Abstract
Exposure of cells to DNA-damaging agents results in a rapid increase in the formation of subnuclear complexes containing Rad51. To date, it has not been determined to what extent DNA damage-induced cytoplasmic to nuclear transport of Rad51 may contribute to this process. We have analyzed subcellular fractions of HeLa and HCT116 cells and found a significant increase in nuclear Rad51 levels following exposure to a modest dose of ionizing radiation (2 grays). We also observed a DNA damage-induced increase in nuclear Rad51 in the Brca2-defective cell line Capan-1. To address a possible Brca2-independent mechanism for Rad51 nuclear transport, we analyzed subcellular fractions for two other Rad51-interacting proteins, Rad51C and Xrcc3. Rad51C has a functional nuclear localization signal, and although we found that the subcellular distribution of Xrcc3 was not significantly affected by DNA damage, there was a damage-induced increase in nuclear Rad51C. Furthermore, RNA interference-mediated depletion of Rad51C in HeLa and Capan-1 cells resulted in lower steady-state levels of nuclear Rad51 as well as a diminished DNA damage-induced increase. Our results provide important insight into the cellular regulation of Rad51 nuclear entry and a role for Rad51C in this process.
Introduction
Cellular surveillance of genome integrity and repair of DNA damage are essential processes that ensure proper development and survival of all organisms. DNA double-strand breaks (DSBs)2 are a particularly deleterious form of genome damage and occur following exposure of cells to exogenous mutagens as well as during normal metabolic processes, e.g. antigen receptor gene rearrangement, restart of stalled replication forks, formation of meiotic DNA crossovers, etc. (1, 2). Mammalian cells use two distinct mechanisms for repair of DSBs, non-homologous end joining and homologous recombination (HR). Processes requiring imprecise DNA repair, such as the creation of antibody diversity, exploit the error-prone non-homologous end joining mechanism. In contrast, HR is an error-free DNA repair pathway and is critical for avoidance of unwanted genetic changes during the meiotic exchange of information between paternal and maternal alleles and for error-free repair of broken chromosomes (3). Rad51 is the central enzymatic component of HR. Upon its regulated recruitment to sites of DNA breaks, Rad51 forms a nucleoprotein filament by polymerizing onto single-stranded DNA at the processed break. This filament catalyzes DNA strand exchange with an undamaged sister chromatid or homologous chromosome, which serves as a template for the restoration of missing genetic information (3, 4).
Visible nuclear Rad51 clusters, or foci, form during S-phase and appear to localize to sites of replicating DNA (5–7). A dramatic increase in the number and size of nuclear Rad51 foci is a hallmark of the early cellular response to genomic insult (7–10). The appearance of DNA damage-induced nuclear Rad51 foci is blocked in cells with deficiencies in several HR-related proteins, including Brca2 and the Rad51 paralogs Rad51B, Rad51C, Rad51D, Xrcc2, and Xrcc3 (11–13). These defects correlate with a decrease in HR efficiency and an increase in chromosome abnormalities and genome instability (12, 13). Interestingly, overexpression of Rad51 leads to increased numbers of nuclear Rad51 foci and formation of higher order Rad51-chromatin complexes in the absence of DNA damage (14–16). This correlates with high levels of HR, genome instability, and increased resistance of cancer cells to radiation and drug treatment (16, 17). Additionally, up-regulation of RAD51 gene expression in mouse embryonic stem cells leads to increased recombination events and genome instability (18). Therefore, the nuclear availability of Rad51 must be carefully regulated to allow appropriate levels of recombination both before and following exposure to DNA damage.
RAD51 gene expression is controlled by a variety of transcriptional activators and repressors (15, 19–21) but is not affected by DNA damage (22, 23). Numerous studies show the presence of significant amounts of cytoplasmic Rad51 in normal cycling cells (23–31), suggesting that the level of nuclear Rad51 is controlled by regulated changes in its subcellular distribution. In fact, exposure of cells to DNA-damaging agents has been shown to elicit movement of numerous DNA damage signaling proteins, cell cycle checkpoint effectors, and DNA-processing enzymes among various cellular compartments (32), including the cytoplasmic to nuclear transport of proteins involved in DNA base excision repair (33, 34) and DNA mismatch repair (35, 36). Although localized nuclear responses to DNA damage by many DSB signaling and repair proteins, including ATM, Chk2, the Mre11-Rad50-Nbs1 complex, MDC1, 53BP1, and Rad51, have been well documented (26, 37–43), the possible contribution of a cytoplasmic to nuclear transport of DSB repair proteins to this response has not been fully considered. In this study, we provide evidence that nuclear transport of Rad51 is an integral part of the cellular response to DNA damage. Additionally, although Brca2 has been implicated in the nuclear transport of Rad51 (12, 25), a Brca2-independent mechanism for nuclear entry of Rad51 clearly exists (40, 44–46). Our data suggest that Rad51C plays an important role in this process.
EXPERIMENTAL PROCEDURES
Reagents
Phenylmethylsulfonyl fluoride, cycloheximide, staurosporine, and DNase I were obtained from Sigma. Protease inhibitor tablets (Complete, with and without EDTA) were obtained from Roche Applied Sciences.
Antibodies
The primary antibodies used were mouse monoclonal antibodies against Rad51 (clone 3C10, Upstate), Rad51B (ab3637, Abcam), Rad51C (NB 100-177, Novus), Rad51D (sc-53432, Santa Cruz Biotechnology), Xrcc2 (ab20253, Abcam), and Xrcc3 (NB 100-180D1, Novus). Cytochrome c was detected with a mouse monoclonal antibody (556433, Pharmingen). For loading control tests on cytosolic samples, we used mouse anti-glyceraldehyde-3-phosphate dehydrogenase antibody (A00084, GenScript); for nuclear samples, mouse anti-Sam68 antibody (610 171, Pharmingen); and for chromatin samples, mouse anti-fibrillarin antibody (mono 38F3, Abcam). The secondary antibody used was a horseradish peroxidase-conjugated goat anti-mouse antibody (12-349, Upstate).
