of notch3, we used a luciferase reporter assay. Cotransfection of cells with the parental luciferase construct (without the notch3 3′-UTR) plus the miR-206 expression plasmid did not change the expression of the reporter (data not shown). However, miR-206 dose-dependently decreased the luciferase activity when cotransfected with the mouse notch3 3′-UTR containing the miR-206 target site (Fig. 1B, left). The inhibitory effect of miR-206 on mouse notch3 3′-UTR activity was partially rescued by coexpression of a full-length notch3 plasmid, suggesting that notch3 was effective in inhibiting the ability of miR-206 to inhibit the expression of the reporter gene containing the miR-206-binding site. The data indicated that notch3 was able to counteract miR-206 function. Expression of notch3 did not fully restore the reporter activity, which may be associated with the lower amount of vector used for overexpression. In addition, suppression of the notch3 3′-UTR by miR-206 was relieved by deletion (Fig. 1B, right) or mutation (supplemental Fig. S2A) of the miR-206-binding site and was markedly recovered by a specific miR-206 inhibitor (Fig. 1C) whose function is to block the cleavage of precursor miR-206 to mature miR-206 (21). In contrast, a non-related miRNA, cel-miR-64, which was not predicted to target notch3, did not show any effect on the expression of luciferase. Furthermore, overexpression of miR-206 markedly decreased the levels of endogenous mouse Notch3 protein (Fig. 1D) in mouse embryonic fibroblasts (MEFs), whereas cel-miR-64 had no effect on mouse Notch33 protein expression. Interestingly, ectopic expression of miR-206 also inhibited mRNA expression of mouse notch3 in MEFs (Fig. 1E). MEFs were used because our pilot study showed that these cells expressed higher levels of notch3, which would provide a better detection system for notch3 down-regulation by miR-206.
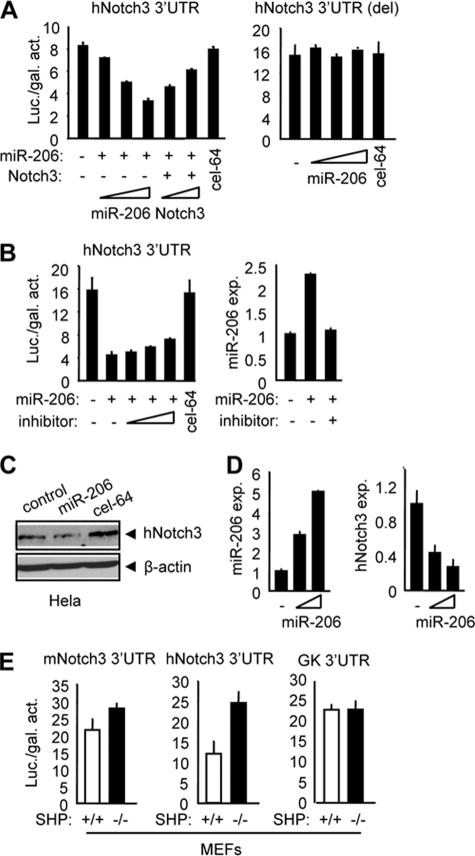
Because the miR-206 target site was also observed in the human notch3 3′-UTR, we tested the effect of miR-206 on human Notch3 expression in HeLa cells. As expected, miR-206 showed a strong inhibitory effect on normal (Fig. 2A, left) but not deleted (Fig. 2A, right) or mutated (supplemental Fig. S2B) human notch3 3′-UTR-luciferase activity. Similarly, forced expression of full-length notch3 largely rescued the repressive effect of miR-206 on human notch3 3′-UTR-luciferase activity (Fig. 2A, left). When the cells were treated with a specific inhibitor of miR-206, the repression of the human notch3 3′-UTR by miR-206 was moderately released (Fig. 2B, left), suggesting that a higher dose of the miR-206 inhibitor would be required to efficiently block the inhibitory effect of miR-206 on the activity of the human notch3 3′-UTR. Real-time PCR analysis confirmed that the expression of miR-206 was induced in HeLa cells transfected with the miR-206 vector, which was completely repressed by a higher concentration of the miR-206 inhibitor (Fig. 2B, right). Next, we determined the inhibitory effect of miR-206 on endogenous human Notch3 protein expression using HeLa cells. As expected, ectopic expression of miR-206 resulted in a significant reduction of human Notch3 protein expression (Fig. 2C and supplemental Fig. S3A). In addition, the level of human notch3 mRNA was also repressed by miR-206 (Fig. 2D). Similar observations were also reported for other miRNAs and their target mRNAs (22–24).
