Abstract
In plants the triple methylation of phosphoethanolamine to phosphocholine catalyzed by phosphoethanolamine N-methyltransferase (PEAMT) is considered a rate-limiting step in the de novo synthesis of phosphatidylcholine. Besides being a major membrane phospholipid, phosphatidylcholine can be hydrolyzed into choline and phosphatidic acid. Phosphatidic acid is widely recognized as a second messenger in stress signaling, and choline can be oxidized within the chloroplast to yield the putative osmoprotectant glycine betaine. Here we describe the cloning and biochemical characterization of a second wheat PEAMT isoform that has a four times higher specific activity than the previously described WPEAMT/TaPEAMT1 enzyme and is less sensitive to product inhibition by S-adenosyl homocysteine, but more sensitive to inhibition by phosphocholine. Both enzymes follow a sequential random Bi Bi mechanism and show mixed-type product inhibition patterns with partial inhibition for TaPEAMT1 and a strong non-competitive component for TaPEAMT2. An induction of TaPEAMT protein expression and activity is observed after cold exposure, ahead of an increase in gene expression. Our results demonstrate direct repression of in vitro enzymatic activities by phosphatidic acid for both enzymes, with TaPEAMT1 being more sensitive than TaPEAMT2 in the physiological concentration range. Other lipid ligands identified in protein-lipid overlays are phosphoinositide mono- as well as some di-phosphates and cardiolipin. These results provide new insights into the complex regulatory circuits of phospholipid biosynthesis in plants and underline the importance of head group biosynthesis in adaptive stress responses.
Introduction
Phosphatidylcholine (PC),4 one of the major plasma membrane phospholipids in most eukaryotes, has been shown to undergo changes in turnover in response to various environmental stresses in plants (for review see Refs. 1, 2) and has a major impact on cell proliferation and apoptosis in mammals (for review see Ref. 3).
In single and multicellular organisms, biosynthetic routes leading to the production of PC are diverse and seem to depend on the availability of precursors for the synthesis of the hydrophilic head group, phosphocholine (P-Cho, see supplemental Fig. S1). Plants use the free phosphobase methylation pathway and can catalyze the conversion of serine to ethanolamine via a unique serine decarboxylase (4). Ethanolamine undergoes phosphorylation followed by the triple methylation of the free phosphobase to P-Cho, catalyzed by PEAMT (5, 6). P-Cho can then be used as the head group to form PC by the two enzymes P-Cho cytidylyltransferase and CDP-choline:diacylglycerol phosphotransferase, that are part of the Kennedy pathway in eukaryotes (7). Genetic evidence was recently presented in Plasmodium that the knockout mutant of the PfPMT gene is unable to synthesize PC in the absence of an external choline supply (8). Plasmodium therefore lacks the enzyme phosphatidylethanolamine methyltransferase found in fungi and mammals. Because higher plants lack that enzyme too and are choline autotrophs, the PEAMT-catalyzed reactions seem likely to be the sole entry point for P-Cho and choline into plant metabolism (9–11). Any choline kinase activity can therefore be seen as a recycling or feedback regulatory function rather than being involved in de novo biosynthesis of P-Cho and PC (12, 13). Plants have evolved two different ways of synthesizing choline: Chenopodiaceae such as spinach and sugarbeet are capable of producing it directly from ethanolamine via ethanolamine kinase, PEAMT, and P-Cho phosphatase catalysis and use it largely for the biosynthesis of glycine betaine (GlyBet (5, 14)). Gramineae, like wheat and barley, however, seem to produce choline, and therefore subsequently GlyBet, through PC hydrolysis (10, 15). One explanation for this could be the low activity of P-Cho phosphatase in these species, for which the identity of the encoding gene is still somewhat elusive (12).