Cultured Cell Lines
Samples were obtained from cultured cells when their passage number was lower than 25. HeLa cells (CCL-2, American Type Culture Collection) were grown at 37 °C and 5% CO2 in Dulbecco's modified Eagle's medium (11995, Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum and 1% penicillin/streptomycin (15140, Invitrogen). HCT116 cells (CCL-247, American Type Culture Collection) were grown in McCoy's 5A medium (16600, Invitrogen) under the same conditions and supplements to the medium. Capan-1 cells (HTB-79, American Type Culture Collection) were grown in Iscove's modified Dulbecco's medium (12440, Invitrogen) with 20% heat-inactivated fetal bovine serum.
Subcellular Fractionation
HeLa, HCT116, and Capan-1 cells were plated on 10-cm dishes and incubated in the dark at 37 °C under an atmosphere of 5% (v/v) CO2 in air. When cells reached ∼90% confluency, selected dishes received cycloheximide (20 μm final concentration). One hour later, a set of plates was exposed to ionizing radiation (IR). Cells were harvested 30 min, 60 min, 2 h, and 8 h following irradiation. Control cells were not irradiated. Subcellular fractionation was performed using modifications to a previously described procedure (29). Briefly, after removing the medium, cells were rinsed with phosphate-buffered saline and then detached by trypsinization, pelleted by centrifugation in 15-ml tubes, transferred to 1.5-ml tubes, and washed with phosphate-buffered saline. Cell pellets were suspended in 10 mm HEPES-KOH (pH 7.1), 50 mm NaCl, 0.3 m sucrose, 0.5% Triton X-100, 0.1 mm EDTA, 1 mm dithiothreitol, and protease inhibitors. After 15 min on ice, samples were centrifuged at 1500 × g for 5 min. Supernatants (cytosolic fraction) were transferred to fresh tubes and stored on ice. Pellets were washed with 10 mm HEPES-NaOH (pH 7.1), 0.1 mm EDTA, 1 mm dithiothreitol, and protease inhibitors, after which they were resuspended in Buffer A (10 mm HEPES-KOH (pH 7.1), 500 mm NaCl, 0.5% Nonidet P-40, 0.1 mm EDTA, 1 mm dithiothreitol, and protease inhibitors) and incubated on ice with occasional vortexing. After 15 min, samples were centrifuged for 10 min at 10,000 × g, and supernatants (nuclear fraction) were transferred to fresh tubes. Pellets were washed with Buffer A and centrifuged briefly, and the supernatant was removed. Pellets were kept on ice until samples for all time points had been collected. Pellets were then washed with 10 mm Tris buffer (pH 8) and resuspended in the same buffer containing DNase I and EDTA-free protease inhibitors. After 30 min at 37 °C, pellets were disrupted by pipetting until fully resuspended and then returned to the 37 °C bath for an additional 30 min. SDS was added to each sample to a final concentration of 1%, and samples were incubated at 65 °C for 20 min. After a brief mixing by vortexing, samples were cooled to room temperature and centrifuged for 2 min at 10,000 × g to remove any undissolved particles. The clear supernatants (chromatin fraction) were transferred to fresh tubes. Protein concentrations were determined (BCA, Pierce), and samples were prepared for gel electrophoresis.
Whole Cell Extracts
Whole cell extracts were prepared by resuspending pelleted cells in lysis buffer (10 mm Tris-HCl (pH 7.2), 1% Triton X-100, 0.1% SDS, 150 mm NaCl, and protease inhibitors) and incubating on ice for 15 min. Protein concentrations were determined using a micro BCA kit (Pierce). Western blot samples were then prepared by adding an equal volume of 2× Laemmli sample buffer and heating to 90 °C for 5 min.
Gel Electrophoresis and Immunoblots
Proteins were separated on 4–12% SDS-polyacrylamide gels and transferred onto Immun-Blot polyvinylidene difluoride membranes (Bio-Rad) at 20 V for 40 min using a semi-dry transfer apparatus. Blots were rinsed for 5 min with blocking buffer (10 mm Tris-HCl (pH 8.0), 300 mm NaCl, and 0.25% Tween 20) and blocked with 15% nonfat milk in blocking buffer for 1 h with slow rocking. Blots were incubated with primary antibody for at least 1 h at room temperature (or overnight at 4 °C), after which they were washed with blocking buffer (3× 5 min) and incubated for 1 h with horseradish peroxidase-conjugated secondary antibody. After a thorough washing with blocking buffer (6× 5 min), specific bands were detected by chemiluminescence on a Fuji LAS-3000 luminescent image analyzer or by exposure to x-ray film. Bands were quantified and normalized to loading controls by analyzing imported tiffs of scanned blots using ImageJ or by using the Fuji LAS-3000 software.
Cytochrome c Release Assay
Cells were seeded into 6-well plates and left untreated, exposed to 2 or 8 grays (Gy) of IR, or treated with 1 μm staurosporine. Cells were then harvested at 2 and 18 h following treatment. Cell lysates were centrifuged at 2100 × g for 10 min, and the resulting supernatants were analyzed by Western blotting for the presence of cytochrome c using a monoclonal antibody.
RNA Interference-mediated Knockdown
Depletion of Rad51C was achieved by treatment of cells with 15 nm small interfering RNA (siRNA) SMARTpool (Dharmacon). For control cells, the equivalent concentration of non-silencing siRNA was used. The transfection lipids were DharmaFECT 1 and 4 (Dharmacon) for HeLa and Capan-1 cells, respectively. Also, Capan-1 cells were trypsinized prior to transfection, which increased significantly the knockdown efficiency. Following transfection, cells were grown for 40–48 h prior to IR treatment. Rad51C depletion was determined to be >90% (data not shown).
Determination of Transcript Levels
Total RNA was isolated from cells using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. cDNA was prepared using the SuperScript first-strand synthesis system (Invitrogen). Changes in gene-specific transcript copy number were evaluated using quantitative PCR with QuantiFast SYBR Green mixture (Qiagen).
Irradiation
Cells were irradiated in a Gammacell 40 Cs137 irradiator (Atomic Energy of Canada Ltd., Ottawa, Canada).