FIGURE 2.
miR-206 targets and regulates human notch3. A: HeLa cells cotransfected with a luciferase reporter construct containing the human notch3 3′-UTR (left) or the miR-206 target site-deleted (del) human notch3 3′-UTR (right), a miR-206 expression plasmid (50, 100, and 200 ng), or a full-length notch3 expression plasmid (50 and 100 ng), and luciferase activities were determined. cel-miR-64 (200 ng) was a non-related miRNA that did not target human notch3 and was used as a negative control. Luc./gal. activity, luciferase/galactosidase activity. B: left, luciferase activities determined in HeLa cells transfected with a human notch3 3′-UTR-reporter in the presence or absence of a miR-206 expression plasmid (200 ng) and a miR-206 inhibitor (final concentrations of 10, 20, and 40 pmol). Right, real-time PCR analysis of miR-206 expression (exp.) in HeLa cells transfected with a control vector (−), miR-206 expression vector (100 ng), or miR-206 inhibitor (final concentration of 40 pmol). C: immunoblot showing that miR-206 (4 μg, 6-cm plate) induced a decrease in endogenous human Notch3 protein in human HeLa cells. β-Actin was used as a loading control. D: real-time PCR analysis of miR-206 and human notch3 expression in HeLa cells overexpressed with a miR-206 expression plasmid (3 and 6 μg, 6-cm plate). E: luciferase activities determined in SHP+/+ (wild-type) and SHP−/− (SHP knock-out) MEFs transfected with the mouse or human notch3 3′-UTR or glucokinase (GK) 3′-UTR. All data are represented as the mean ± S.E.
We have shown recently that the nuclear receptor SHP activates miR-206 gene expression via a cascade dual inhibitory mechanism (25). Endogenous miR-206 expression was ∼5-fold lower in SHP−/− relative to SHP+/+ MEFs (supplemental Fig. S3B). We transfected both mouse and human notch3 3′-UTRs into MEFs without coexpression of miR-206 and predicted that the basal luciferase activities of mouse and human notch3 3′-UTRs would be higher in SHP−/− compared with SHP+/+ cells. Indeed, an elevated expression of luciferase was observed in SHP−/− cells transfected with the mouse (Fig. 2E, left) and human (Fig. 2E, middle) notch3 3′-UTRs but not with the non-related 3′-UTR of glucokinase (Fig. 2E, right). Overall, the above results identified notch3 as a novel target gene of miR-206.
miR-206 Activates Apoptotic Cell Death via Cross-talk with notch3
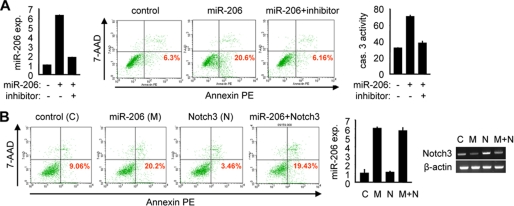
notch3 was reported to resist Fas ligand-induced apoptosis in smooth muscle cells (26), suggesting a potential role of notch3 in regulating apoptosis signaling. We investigated whether miR-206 may be an active component of the apoptotic pathway via cross-talk with notch3. Forced expression of miR-206 (Fig. 3A, left) resulted in an ∼3-fold increase in apoptotic cell death in HeLa cells, and the percentage of apoptotic cells induced by miR-206 was decreased to the basal level when the cells were treated with the specific miR-206 inhibitor (Fig. 3A, middle), demonstrating the potent pro-apoptotic effect of miR-206. Consistent with this, caspase 3 activity in cells was increased by miR-206, and the activation was blocked by the miR-206 inhibitor (Fig. 3A, right).