GlyBet is thought to act as a compatible solute or osmoprotectant under various abiotic stresses ranging from drought and salinity to cold (16–18). The ability of different genotypes to withstand these conditions has been shown to correlate with their ability to synthesize GlyBet (19–21). This compound acts in the chloroplast to protect photosystem II and other protein complexes, as well as to maintain membrane integrity and scavenge reactive oxygen species, but also to reduce the extent of K+ loss in response to salinity and oxidative stress (see Ref. 22 for review). Interestingly, some of these functions overlap with those proposed for alterations in cell membrane phospholipid composition that have been observed under cold or drought stress (23–26). It is therefore not entirely clear whether GlyBet accumulation is a protective mechanism in itself, a symptom of stress susceptibility (27), or merely a consequence of increased phospholipid turnover (10, 25).
Due to its central position in the phospholipid and GlyBet synthetic pathways the role of PEAMT in plants' response to cold or salinity has been intensively studied (14, 28, 29). Although PEAMT has been proposed to be one important bottleneck for the de novo synthesis of choline, GlyBet, and PC (30), other enzymes such as S-adenosylmethionine (SAM) synthetase, S-adenosylhomocysteine (SAH) hydrolase, choline kinase, and CTP:P-Cho cytidylyltransferase have also been shown to have increased activities under these abiotic stress conditions (13, 29, 31, 32).
Individual PEAMT enzymes from spinach, wheat, and Arabidopsis have been characterized at the molecular level (28, 33, 34). They are all putative cytosolic enzymes composed of two methyltransferase domains. In the spinach SoPEAMT and wheat WPEAMT/TaPEAMT1 enzymes the N-terminal domain catalyzes the first methylation step, whereas the C-terminal half is only capable of catalyzing the second and third methylations. Although more than one PEAMT isoform has been identified in maize, rice, and Arabidopsis (13, 35), no detailed genomic or biochemical comparison of those isoforms in any species has been undertaken so far.
Here we report the molecular and biochemical characterization of two phosphoethanolamine N-methyltransferase isoforms in wheat. They show differential transcriptional regulation in response to cold acclimation and pronounced differences in their enzymatic properties that enable them to operate under different metabolic conditions. The consequences of these differences in enzymatic function for the plant's adaptability to different environments as well as for genetic engineering will be discussed.
EXPERIMENTAL PROCEDURES
Materials
All reagents, unless stated otherwise, were purchased from Sigma-Aldrich.
Plant Material
Triticum aestivum cv. Egret plants were grown in sterile vermiculite grade 3 under a 16-h photoperiod at 270 μmol m−2 s−1 photon flux density, 24 °C (21 °C at night), and 80% relative humidity. Plants were watered daily with one-third Hoagland's solution (36) by replacing the amount of water lost through transpiration and direct evaporation. For the cold treatment 7-day-old seedlings were transferred to a cabinet set up to match the same conditions as above, but running at 7 °C (day) and 4 °C (night). 1, 2, and 6 days later roots and shoots were harvested from both controls and cold-treated plants, immediately frozen in liquid N2, and then stored at −80 °C.
Expression and Purification of His6-tagged Wheat PEAMT Proteins
Following PCR amplification on a wheat cv. Egret cDNA library, the wheat TaPEAMT1 open reading frame was cloned into the Escherichia coli expression vector pET28a (Novagen/Merck, Kilsyth, Victoria, Australia) using the following primers: 5′-AACATATGGACACCATCACCGTCGTC-3′ containing a 5′ NdeI restriction site and 5′-AAGGATCCTCACTTGGTCGCGATGAACAG-3′ containing a 5′ BamHI restriction site, while the wheat TaPEAMT2 open reading frame was cloned using primers 5′-ATGGATCCGACGCCTCCGCCGCC-3′ containing a 5′ BamHI restriction site and 5′-ATGTCGACTCACTTGGTCCCGATGAACAGC-3′ containing a 5′ SalI restriction site. The resulting overexpression plasmids were sequenced and transformed into E. coli Rosetta (DE3)pLysS (Novagen). Recombinant His-tagged PEAMT proteins were purified to near-homogeneity by nickel-nitrilotriacetic acid affinity chromatography (see supplemental Fig. S2A). Protein purity was estimated by SDS-PAGE followed by Coomassie staining, and protein concentration was determined using Bradford reagent (Bio-Rad). Aliquots of the purified recombinant PEAMT proteins were stored at −80 °C.