RESULTS
DNA Damage Induces an Increase in Nuclear Levels of Rad51
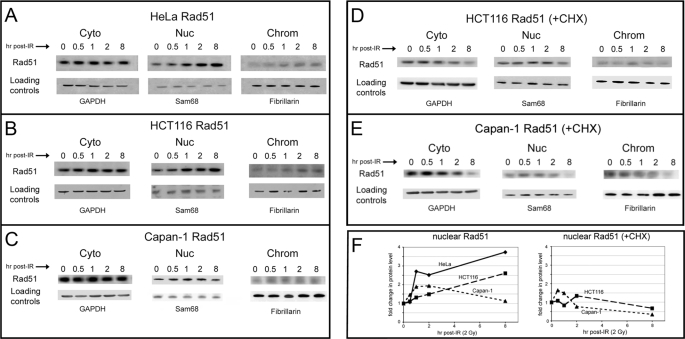
Significant levels of cytoplasmic Rad51 have been observed by several groups (24–31), and we have also described a DNA damage-dependent accumulation of cytoplasmic Rad51 at the nuclear periphery (30). Therefore, we asked whether this pool of cytoplasmic protein contributes to an increase in nuclear levels of Rad51 in response to DNA damage. Indeed, we found that Rad51 levels increased in the nucleoplasm of HeLa cells in response to DNA damage (2 Gy of IR) (Fig. 1A). During this time course, we also observed an increase in chromatin-associated Rad51 (Fig. 1A). Experiments performed using HCT116 cells also showed a DNA damage-induced increase in nucleoplasmic and chromatin-associated Rad51 (Fig. 1B). The Western blots in Fig. 1, as well as all subsequent blots, are representative of over three repetitions of complete experiments. From these analyses, we calculated overall 3.7- and 2.6-fold increases in HeLa and HCT116 nucleoplasmic Rad51 at 8 h following damage, respectively (Fig. 1F and supplemental Table 1). As we have done previously (30, 47), RNA interference-mediated knockdown of Rad51, Rad51C, and Xrcc3 was performed to demonstrate specificity of the Western blot signals (data not shown).
FIGURE 1.
DNA damage induces an increase in nuclear levels of Rad51 in Brca2-proficient and Brca2-deficient cells. HeLa (A), HCT116 (B), and Capan-1 (C) cells grown at 37 °C were harvested at the indicated times following exposure to 2 Gy of IR and fractionated as described under “Experimental Procedures” to yield cytoplasmic (Cyto), nucleoplasmic (Nuc), and chromatin (Chrom) samples. D and E, HCT116 and Capan-1 cells, respectively, were treated with cycloheximide (CHX; 20 μm) 1 h prior to exposure to 2 Gy of IR. A portion of each fraction (30 μg of total protein) was loaded onto 4–12% SDS-polyacrylamide gels, and Western blots were developed using a mouse anti-Rad51 monoclonal antibody. Blots were also developed using the following markers as loading controls: glyceraldehyde-3-phosphate dehydrogenase (GAPDH; cytoplasmic), Sam68 (nucleoplasmic), and fibrillarin (chromatin). F, changes in levels of nuclear Rad51 as a function of time after IR treatment in A–E were quantified as described under “Experimental Procedures.” The data shown are representative of the results of at least three separate experiments, and the S.D. observed with quantification was <20%.
DNA Damage Induces an Increase in Nuclear Levels of Rad51 in Brca2-deficient Capan-1 Cells
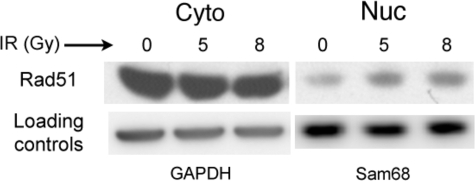
Although it has been proposed that the Brca2 protein may be directly involved in nuclear transport of Rad51 (25), several studies suggest a Brca2-independent mechanism for Rad51 nuclear entry both before and after DNA damage (25, 40, 45). To address the requirement for Brca2 in the DNA damage-induced increase in nuclear Rad51, we used the pancreatic cancer cell line Capan-1 to perform analyses of subcellular fractions before and after IR exposure. Capan-1 cells carry a single BRCA2 allele that encodes a truncated protein missing the functional C-terminal nuclear localization signals (NLSs) and the Cdk-regulated C-terminal Rad51 interaction domain but that maintains six of eight Rad51-binding BRC domains (48–51). We confirmed the 6174ΔT mutation in the Capan-1 line used in our laboratory (48, 49). Although native Brca2 protein localizes to both the cytoplasm and nucleus (52), Capan-1 cells show a significant decrease in nuclear levels of the truncated Brca2 protein (25, 52) and a decrease in nuclear Rad51 (25). As expected, in undamaged Capan-1 cells, we found a greater ratio of cytoplasmic to nuclear Rad51 (Fig. 1C) relative to that seen in HeLa and HCT116 cells (Fig. 1, A and B). However, we observed a distinct DNA damage-induced increase in Capan-1 nuclear Rad51 levels at 1 and 2 h following IR treatment (2 Gy), approaching a 2-fold change (Fig. 1, C and F, and supplemental Table 1). Additionally, chromatin-associated Rad51 showed an approximate 2-fold increase following IR exposure (Fig. 1C). To ensure that this increased nuclear Rad51 reflects a true response to DNA damage, we repeated these experiments using 5- and 8-Gy exposures of Capan-1 cells and observed a reproducible increase in nuclear Rad51 at 2 h post-treatment, approximating a 2-fold change (Fig. 2 and supplemental Table 1).
FIGURE 2.
Levels of Capan-1 nuclear Rad51 increase in an IR dose-dependent manner. Capan-1 cells exposed to 5 or 8 Gy of IR were grown at 37 °C for 2 h. Cells were harvested and fractionated as described under “Experimental Procedures,” and portions of the cytoplasmic (Cyto) and nucleoplasmic (Nuc) fractions (25 μg of total protein) were loaded onto 4–12% SDS-polyacrylamide gels. Western blots were developed using a mouse anti-Rad51 monoclonal antibody, and levels of cytoplasmic and nuclear Rad51 were quantified as described under “Experimental Procedures” (supplemental Table 1). The blot shown is representative of four separate experiments. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Because elevated levels of DNA damage can induce an apoptotic response in various cell types, we asked whether the DNA damage used in these experiments leads to the occurrence of any possible apoptotic events. We performed experiments in which release of mitochondrial cytochrome c, a benchmark early apoptotic event, was analyzed following exposure of cells to both low and moderate doses of IR. Exposure of HeLa cells to either 2 or 8 Gy of IR resulted in levels of cytosolic cytochrome c similar to those in untreated cells, even at 18 h following IR treatment (supplemental Fig. 1). In contrast, treatment of cells with 1 μm staurosporine resulted in a significant increase in cytosolic cytochrome c at both 2 and 18 h following treatment. Other groups (53) have seen levels of cytosolic cytochrome c in negative and positive controls similar to what we observed. Therefore, we found no evidence for DNA damage-induced apoptosis under the conditions used in our study.