FIGURE 3.
miR-206 activates apoptosis. A: left, real-time PCR analysis of miR-206 expression (exp.). Middle, annexin V staining and flow cytometry analysis of cell death. The lower right quadrant of each plot contains early apoptotic cells. This experiment was repeated three independent times, and similar results were obtained each time. Right, caspase 3 (cas. 3) activity assays. 7-AAD, 7-aminoactinomycin; PE, phycoerythrin. B: left, annexin V staining and flow cytometry analysis of cell death. Middle, real-time PCR analysis of miR-206 expression. Right, semiquantitative PCR analysis of notch3 mRNA expression. All experiments were performed in HeLa cells (1 × 106, 6-cm plate) transfected with a miR-206 expression vector (6 μg) in the absence or presence of its inhibitor (final concentration of 40 pmol) or with a full-length notch3 (N) expression vector (6 μg) alone or in combination with miR-206 (M). C, control.
The above studies showed that the expression of notch3 was decreased by miR-206; thus, we predicted that miR-206 might be involved in the negative regulation of anti-apoptosis signaling by notch3. As expected, cells overexpressing miR-206 showed increased apoptotic cell death (Fig. 3B, left). In contrast, expression of notch3 exhibited the anti-apoptotic effect by decreasing the basal cellular apoptosis. When miR-206 and notch3 were coexpressed in cells, the apoptosis inhibited by Notch3 was largely rescued by miR-206. The expression of miR-206 and notch3 was confirmed by real-time and semiquantitative PCR analysis, respectively (Fig. 3B, middle and right). Overexpression of notch3 did not alter the level of miR-206 (Fig. 3B, middle), whereas overexpression of miR-206 decreased the mRNA expression of notch3 (Fig. 3B, left). Thus, there existed a cross-talk between miR-206 and notch3 in regulating apoptosis signaling.
miR-206 Represses Tumor Cell Migration and Focus Formation via Cross-talk with notch3
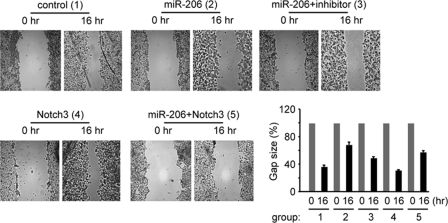
The mechanism of cancer formation is associated with the inhibition of tumor cell apoptosis or activation of cell proliferation. Our preliminary studies did not reveal a marked effect of miR-206 on cellular growth (data not shown). Because miR-206 strongly activated apoptosis, we tested the tumor inhibitory effect of miR-206. We first determined the effect of miR-206 on the inhibition of HeLa cell migration using wound healing assays. As shown in Fig. 4, overexpression of miR-206 resulted in an ∼50% decrease in the cell migration rate compared with the control group, and the effect of miR-206 was attenuated by the miR-206 inhibitor. Expression of notch3 moderately decreased cell migration under basal conditions, which was largely inhibited by coexpression of miR-206. Because the transfection efficiency was ∼60–70% and not all cells were transfected with the vectors, the data were thus considered significant.
FIGURE 4.
miR-206 inhibits tumor cell migration. A wound healing assay was performed on HeLa cells (1 × 106, 6-cm plate) transfected with either an empty vector (control) or a miR-206 expression vector (6 μg) in the absence or presence of the miR-206 inhibitor (final concentration of 40 pmol) or with a full-length notch3 expression vector (6 μg) alone or in combination with miR-206. The experiments were repeated three times with similar results, and one representative experiment is shown. Wound closure was photographed and quantified as shown. The residual gap between the migrating cells from the opposing wound edge is expressed as a percentage of the initial scraped area.
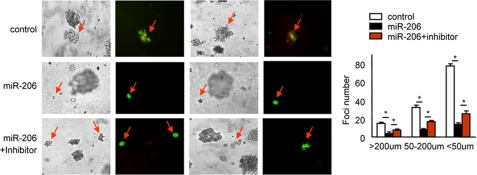
We next determined whether miR-206 inhibits HeLa cells to form foci. As expected, expression of miR-206 significantly prevented the formation of foci by HeLa cells in soft agar, as indicated by the markedly decreased number and size of foci in miR-206-overexpressing cells. The inhibitory effect of miR-206 was blocked, although not fully but significantly, by the miR-206 inhibitor (Fig. 5), demonstrating the specific effect of miR-206. It was noted that the efficacy of the miR-206 inhibitor to reverse the effect of miR-206 was not as strong as in prior studies. Because focus formation assays took >2 weeks to complete compared with other studies that took only a few days, it was likely that the efficiency of the miR-206 inhibitor was decreased with time when the foci were formed. Nonetheless, miR-206 exhibited a striking and impressive effect in suppressing focus formation.