Antibody Production
Recombinant TaPEAMT1 protein was eluted from the HiTrap column as before and exchanged into 1× phosphate-buffered saline using a PD-10 column. This antigen solution (1 mg/ml) was injected into rabbits using standard protocols for antibody production. The α-TaPEAMT1 polyclonal antibody detects recombinant TaPEAMT1 and TaPEAMT2 proteins with comparable affinity (data not shown).
Protein Extraction and Immunoblotting
Protein extraction and immunoblotting were carried out as described previously (37). α-TaPEAMT1, α-RbcL, and α-TCTP (37) primary antibodies were used as 1:1,000, 1:10,000, and 1:5,000 dilutions, respectively. Anti-rabbit IgG alkaline phosphatase conjugate (Promega, Madison, WI) was used in a 1:10,000 dilution. Chemiluminescence detection was performed using the Western Star Immunodetection System according to the manufacturer's instructions (Applied Biosystems, Scoresby, Victoria, Australia) and a charge-coupled device camera system (VersaDoc 3000, Bio-Rad).
Quantitative Real-time Reverse transcription-PCR
mRNA isolation and reverse transcription using magnetic oligo(dT)25-coated beads (Dynabeads®, Invitrogen) were carried out as described previously (38). Quantitative PCR and Ct value determinations were carried out using a Prism® 7900HT Sequence Detection System (Applied Biosystems) and accompanying software according to the manufacturer's instructions. Reactions contained 2.5 μl of bead-bound cDNA (0.5 ng of mRNA equivalent), 2.5 μl of primer mix (1.2 μm each), and 5 μl of 2× Power Sybr® Green PCR Master Mix (Applied Biosystems). After each run a melting curve analysis was performed to verify target-specific product amplification. Primers used for specific amplification of target transcripts were as follows: TaPEAMT1 (accession number AY065971), 5′-GACCGCACCGACCAGTTCCTGA-3′ and 5′-GCGCTCCAGCCGTTGACGAT-3′; TaPEAMT2 (accession number FJ803924), 5′-AAAACCTGGGGGTAAGGTCCTAATCAGT-3′ and 5′-CTTAATGTATGCCGCAAACTCTTCAGATG-3′. The following primers were used to detect reference gene expression: TaAPT1 (accession number U22442), 5′-GCAGCCCAATGACCGAGTCCTTA-3′ and 5′-GGCCCTTCAGCTCTGGCAGTTC-3′; TaPDF2 (accession number BT009473), 5′-CCCTTCAGGCGTGTGACCAGAT-3′ and 5′-TAAATTACTGGGCTAGAAAGAACTCTCAGACTCT-3′; and TaEF1α (accession number M90077), 5′-AAATGAGGGGCTTACCTGAATCCATCTA-3′ and 5′-CGCATATCACACGGCGCTAACAG-3′.
Preparation of Lipid Vesicles
Liposomes were prepared according to Potocky et al. (39). 3-sn-Phosphatidic acid from egg yolk or l-α-phosphatidylcholine from soybean (Sigma- Aldrich) were dissolved in chloroform/methanol (2:1, v/v). After evaporation under a dry nitrogen stream the lipid film was hydrated in MilliQ water for 1 h followed by 30-min sonication in a water bath at a temperature above the gel-liquid crystal transition temperature of the individual lipid. Aliquots of 100 mm stock solutions of these small, unilamellar vesicles were stored in glass vials at −20 °C.