Our use of several different cell types strongly supports the idea that a DNA damage-induced increase in nuclear Rad51 is a general cellular response to genomic insult and that this occurs at least in part in a Brca2-independent manner. To further characterize this response, we next asked to what extent pre-existing pools of cytoplasmic Rad51 contribute to the DNA damage-induced nuclear transport relative to Rad51 derived from ongoing protein synthesis.
DNA Damage-induced Nuclear Transport of Rad51 Involves Both Pre-existing Pools of Cytoplasmic Rad51 and Protein Derived from Ongoing Synthesis
Analyses of subcellular fractions were performed as described above, but protein synthesis was inhibited by addition of cycloheximide. A DNA damage-induced increase in HCT116 nucleoplasmic Rad51 was still observed, which approximated a 1.5-fold change at 1 h following treatment (Fig. 1, D and F, and supplemental Table 1). Using Capan-1 cells in the presence of cycloheximide, we observed a reproducible increase in nuclear Rad51 at 0.5 and 1 h after IR treatment, followed by a decrease in Rad51 levels in all subcellular fractions at 2 and especially 8 h after IR treatment (Fig. 1, E and F, and supplemental Table 1). Similar changes were observed using HeLa and HEK293 cells (data not shown). In all cell types, Rad51 levels in each subcellular fraction showed a significant decrease at 8 h after IR treatment (Fig. 1, D–F). DNA damage does not influence the level of RAD51 gene expression (22, 23), and although an increase in nuclear Rad51 in the presence of cycloheximide indicates that pre-existing pools of cytoplasmic protein are involved in this process, the additional increase in nuclear Rad51 in cells not grown in cycloheximide (Fig. 1, A–C) suggests that protein derived from ongoing RAD51 gene expression also contributes.
Possible Brca2-independent Mechanism for DNA Damage-induced Rad51 Nuclear Transport
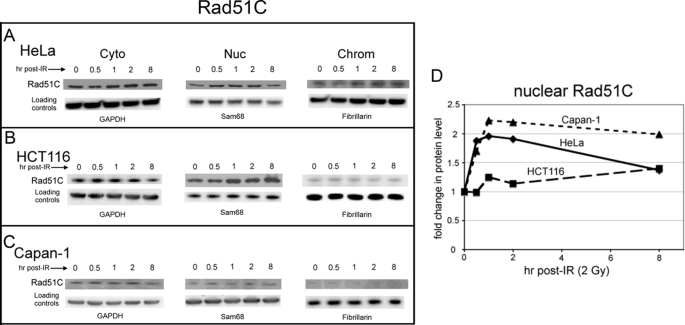
Nuclear entry of the Rad51 protein likely requires its interaction with another protein that contains a functional NLS. Rad51 itself has no detectable NLS (analyzed using PredictNLS and PSORTII programs), and nuclear pore complexes do not allow free diffusion of molecules of this size (54–56). Therefore, given our observation of Brca2-independent nuclear entry by Rad51 following DNA damage using Capan-1 cells, we asked whether other Rad51-interacting proteins show patterns of DNA damage-induced redistribution similar to Rad51. Several proteins known to interact directly with Rad51 also influence Rad51-mediated recombination and DNA repair (see “Discussion”), and our initial analyses focused on the Rad51 paralog proteins Rad51C and Xrcc3 (57, 58). Subcellular fractions were probed for Xrcc3 and Rad51C at various times following IR exposure. For Xrcc3, we found no significant DNA damage-dependent changes in the cytoplasmic, nuclear, or chromatin-associated levels of protein in HeLa, HCT116, and Capan-1 cells (supplemental Fig. 2 and Table 1). In contrast, nucleoplasmic levels of Rad51C increased 1.5–2.2-fold in HeLa, HCT116, and Capan-1 cells following exposure of cells to 2 Gy of IR (Fig. 3 and supplemental Table 1).
FIGURE 3.
DNA damage induces an increase in nuclear levels of Rad51C in HeLa, HCT116, and Capan-1 cells. HeLa (A), HCT116 (B), and Capan-1 (C) cells grown at 37 °C were harvested at the indicated times following exposure to 2 Gy of IR and fractionated as described under “Experimental Procedures” to yield cytoplasmic (Cyto), nucleoplasmic (Nuc), and chromatin (Chrom) samples. A portion of each fraction (30 μg of total protein) was loaded onto 4–12% SDS-polyacrylamide gels, and Western blots were developed using a mouse anti-Rad51C monoclonal antibody. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. D, changes in levels of nuclear Rad51C as a function of time after IR treatment in A–C were quantified as described under “Experimental Procedures.” The blot shown is representative of the results of at least three separate experiments.
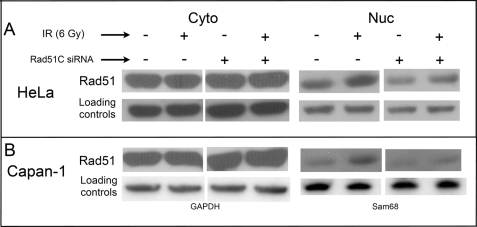
To investigate the possibility that Rad51C plays a role in regulating nuclear entry of Rad51, cells were transfected with Rad51C siRNAs and analyzed for the subcellular distribution of Rad51 before and after IR treatment. In mock-transfected HeLa cells, we observed the expected DNA damage-induced increase in nuclear Rad51 at 2 h following treatment of cells with 6 Gy of IR (Fig. 4A). However, in cells depleted of Rad51C, there was a decrease in the steady-state level of nuclear Rad51 as well as a diminished DNA damage-induced increase in nuclear Rad51. Similar experiments were performed using Capan-1 cells. In mock-transfected cells, we observed a 2-fold increase in nuclear Rad51 at 2 h following treatment with 6 Gy of IR, but this increase was significantly diminished in cells depleted of Rad51C (Fig. 4B and supplemental Table 1). As in HeLa cells, steady-state levels of Capan-1 nuclear Rad51 were also lower.