FIGURE 5.
miR-206 inhibits focus formation. Left, focus formation assay performed on HeLa cells (0.5 × 106, 35-mm plate) transfected with either a GFP-control vector or a GFP-miR-206 expression vector (3 μg) in the absence or presence of the miR-206 inhibitor (final concentration of 40 pmol). GFP signal indicates foci formed from cells transfected with the GFP-control or GFP-miR-206 vector. Right, statistical analysis of foci formed. Data are represented as the mean ± S.E. (n = three fields/plate of three plates). *, p < 0.01.
We could not examine the counteracting effect of miR-206 with notch3 in focus formation because the notch3 expression vector did not have GFP or another fluorescence tag as a marker to indicate cells that were transfected with the vector. Based on the results from the above experiments, it is tempting to predict that such a counteracting effect between miR-206 and notch3 would be observed.
DISCUSSION
Despite increasing evidence pointing to a role for miRNAs in apoptotic cell signaling and tumor progression, the tumor-suppressive or oncogenic effect of miRNAs has not been explored extensively. In this study, we identified notch3 as a target of miR-206 and revealed a novel function of miR-206 in activating cellular apoptosis and suppressing tumor growth via cross-talk with notch3.
A major finding of this study is the identification of notch3 as a target of miR-206. Although it is generally believed that miRNAs regulate gene expression through decreased translation or increased degradation of the target message (1), our study shows that both mouse and human notch3 are down-regulated at both the protein and mRNA levels by miR-206. This suggests that miR-206 regulation of its target gene is not restricted at the post-transcriptional level but also occurs at the transcriptional level. Inhibition of the Notch3 protein level by miR-206 is via direct miR-206 binding to the 3′-UTR of notch3. The mechanism of miR-206 inhibition of notch3 mRNA expression is currently unclear. Lim et al. (22) reported that some miRNAs down-regulate large numbers of target mRNAs. Another group reported that the mRNA and protein levels of transforming growth factor-β and SMAD3 are decreased by a virus-encoded microRNA, miR-LAT (23). The mRNA and protein levels of Bcl-2 are also down-regulated by miR-1 (24). Thus, our result is not unusual and provides new evidence to support the conclusion that miRNAs regulate not only the stability and translation but also the transcription of target mRNAs; the latter is most likely via an indirect mechanism.
Another major finding of this study is the identification of miR-206 as a potent inducer of apoptosis, which is associated with its tumor suppressor function. Although the function of miR-206 has been well established in skeletal muscle physiology (10–12), the role of miR-206 in other tissues, particularly in cancer progression, has largely remained unstudied. Our study provides several lines of evidence for a tumor suppressor function of miR-206. We have shown that miR-206 functions as a potent pro-apoptotic inducer, suggesting a general function of miR-206 in activating apoptosis signaling. miR-206-mediated inhibition of notch3 expression and the antagonizing effect of miR-206 against the anti-apoptotic effect of notch3 strongly suggest a pivotal role for miR-206 in regulating the oncogenic function of notch3 and its involvement in the pathogenesis of cancer.
We propose that miR-206 functions as a tumor suppressor and is a central component of apoptosis signaling and that up-regulation of miR-206 inhibits the tumorigenic progression of cancer. Indeed, elevated levels of miR-206 inhibited tumor cell migration and focus formation. Although further work is needed to fully characterize the mechanism by which miR-206 activates apoptosis and inhibits tumor growth in vivo, these data suggest that miR-206 serves as a potential tumor suppressor and a critical factor in regulating tumorigenesis.
Supplementary Material
Acknowledgments
We thank Dr. Curt Hagedorn for critically reading the manuscript and Dr. Michael Wang for the full-length notch3 expression vector.
This work wasThis work performed by the Flow Cytometry Core was supported by National Institutes of Health Grant P30 CA042014 awarded to the Huntsman Cancer Institute.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Table S1.
- miRNA
- microRNA
- 3′-UTR
- 3′-untranslated region
- TBS
- Tris-buffered saline
- GFP
- green fluorescent protein
- MEF
- mouse embryonic fibroblast.