Phosphoethanolamine N-Methyltransferase Assay
Unless stated otherwise, all purification steps were carried out at 4 °C. 100 mg of plant tissue ground under liquid N2 was homogenized in 1.5 volumes of 50 mm HEPES, pH 7.8, 10 mm KCl, 1 mm EDTA, 1 mm EGTA, 10% (v/v) glycerol, 5 mm dithiothreitol, and 0.5 mm phenylmethylsulfonyl fluoride buffer. After centrifugation, 120 μl of supernatant was passed through a desalting spin column (Pierce/Thermo Fisher Scientific Inc.) pre-equilibrated in 4 volumes of 50 mm HEPES, pH 7.8, 1 mm EDTA, 10% (v/v) glycerol, 5 mm dithiothreitol, and 0.5 mm phenylmethylsulfonyl fluoride. Protein amounts were determined using Bradford reagent (Bio-Rad). 80–160 μg of leaf protein extract and 60 μg of root protein extract were incubated in 50 mm HEPES, pH 8.6, 1 mm EDTA, 800 μm phosphoethanolamine (P-EA), and 2000 μm SAM (60:1 ratio of cold SAM over [14C]SAM/S-adenosyl-l-[methyl-14C]methionine with an average specific activity of 2 GBq/mmol, Amersham Biosciences). For control assays P-EA was omitted. Samples were incubated at 30 °C for 30 min, and reactions were stopped by shock-freezing in liquid N2. To separate unincorporated 14C-labeled SAM from methylated products, reaction mixes were passed through 2-ml AG 50W-X8 (H+) cation exchanger columns (Bio-Rad) as described by Charron et al. (28) except that samples were eluted twice in 5 ml of 1 n HCl. 1 ml of combined eluates was added to 3 ml of Ultima Gold XR scintillation liquid (Packard BioScience/PerkinElmer Life Sciences, Rowville, Victoria, Australia) and counted for 20 min in a Tri-Carb 2800TR liquid scintillation analyzer (PerkinElmer Life Sciences) using a 14C quench correction for the amount of HCl in each sample. Elution efficiency of methylated phosphobases was estimated by comparing samples with and without addition of cold SAM.
Enzyme Kinetics
In all experiments the reactions were only allowed to proceed to a negligible extent (6–7% for TaPEAMT1 and 9–14% for TaPEAMT2 at saturating substrate concentrations), and product formation was linear in response to protein concentration and time. Unless stated otherwise, 1 μg of recombinant TaPEAMT protein was used. The assay conditions and ion-exchange chromatography procedures were as mentioned above. To ensure that the column's cation exchanger binding capacity was not exceeded, assay reactions using high substrate amounts were diluted prior to column loading. For initial velocity studies both substrates were varied in a 5 × 5 matrix according to Eisenthal and Danson (40) (see Table 1). In control reactions either P-EA or SAM were omitted. For product inhibitor studies the corresponding substrate-product pairs, i.e. P-EA and P-Cho or SAM and SAH, were varied accordingly, with the concentration of the second substrate kept at saturation (see Table 1). For phosphatidic acid and phosphocholine inhibition plots, phospholipid concentrations were varied while both substrates were used at saturating concentrations. Initial velocity data for the two-substrate kinetics or product inhibition studies were fitted to the general velocity equations using the Enzyme Kinetics Module for SigmaPlot (SYSTAT, see supplemental Fig. S4A for equations).
TABLE 1.
Summary of the kinetic parameters obtained from the biochemical analysis of the two wheat PEAMT proteins
Data were fitted to Equations 1–3 given in supplemental Fig. S4A, which describe the sequential random Bi Bi (supplemental Fig. 4B) and partial/full mixed inhibition mechanisms (supplemental Fig. 4C).