FIGURE 4.
Rad51C depletion decreases the steady-state level of nuclear Rad51 and diminishes its DNA damage-induced nuclear transport in Brca2-proficient and Brca2-deficient cells. HeLa (A) and Capan-1 (B) cells were transfected with a nonspecific (−) or Rad51C-specific (+) siRNA pool (SMARTpool), grown for 42 h at 37 °C, exposed to 6 Gy of IR, and harvested 2 h later. Cytoplasmic (Cyto) and nuclear (Nuc) fractions were analyzed by Western blotting using an anti-Rad51 monoclonal antibody. Changes in levels of cytoplasmic and nuclear Rad51 were quantified as described under “Experimental Procedures.” GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Rad51 has been shown to interact with two paralog protein complexes, the Rad51C/Xrcc3 heterodimer and tetrameric Rad51B-C-D/Xrcc2 (59). Previous studies also suggest that the stability of other Rad51 paralog proteins is dependent on Rad51C (60). We found that depletion of Rad51C led to a significant reduction in cellular levels of Xrcc3, as was observed previously (60), yet levels of Rad51B, Rad51D, and Xrcc2 remained unaffected (supplemental Fig. 3A). Analysis of transcript levels revealed that this loss of Xrcc3 in Rad51C-depleted cells resulted from destabilization of the Xrcc3 protein rather than fortuitous off-site targeting of Xrcc3 mRNA by the Rad51C siRNA (supplemental Fig. 3B). The observations that nuclear levels of Rad51C but not Xrcc3 increased in response to IR and that Rad51C depletion affected the stability of only Xrcc3 and not the other paralog proteins support the idea that regulation of Rad51 nuclear levels is a function specific to Rad51C and independent of the other paralogs.
DISCUSSION
The subcellular distribution of a number of DNA damage signaling and repair proteins changes in response to DNA damage (32–36). Results in this study have demonstrated a specific DNA damage-induced cytoplasmic to nuclear transport of two proteins involved in the catalysis of HR-mediated DNA repair, Rad51 and Rad51C. We found that levels of each protein in both the nucleoplasmic and chromatin fractions increased during an 8-h time course following exposure to a modest dose of IR (2 Gy). We also observed increases in Rad51 levels in these subcellular fractions when protein synthesis was inhibited, supporting a model in which both pre-existing pools of Rad51 and newly synthesized protein contribute to the observed DNA damage-dependent nuclear increases. Rad51 protein levels are regulated by a complex interplay between transcription repression, activation (19–21), and protein turnover (30, 61). Additionally, although RAD51 gene expression is not affected by DNA damage (22, 23), protein levels are higher in late S, G2, and M relative to G1 (19, 22). Therefore, in the asynchronous cell populations used in our studies, newly synthesized Rad51 following DNA damage likely arises from ongoing steady-state synthesis, and cellular levels may be slightly exaggerated in some cells that arrest in S, G2, or M in response to DNA damage. It is not known if Rad51 degradation is regulated as a function of cell cycle or DNA damage, but under the conditions used in our studies, the balance between Rad51 synthesis and degradation permits a view into the DNA damage-dependent mobility of both pre-existing pools of Rad51 and newly synthesized protein among various subcellular compartments. Further studies are aimed at understanding how the combined processes of Rad51 synthesis, degradation, and subcellular redistribution in response to DNA damage regulate nuclear levels of Rad51. Given that Rad51 is the central catalyst of HR, careful regulation of its nuclear levels is required to prevent the deleterious effects resulting from either too little or too much nuclear Rad51, both of which can contribute to the increased genome instability observed in various cancers (14–17).
Although the transport of Rad51 into the nucleus has been attributed to its direct interaction with the Brca2 protein (25), other evidence demonstrates that Rad51 can enter the nucleus independently of Brca2 both before and after genomic stress (25, 40, 45). Studies by Yu et al. (40) are particularly revealing. Using a series of mutant Rad51 proteins, they found that those with combined defects in self-association and Brca2 binding achieved nuclear entry, whereas those with a specific defect only in Brca2 binding were blocked from nuclear entry. Thus, an interaction between Rad51 and Brca2 is not required for nuclear transport of Rad51. Rather, these data support the idea that an important function of Brca2 may be to prevent formation of Rad51 filaments in the cytoplasm, which could otherwise compromise nuclear entry by a large Rad51 polymer. The observation that Capan-1 cells show a decrease in the overall level of nuclear Rad51 relative to BRCA2+/+ pancreatic cells (25), similar to what we see in this study, is likely due to the fact that the truncated Brca2 protein in Capan-1 cells maintains its ability to bind Rad51 but is no longer capable of moving into the nucleus due to the lack of its functional C-terminal NLS domains (52). Thus, most cellular Rad51 in Capan-1 cells will be sequestered in the cytoplasm (44). This undoubtedly contributes to the diminished DNA damage-induced increase in nuclear Rad51 we observed in Capan-1 cells relative to HeLa and HCT116 cells.
A more clearly defined function for Brca2 involves its mediator activity for Rad51 nucleoprotein filament assembly at the site of a DNA break (12, 62–66). Following exposure of Capan-1 cells to IR, we observed a modest increase in chromatin-associated Rad51 (Fig. 1C). However, previous studies using Capan-1 cells show little to no increase in the number of cells with DNA damage-induced nuclear Rad51 foci and no increase in the size the foci (45). This likely reflects a defect in the ability of the truncated Brca2 protein to mediate loading of Rad51 at the sites of IR-induced DSBs. The increase in chromatin-associated Rad51 observed in our studies may represent increased nonspecific binding due to the DNA damage-induced increased levels of Capan-1 nuclear Rad51. Although Brca2 may indeed play a role in the nuclear entry of Rad51, the work presented here, together with other studies (40, 45), demonstrates a Brca2-independent mechanism for Rad51 nuclear import. The fact that Rad51 has no discernible NLS suggests that it must interact with another protein(s) containing a functional NLS. Data in this study support a model in which Rad51C contributes to this process.