REFERENCES
- 1.Filipowicz W., Bhattacharyya S. N., Sonenberg N. (2008) Nat. Rev. Genet. 9, 102–114 [DOI] [PubMed] [Google Scholar]
- 2.Bartel D. P. (2004) Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 3.Du T., Zamore P. D. (2005) Development 132, 4645–4652 [DOI] [PubMed] [Google Scholar]
- 4.Wienholds E., Plasterk R. H. (2005) FEBS Lett. 579, 5911–5922 [DOI] [PubMed] [Google Scholar]
- 5.Park S. M., Peter M. E. (2008) Cytokine Growth Factor Rev. 19, 303–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cimmino A., Calin G. A., Fabbri M., Iorio M. V., Ferracin M., Shimizu M., Wojcik S. E., Aqeilan R. I., Zupo S., Dono M., Rassenti L., Alder H., Volinia S., Liu C. G., Kipps T. J., Negrini M., Croce C. M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 13944–13949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang T. C., Wentzel E. A., Kent O. A., Ramachandran K., Mullendore M., Le K. H., Feldmann G., Yamakuchi M., Ferlito M., Lowenstein C. J., Arking D. E., Beer M. A., Maitra A., Mendell J. T. (2007) Mol. Cell 26, 745–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raver-Shapira N., Marciano E., Meiri E., Spector Y., Rosenfeld N., Moskovits N., Bentwich Z., Oren M. (2007) Mol. Cell 26, 31–43 [DOI] [PubMed] [Google Scholar]
- 9.Esquela-Kerscher A., Slack F. J. (2006) Nat. Rev. Cancer 6, 259–269 [DOI] [PubMed] [Google Scholar]
- 10.Kim H. K., Lee Y. S., Sivaprasad U., Malhotra A., Dutta A. (2006) J. Cell Biol. 174, 677–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson C., Catoe H., Werner R. (2006) Nucleic Acids Res. 34, 5863–5871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rao P. K., Kumar R. M., Farkhondeh M., Baskerville S., Lodish H. F. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 8721–8726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adams B. D., Furneaux H., White B. A. (2007) Mol. Endocrinol. 21, 1132–1147 [DOI] [PubMed] [Google Scholar]
- 14.Kondo N., Toyama T., Sugiura H., Fujii Y., Yamashita H. (2008) Cancer Res. 68, 5004–5008 [DOI] [PubMed] [Google Scholar]
- 15.Negrini M., Calin G. A. (2008) Breast Cancer Res. 10, 203–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen T., Olsen L., Lindow M., Jakobsen K. D., Ullum H., Jonsson E., Andreassen O. A., Djurovic S., Melle I., Agartz I., Hall H., Timm S., Wang A. G., Werge T. (2007) PLoS ONE 2, e873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walden T. B., Timmons J. A., Keller P., Nedergaard J., Cannon B. (2009) J. Cell. Physiol. 218, 444–449 [DOI] [PubMed] [Google Scholar]
- 18.Roccaro A. M., Sacco A., Chen C., Runnels J., Leleu X., Azab F., Azab A. K., Jia X., Ngo H. T., Melhem M. R., Burwick N., Varticovski L., Novina C. D., Rollins B. J., Anderson K. C., Ghobrial I. M. (2009) Blood 113, 4391–4402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song G., Wang L. (2008) Nucleic Acids Res. 36, 5727–5735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He N., Park K., Zhang Y., Huang J., Lu S., Wang L. (2008) Gastroenterology 134, 793–802 [DOI] [PubMed] [Google Scholar]
- 21.Vermeulen A., Robertson B., Dalby A. B., Marshall W. S., Karpilow J., Leake D., Khvorova A., Baskerville S. (2007) RNA 13, 723–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lim L. P., Lau N. C., Garrett-Engele P., Grimson A., Schelter J. M., Castle J., Bartel D. P., Linsley P. S., Johnson J. M. (2005) Nature 433, 769–773 [DOI] [PubMed] [Google Scholar]
- 23.Gupta A., Gartner J. J., Sethupathy P., Hatzigeorgiou A. G., Fraser N. W. (2006) Nature 442, 82–85 [DOI] [PubMed] [Google Scholar]
- 24.Tang Y., Zheng J., Sun Y., Wu Z., Liu Z., Huang G. (2009) Int. Heart J. 50, 377–387 [DOI] [PubMed] [Google Scholar]
- 25.Song G., Wang L. (2009) PLoS ONE 4, e6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang W., Prince C. Z., Mou Y., Pollman M. J. (2002) J. Biol. Chem. 277, 21723–21729 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.