| TaPEAMT1 | TaPEAMT2 | |
|---|---|---|
| Initial velocity study | ||
| P-EA (μm) | 100–2,000 | 50–1,000 |
| SAM (μm) | 200–4,000 | 100–4,000 |
| Vmax (nmol/(mg*min)) | 552 ± 29 | 1,351 ± 82 |
| Km(app., P-EA) (μm) | 390 ± 120 | 185 ± 49 |
| Km(app., SAM) (μm) | 601 ± 178 | 1,060 ± 295 |
| α | 1.1 | 1.2 |
| Mechanism | Random Bi Bi sequential | Random Bi Bi sequential |
| R2 (n = 2) | 0.975 | 0.981 |
| P-Cho feedback inhibition | ||
| P-EA (μm) | 100–4,000 | 200–4,000 |
| SAM (μm) | 2,000 | 4,000 |
| P-Cho (μm) | 1000–10,000 | 50–1,000 |
| Vmax (nmol/(mg*min)) | 430 ± 12 | 1231 ± 35 |
| Km(app., P-EA) (μm) | 328 ± 38 | 263 ± 34 |
| Ki(P-Cho) (μm) | 306 ± 57 | 137 ± 38 |
| α | 5.5 | 2.5 |
| β | 0.32 | |
| Mechanism | Mixed (partial) | Mixed (full/partial) |
| R2 (n = 2) | 0.971 | 0.975 |
| SAH feedback inhibition | ||
| P-EA (μm) | 2,000/4,000 | 2,000 |
| SAM (μm) | 300–4,000 | 300–4,000 |
| SAH (μm) | 50–500 | 25–500 |
| Vmax (nmol/(mg*min)) | 436 ± 24 | 1495 ± 63 |
| Km(app., SAM) (μm) | 655 ± 112 | 754 ± 96 |
| Ki(SAH) (μm) | 31 ± 8 | 70 ± 16 |
| α | 3.5 | 5.7 |
| β | 0.47 | 0.07 |
| Mechanism | Mixed (partial) | Mixed (partial/full) |
| R2 (n = 2) | 0.943 | 0.966 |
Lipid-Protein Overlay Assay
Binding of recombinant PEAMT protein to phospholipids was analyzed according to Dowler et al. (41) using 1 mm stocks of 1,2-dipalmitoyl-sn-glycero-3-phosphocholine or -phosphate (Echelon Biosciences Inc., Salt Lake City, UT). Spotted membranes as well as commercial membrane lipids and phosphoinositide (PIP) strips (Echelon Biosciences Inc.) were blocked for 4 h in 0.2% I-block in 1× phosphate-buffered saline with 0.1% Tween-20. Blots were then incubated with 40–80 nm recombinant TaPEAMT protein overnight at 4 °C, before lipid-bound protein was detected using a 1:2,000 dilution of α-TaPEAMT1 antibody (see above) and 1:10,000 dilution of α-rabbit IgG alkaline phosphatase conjugate (Promega) followed by chemiluminescence detection according to the manufacturer's instructions (Applied Biosystems).
RESULTS
Identification of a Second Wheat PEAMT cDNA
Through sequence comparison of the published WPEAMT/TaPEAMT1 cDNA sequence (28) (accession no. AY065971, chromosome 1) with expressed sequence tag data available on-line (Gramene: A Resource for Comparative Grass Genomics, and NCBI, the UniGene expressed sequence tag collection from the National Center for Biotechnology Information), we identified a second set of expressed sequence tag sequences distinctly different from the homologous classes of WPEAMT/TaPEAMT1 contigs. This new set of sequences fell into three contig classes likely representing homeologs from the three wheat genomes. We used the most prominent class of expressed sequence tag sequences to deduce an in silico TaPEAMT2 cDNA sequence prediction and to design primers to amplify the cDNA for subsequent analyses. The cloned open reading frame sequence for TaPEAMT2 shows 77% identity to the WPEAMT/TaPEAMT1 open reading frame and has been submitted to GenBank®, accession number FJ803924. Both cDNAs encode 57-kDa proteins that seem to lack N-terminal signal peptide sequences and therefore most likely localize to the cytoplasm.