The Rad51C protein was originally identified through sequence comparisons with Rad51 and Xrcc3 (67), the latter identified earlier as a Rad51 family member in complementation screens (57, 68). Rad51C has important roles in promoting Rad51-mediated DNA repair and genome stability (69–71) and appears to act early in the HR pathway. Rad51C defects prevent DNA damage-induced Rad51 nuclear focus formation (13) and cause developmental arrest of mouse spermatocytes in the early stages of meiotic prophase I (72), both of which are consistent with an early HR function. A role late in HR for human Rad51C had also been suggested through its association with a Holliday junction resolvase activity (73), but with the recent demonstration that it does not associate with the human crossover resolvase GEN1, the specific contribution of Rad51C to this late HR activity remains to be clarified (74). Rad51C also shows a complex pattern of protein-protein associations as it is present in at least three distinct complexes containing one or more of the other four members of the Rad51 paralog group, Rad51B/Rad51C/Rad51D/Xrcc2, Rad51B/Rad51C, and Rad51C/Xrcc3 (75–77). Independent of its existence in these complexes, Rad51C is only one of two paralog proteins that specifically interact with Rad51 (58). Rad51C also contains a functional C-terminal NLS (71), and given that the Brca2-defective Capan-1 cell line shows some level of nuclear Rad51 prior to DNA damage (this study and Ref. 45) and a distinct DNA damage-induced increase in nuclear Rad51 (Figs. 1, 2, and 4), we suggest that Rad51C plays a role in the nuclear transport of Rad51 in cells both before and following exposure to DNA damage. This idea is supported by our finding that nuclear levels of Rad51C also increase in response to DNA damage in both Brca2-proficient and Brca2-defective cells and especially by data showing that depletion of Rad51C significantly diminishes levels of nuclear Rad51 before and following DNA damage. Depletion of Rad51C has been shown to reduce IR-induced S-phase arrest (78), which could be thought to contribute to lower levels of nuclear Rad51 by virtue of having fewer cells in a phase with higher overall levels of Rad51. However, this same study showed an increased number of Rad51C-depleted cells with higher G2/M DNA content, which are also stages with greater overall levels of Rad51 (19, 22). Therefore, although changes in cell cycle progression resulting from Rad51C depletion may contribute to diminished levels of nuclear Rad51, data presented in our study support a more direct role for Rad51C in regulating levels of nuclear Rad51.
Additionally, other Rad51-interacting proteins, such as Rad51AP1/Pir51 (79), Rad51AP2 (80), Rad52 (81), and Rad54 (82), have NLS domains. However, Rad51AP1 has been shown to be exclusively nuclear (23), Rad51AP2 is expressed only in meiotic cells (80), and vertebrate Rad52 appears to have little effect on Rad51-mediated HR (83, 84). Therefore, these are unlikely candidates for assisting nuclear entry of Rad51. Rad54 may assist in the cytoplasmic to nuclear transport of Rad51, but to date, its subcellular distribution before and after DNA damage has not been examined.
This study offers important insight into a novel role for Rad51C and correlates with other studies suggesting that it functions early in the HR pathway. Rad51 nuclear entry likely involves its interaction with several proteins, including Brca2 and Rad51C, and further studies will be designed to identify other possible factors contributing to this process and how this aspect of the cellular response to genomic stress is regulated.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants GM044772 and GM065851 (to K. L. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3 and Table 1.
- DSB
- double-strand break
- HR
- homologous recombination
- IR
- ionizing radiation
- Gy
- grays
- siRNA
- small interfering RNA
- NLS
- nuclear localization signal.
REFERENCES
- 1.Wyman C., Kanaar R. (2004) Curr. Biol. 14, R629–R631 [DOI] [PubMed] [Google Scholar]
- 2.Lieber M. R. (2008) J. Biol. Chem. 283, 1–5 [DOI] [PubMed] [Google Scholar]
- 3.San Filippo J., Sung P., Klein H. (2008) Annu. Rev. Biochem. 77, 229–257 [DOI] [PubMed] [Google Scholar]
- 4.Li X., Heyer W. D. (2008) Cell Res. 18, 99–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tashiro S., Kotomura N., Shinohara A., Tanaka K., Ueda K., Kamada N. (1996) Oncogene 12, 2165–2170 [PubMed] [Google Scholar]
- 6.Scully R., Chen J., Plug A., Xiao Y., Weaver D., Feunteun J., Ashley T., Livingston D. M. (1997) Cell 88, 265–275 [DOI] [PubMed] [Google Scholar]
- 7.Haaf T., Raderschall E., Reddy G., Ward D. C., Radding C. M., Golub E. I. (1999) J. Cell Biol. 144, 11–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haaf T., Golub E. I., Reddy G., Radding C. M., Ward D. C. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 2298–2302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raderschall E., Golub E. I., Haaf T. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 1921–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tashiro S., Walter J., Shinohara A., Kamada N., Cremer T. (2000) J. Cell Biol. 150, 283–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen J. J., Silver D., Cantor S., Livingston D. M., Scully R. (1999) Cancer Res. 59, 1752s–1756s [PubMed] [Google Scholar]
- 12.Yuan S. S., Lee S. Y., Chen G., Song M., Tomlinson G. E., Lee E. Y. (1999) Cancer Res. 59, 3547–3551 [PubMed] [Google Scholar]
- 13.Takata M., Sasaki M. S., Tachiiri S., Fukushima T., Sonoda E., Schild D., Thompson L. H., Takeda S. (2001) Mol. Cell. Biol. 21, 2858–2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raderschall E., Bazarov A., Cao J., Lurz R., Smith A., Mann W., Ropers H. H., Sedivy J. M., Golub E. I., Fritz E., Haaf T. (2002) J. Cell Sci. 115, 153–164 [DOI] [PubMed] [Google Scholar]
- 15.Hannay J. A., Liu J., Zhu Q. S., Bolshakov S. V., Li L., Pisters P. W., Lazar A. J., Yu D., Pollock R. E., Lev D. (2007) Mol. Cancer Ther. 6, 1650–1660 [DOI] [PubMed] [Google Scholar]
- 16.Klein H. L. (2008) DNA Repair 7, 686–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raderschall E., Stout K., Freier S., Suckow V., Schweiger S., Haaf T. (2002) Cancer Res. 62, 219–225 [PubMed] [Google Scholar]
- 18.Richardson C., Stark J. M., Ommundsen M., Jasin M. (2004) Oncogene 23, 546–553 [DOI] [PubMed] [Google Scholar]
- 19.Iwanaga R., Komori H., Ohtani K. (2004) Oncogene 23, 8581–8590 [DOI] [PubMed] [Google Scholar]
- 20.Hasselbach L., Haase S., Fischer D., Kolberg H. C., Stürzbecher H. W. (2005) Eur. J. Gynaecol. Oncol. 26, 589–598 [PubMed] [Google Scholar]
- 21.Arias-Lopez C., Lazaro-Trueba I., Kerr P., Lord C. J., Dexter T., Iravani M., Ashworth A., Silva A. (2006) EMBO Rep. 7, 219–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen F., Nastasi A., Shen Z., Brenneman M., Crissman H., Chen D. J. (1997) Mutat. Res. 384, 205–211 [DOI] [PubMed] [Google Scholar]
- 23.Henson S. E., Tsai S. C., Malone C. S., Soghomonian S. V., Ouyang Y., Wall R., Marahrens Y., Teitell M. A. (2006) Mutat. Res. 601, 113–124 [DOI] [PubMed] [Google Scholar]
- 24.Yoshikawa K., Ogawa T., Baer R., Hemmi H., Honda K., Yamauchi A., Inamoto T., Ko K., Yazumi S., Motoda H., Kodama H., Noguchi S., Gazdar A. F., Yamaoka Y., Takahashi R. (2000) Int. J. Cancer 88, 28–36 [PubMed] [Google Scholar]
- 25.Davies A. A., Masson J. Y., McIlwraith M. J., Stasiak A. Z., Stasiak A., Venkitaraman A. R., West S. C. (2001) Mol. Cell 7, 273–282 [DOI] [PubMed] [Google Scholar]
- 26.Essers J., Houtsmuller A. B., van Veelen L., Paulusma C., Nigg A. L., Pastink A., Vermeulen W., Hoeijmakers J. H., Kanaar R. (2002) EMBO J. 21, 2030–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kraakman-van der Zwet M., Overkamp W. J., van Lange R. E., Essers J., van Duijn-Goedhart A., Wiggers I., Swaminathan S., van Buul P. P., Errami A., Tan R. T., Jaspers N. G., Sharan S. K., Kanaar R., Zdzienicka M. Z. (2002) Mol. Cell. Biol. 22, 669–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forget A. L., Bennett B. T., Knight K. L. (2004) J. Cell. Biochem. 93, 429–436 [DOI] [PubMed] [Google Scholar]
- 29.Liu N., Lim C. S. (2005) J. Cell. Biochem. 95, 942–954 [DOI] [PubMed] [Google Scholar]
- 30.Bennett B. T., Knight K. L. (2005) J. Cell. Biochem. 96, 1095–1109 [DOI] [PubMed] [Google Scholar]
- 31.Mladenov E., Anachkova B., Tsaneva I. (2006) Genes Cells 11, 513–524 [DOI] [PubMed] [Google Scholar]
- 32.Tembe V., Henderson B. R. (2007) Cell. Signal. 19, 1113–1120 [DOI] [PubMed] [Google Scholar]
- 33.Conlon K. A., Zharkov D. O., Berrios M. (2003) DNA Repair 2, 1337–1352 [DOI] [PubMed] [Google Scholar]
- 34.Conlon K. A., Zharkov D. O., Berrios M. (2004) DNA Repair 3, 1601–1615 [DOI] [PubMed] [Google Scholar]
- 35.Brieger A., Plotz G., Raedle J., Weber N., Baum W., Caspary W. F., Zeuzem S., Trojan J. (2005) Mol. Carcinog. 43, 51–58 [DOI] [PubMed] [Google Scholar]
- 36.Knudsen N. Ø., Nielsen F. C., Vinther L., Bertelsen R., Holten-Andersen S., Liberti S. E., Hofstra R., Kooi K., Rasmussen L. J. (2007) Nucleic Acids Res. 35, 2609–2619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mirzoeva O. K., Petrini J. H. (2001) Mol. Cell. Biol. 21, 281–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lukas C., Falck J., Bartkova J., Bartek J., Lukas J. (2003) Nat. Cell Biol. 5, 255–260 [DOI] [PubMed] [Google Scholar]
- 39.Bakkenist C. J., Kastan M. B. (2003) Nature 421, 499–506 [DOI] [PubMed] [Google Scholar]
- 40.Yu D. S., Sonoda E., Takeda S., Huang C. L., Pellegrini L., Blundell T. L., Venkitaraman A. R. (2003) Mol. Cell 12, 1029–1041 [DOI] [PubMed] [Google Scholar]
- 41.van Veelen L. R., Cervelli T., van de Rakt M. W., Theil A. F., Essers J., Kanaar R. (2005) Mutat. Res. 574, 22–33 [DOI] [PubMed] [Google Scholar]
- 42.Bekker-Jensen S., Lukas C., Melander F., Bartek J., Lukas J. (2005) J. Cell Biol. 170, 201–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bekker-Jensen S., Lukas C., Kitagawa R., Melander F., Kastan M. B., Bartek J., Lukas J. (2006) J. Cell Biol. 173, 195–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Orelli B. J., Bishop D. K. (2001) Breast Cancer Res. 3, 294–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tarsounas M., Davies D., West S. C. (2003) Oncogene 22, 1115–1123 [DOI] [PubMed] [Google Scholar]
- 46.Lee S. A., Roques C., Magwood A. C., Masson J. Y., Baker M. D. (2009) DNA Repair 8, 170–181 [DOI] [PubMed] [Google Scholar]
- 47.Bennett B. T., Bewersdorf J., Knight K. L. (2009) Methods 48, 63–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goggins M., Schutte M., Lu J., Moskaluk C. A., Weinstein C. L., Petersen G. M., Yeo C. J., Jackson C. E., Lynch H. T., Hruban R. H., Kern S. E. (1996) Cancer Res. 56, 5360–5364 [PubMed] [Google Scholar]
- 49.Abbott D. W., Freeman M. L., Holt J. T. (1998) J. Natl. Cancer Inst. 90, 978–985 [DOI] [PubMed] [Google Scholar]