Phylogenetic Analysis of PEAMT Proteins
Higher plant PEAMT proteins form two distinct clades for dicotyledonous and monocotyledonous species, clearly separated from the protein sequence for the moss Physcomitrella patens and the rather isolated protein sequence from purple sea urchin as well as from the other two clades, one containing three vertebrate proteins from Xenopus laevis, pufferfish, and zebrafish, the other containing the protein from the apicomplexan parasite Plasmodium falciparum and two Caenorhabditis elegans proteins, CePMT-1 and -2 (see supplemental Fig. S3). The proteins in the vertebrate clade are only putative PEAMT orthologs and have not been characterized in any detail yet. The higher plant proteins fall into several distinct groups with the monocot clade forming two branches, one for each of the two isoforms that can be found in nearly all species analyzed to date, including the newly identified TaPEAMT2 sequence characterized in this study. This could be interpreted as a sign of some specialization of enzymatic function within monocotyledons. The dicot clade is structured quite differently, with only one isoform identified for most species to date, with the exception of Arabidopsis thaliana (three isoforms, NMT1–3), Medicago truncatula, and Vitis vinifera (two isoforms). AtNMT1 and AtNMT2 are located on duplicated segments of chromosomes 3 and 1, respectively, indicating that they have arisen from a very recent gene duplication event (42). A very strong separate branch is formed by known Chenopodiacean proteins, indicating that some enzyme specialization occurred for these GlyBet accumulators (5, 9, 14). There is, however, no general trend for protein sequences of GlyBet accumulators to group together, and the dicotyledonous clade is organized instead according to taxonomic groups.
Response to Cold Acclimation
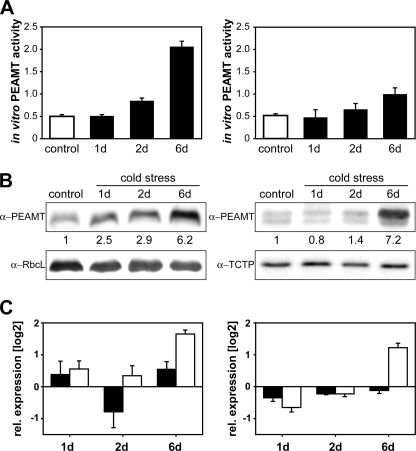
After the identification of a second PEAMT isoform in wheat we probed for functional differences using cold treatments that have been shown to induce PEAMT gene expression in wheat and Arabidopsis (13, 28). As expected from these published results we were able to detect increases in PEAMT protein and activity levels in leaves within 24–48 h after transfer to cold, with a 6-fold increase in total PEAMT protein amounts and a 4-fold increase of in vitro activity observed after 6-day exposure (Fig. 1, A and B). No PEAMT activity or protein expression has previously been detected in roots (28), but in this study total protein and activity levels in roots were comparable to those in leaves, and an increase in total root PEAMT protein (7-fold) and activity (2-fold) was recorded after 6 days of cold acclimation. Analysis by gene-specific quantitative reverse transcription-PCR showed a differential response of the two TaPEAMT genes with a clear induction of TaPEAMT2 being detectable 6 days after transfer, in both leaves and roots, whereas changes in TaPEAMT1 transcript remained within noise level (Fig. 1C). It is likely then that the large induction of PEAMT protein expression and in vitro activity at that time primarily reflects the induction of the TaPEAMT2 isoform. The absence of TaPEAMT1 gene induction by cold contrasts with an earlier report by Charron et al. (28) based on Northern hybridization experiments. Differential expression of PEAMT genes has also been reported in Arabidopsis under salt stress (13). The absence of significant changes in either PEAMT gene expression at the 2-day time point in our experiment when protein expression was already significantly increased in leaves (Fig. 1B, left panel) is indicative of post-transcriptional or post-translational regulation of PEAMT. To assess the physiological significance of these results, we decided to examine the kinetic properties of the two wheat PEAMT isoforms.
FIGURE 1.