- 50.Chen P. L., Chen C. F., Chen Y., Xiao J., Sharp Z. D., Lee W. H. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 5287–5292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Esashi F., Christ N., Gannon J., Liu Y., Hunt T., Jasin M., West S. C. (2005) Nature 434, 598–604 [DOI] [PubMed] [Google Scholar]
- 52.Spain B. H., Larson C. J., Shihabuddin L. S., Gage F. H., Verma I. M. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 13920–13925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Plescia J., Salz W., Xia F., Pennati M., Zaffaroni N., Daidone M. G., Meli M., Dohi T., Fortugno P., Nefedova Y., Gabrilovich D. I., Colombo G., Altieri D. C. (2005) Cancer Cell 7, 457–468 [DOI] [PubMed] [Google Scholar]
- 54.Paine P. L., Moore L. C., Horowitz S. B. (1975) Nature 254, 109–114 [DOI] [PubMed] [Google Scholar]
- 55.Panté N., Kann M. (2002) Mol. Biol. Cell 13, 425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fried H., Kutay U. (2003) Cell. Mol. Life Sci. 60, 1659–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu N., Lamerdin J. E., Tebbs R. S., Schild D., Tucker J. D., Shen M. R., Brookman K. W., Siciliano M. J., Walter C. A., Fan W., Narayana L. S., Zhou Z. Q., Adamson A. W., Sorensen K. J., Chen D. J., Jones N. J., Thompson L. H. (1998) Mol. Cell 1, 783–793 [DOI] [PubMed] [Google Scholar]
- 58.Schild D., Lio Y. C., Collins D. W., Tsomondo T., Chen D. J. (2000) J. Biol. Chem. 275, 16443–16449 [DOI] [PubMed] [Google Scholar]
- 59.Rodrigue A., Lafrance M., Gauthier M. C., McDonald D., Hendzel M., West S. C., Jasin M., Masson J. Y. (2006) EMBO J. 25, 222–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lio Y. C., Schild D., Brenneman M. A., Redpath J. L., Chen D. J. (2004) J. Biol. Chem. 279, 42313–42320 [DOI] [PubMed] [Google Scholar]
- 61.Kanamoto T., Hellman U., Heldin C. H., Souchelnytskyi S. (2002) EMBO J. 21, 1219–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang H., Li Q., Fan J., Holloman W. K., Pavletich N. P. (2005) Nature 433, 653–657 [DOI] [PubMed] [Google Scholar]
- 63.San Filippo J., Chi P., Sehorn M. G., Etchin J., Krejci L., Sung P. (2006) J. Biol. Chem. 281, 11649–11657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Davies O. R., Pellegrini L. (2007) Nat. Struct. Mol. Biol. 14, 475–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Esashi F., Galkin V. E., Yu X., Egelman E. H., West S. C. (2007) Nat. Struct. Mol. Biol. 14, 468–474 [DOI] [PubMed] [Google Scholar]
- 66.Thorslund T., West S. C. (2007) Oncogene 26, 7720–7730 [DOI] [PubMed] [Google Scholar]
- 67.Dosanjh M. K., Collins D. W., Fan W., Lennon G. G., Albala J. S., Shen Z., Schild D. (1998) Nucleic Acids Res. 26, 1179–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tebbs R. S., Zhao Y., Tucker J. D., Scheerer J. B., Siciliano M. J., Hwang M., Liu N., Legerski R. J., Thompson L. H. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 6354–6358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.French C. A., Masson J. Y., Griffin C. S., O'Regan P., West S. C., Thacker J. (2002) J. Biol. Chem. 277, 19322–19330 [DOI] [PubMed] [Google Scholar]
- 70.Godthelp B. C., Wiegant W. W., van Duijn-Goedhart A., Schärer O. D., van Buul P. P., Kanaar R., Zdzienicka M. Z. (2002) Nucleic Acids Res. 30, 2172–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.French C. A., Tambini C. E., Thacker J. (2003) J. Biol. Chem. 278, 45445–45450 [DOI] [PubMed] [Google Scholar]
- 72.Kuznetsov S., Pellegrini M., Shuda K., Fernandez-Capetillo O., Liu Y., Martin B. K., Burkett S., Southon E., Pati D., Tessarollo L., West S. C., Donovan P. J., Nussenzweig A., Sharan S. K. (2007) J. Cell Biol. 176, 581–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu Y., Masson J. Y., Shah R., O'Regan P., West S. C. (2004) Science 303, 243–246 [DOI] [PubMed] [Google Scholar]
- 74.Ip S. C., Rass U., Blanco M. G., Flynn H. R., Skehel J. M., West S. C. (2008) Nature 456, 357–361 [DOI] [PubMed] [Google Scholar]
- 75.Masson J. Y., Stasiak A. Z., Stasiak A., Benson F. E., West S. C. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 8440–8446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wiese C., Collins D. W., Albala J. S., Thompson L. H., Kronenberg A., Schild D. (2002) Nucleic Acids Res. 30, 1001–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Miller K. A., Yoshikawa D. M., McConnell I. R., Clark R., Schild D., Albala J. S. (2002) J. Biol. Chem. 277, 8406–8411 [DOI] [PubMed] [Google Scholar]
- 78.Badie S., Liao C., Thanasoula M., Barber P., Hill M. A., Tarsounas M. (2009) J. Cell Biol. 185, 587–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kovalenko O. V., Golub E. I., Bray-Ward P., Ward D. C., Radding C. M. (1997) Nucleic Acids Res. 25, 4946–4953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kovalenko O. V., Wiese C., Schild D. (2006) Nucleic Acids Res. 34, 5081–5092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shen Z., Cloud K. G., Chen D. J., Park M. S. (1996) J. Biol. Chem. 271, 148–152 [DOI] [PubMed] [Google Scholar]
- 82.Golub E. I., Kovalenko O. V., Gupta R. C., Ward D. C., Radding C. M. (1997) Nucleic Acids Res. 25, 4106–4110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yamaguchi-Iwai Y., Sonoda E., Buerstedde J. M., Bezzubova O., Morrison C., Takata M., Shinohara A., Takeda S. (1998) Mol. Cell. Biol. 18, 6430–6435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rijkers T., Van Den Ouweland J., Morolli B., Rolink A. G., Baarends W. M., Van Sloun P. P., Lohman P. H., Pastink A. (1998) Mol. Cell. Biol. 18, 6423–6429 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.