PEAMT expression profiles and enzymatic activities in cold acclimated wheat cv. Egret. Seedlings were germinated in sterile vermiculite and watered daily with one-third Hoagland solution. They were transferred from 24 °C to 7 °C at the indicated time points and harvested when 2 weeks old. A, total in vitro PEAMT activity in desalted leaf and root protein extracts (averaged measurements with 70 and 140 μg of total leaf protein, left panel, or 20 and 30 μg of total root protein, right panel) after 1, 2, and 6 days of cold exposure (black bar) or continued growth at ambient temperature (open bar) plotted as mean ± S.E. nanomoles of SAM/(mg of protein*min), n ≥ 3. B, Western analysis of total soluble leaf proteins (10 μg, left panel) and root proteins (4 μg, right panel). A quantification of band intensities relative to controls is given below the α-PEAMT blots. Separation of bands in leaf extracts might be impeded by the RbcL band. Alternatively the second, lower band visible in root extracts could represent an additional PEAMT isoform or hint at a root-specific post-translational modification. C, real-time PCR analysis of TaPEAMT1 (solid bars) and TaPEAMT2 (open bars) expression in cDNA isolated from leaves (left panel) and roots (right panel). Data are relative expression ratios of cold-treated versus control samples, normalized to three control genes (mean ± S.E., n = 3).
Biochemical Characterization
To date only one isoform from spinach, Arabidopsis, and wheat has been biochemically characterized (6, 28, 33, 34). These experiments used only partially purified proteins, either isolated from plant material or recombinantly expressed in E. coli or yeast. In this study we compared the kinetic properties of two different PEAMT isoforms within a plant species using highly purified recombinant proteins (supplemental Fig. S2A). After correcting for residual contaminant E. coli proteins in the final desalted protein extracts, the protein dependence of PEAMT activity was determined for both isoforms. Specific P-base N-methylation activities were determined from the linear phase of the two curves in supplemental Fig. S2B. Using physiologically relevant substrate concentrations of 200 μm P-EA and SAM as described earlier (6), we found that TaPEAMT2 is four times more active than TaPEAMT1 with specific activities of 2353 ± 122 and 570 ± 62 pmol/s/mg of protein, respectively. The TaPEAMT2 specific activity is quite similar to that reported for the purified native spinach enzyme (6), whereas the value we obtained for recombinant TaPEAMT1 is ∼90-fold higher than reported previously for recombinant WPEAMT from crude E. coli extracts (28).
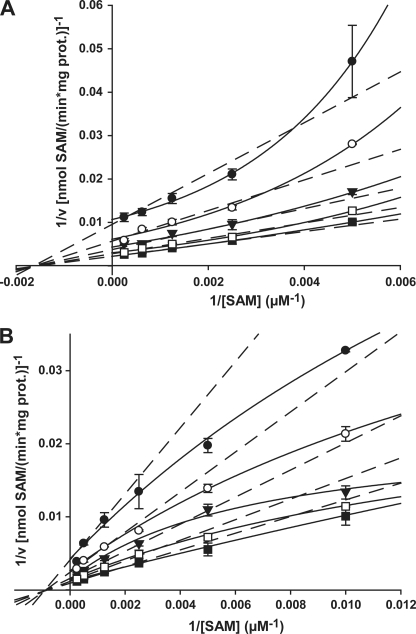
Initial Velocity Studies
Although apparent Km values for the two substrates P-EA and SAM have been reported for one recombinant wheat and one recombinant spinach PEAMT enzyme in crude extracts (28, 33), no detailed mechanistic study of any plant enzyme has been performed to date. When we used a similar substrate concentration range as in these published analyses in our experiments, apparent Km values were of the same order as those reported for the recombinant spinach enzyme with 198 ± 16 μm (TaPEAMT1) and 111 ± 12 μm (TaPEAMT2) for P-EA and 396 ± 38 μm (TaPEAMT1) and 212 ± 43 μm (TaPEAMT2) for SAM and revealed higher affinities of the second wheat isoform for both substrates. It was obvious, however, from these initial measurements that no true substrate saturation had been achieved (data not shown). We therefore undertook a more detailed mechanistic study by varying both substrates simultaneously over a wider concentration range (Fig. 2 and Table 1). Although for both enzymes the Km values for P-EA increased by less than 2-fold, there was a 5-fold decrease in TaPEAMT2 affinity for SAM under saturating compared with non-saturating conditions (data not shown). For TaPEAMT1, however, the affinity for SAM decreased only ∼1.5-fold. The favored kinetic model for both enzymes is that of a sequential random Bi Bi mechanism (R2 = 0.98, see Table 1 and supplemental Fig. S4B). For TaPEAMT1 the double reciprocal plot for P-EA fits quite well with the predicted model (data not shown), whereas for SAM the data points diverge from the expected linear behavior and appear to fit a parabolic relationship (see Fig. 2A). The enzyme therefore exhibits strongly cooperative substrate binding for SAM with an apparent specific binding site concentration or napp of 2 (43). This behavior is most likely due to the presence of two separate SAM binding sites, as predicted from the presence of two separate methyltransferase domains in the N- and C-terminal halves of the wheat (and spinach) PEAMT enzymes (28, 33). For TaPEAMT2 the double reciprocal plots diverge from the expected set of straight lines for both substrates, showing a more hyperbolic pattern, especially when SAM concentration is varied at different constant P-EA concentrations (Fig. 2B). This indicates negative cooperativity between the two SAM binding sites (43). This behavior would explain the rather low apparent Km observed for SAM at physiological substrate concentrations (see above), compared with the huge increase in the Km for SAM observed at substrate saturation as a result of marked substrate inhibition under these conditions. This might also explain why TaPEAMT2 is 4-fold more active than TaPEAMT1 under a physiological substrate concentration range (200 μm P-EA and 200 μm SAM), while there is only a 2.5-fold difference in Vmax between the two enzymes closer to substrate saturation (see Table 1).
FIGURE 2.
Initial velocity plots for TaPEAMT1 and TaPEAMT2. Both enzymes follow a sequential random Bi Bi mechanism (R2 = 0.975/0.981 for TaPEAMT1/2). A, double reciprocal plot of 1/v versus 1/[SAM] for TaPEAMT1 generated at five fixed P-EA concentrations of 100 (●), 200 (○), 400 (▼), 800 (□), and 2000 μm (■) using 1 μg of recombinant protein (mean ± S.E., n = 2). Whereas plots against 1/[P-EA] are linear (not shown), the plots against 1/[SAM] deviate from a linear relationship (dashed lines) and appear to be parabolic (solid lines). When 1/v is plotted against 1/[SAM]2, the linear relationship is restored confirming that PEAMT has two separate binding sites (napp = 2), which exhibit strong cooperative substrate binding at high [SAM]. B, double reciprocal plot 1/v versus 1/[SAM] for TaPEAMT2 at five fixed P-EA concentrations of 50 (●), 100 (○), 200 (▼), 400 (□), and 1000 μm (■) using 1 μg of recombinant protein (mean ± S.E., n = 2). Both double reciprocal plots show a more hyperbolic scattering of data points, with the deviation from a linear relationship being more pronounced for the plot of 1/v against 1/[SAM]. This suggests a partial negative cooperativity of substrate binding, especially at high SAM concentrations. No forcing or weighting of data points was applied.
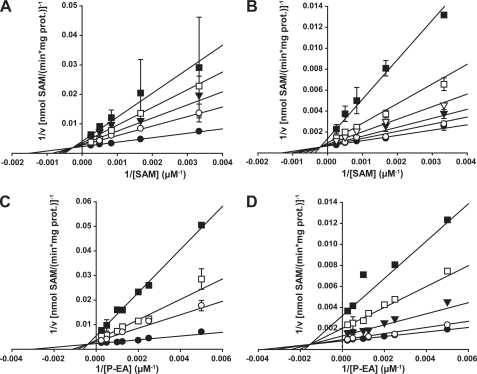
Product Inhibition
To further elucidate the kinetic mechanisms for both wheat PEAMT isoforms the two reaction products P-Cho and SAH were varied at different fixed concentrations of P-EA and SAM, respectively. The corresponding cosubstrate was kept at saturation (Fig. 3). For TaPEAMT1 apparent Km values for P-EA and SAM as well as Vmax were comparable to the ones obtained from the initial velocity study (see Table 1). Both double reciprocal replots show a set of straight lines intersecting above the abscissa and to the left side of the ordinate, pointing to a mixed type of inhibition (see Fig. 3, A and B). The best R2 values in both studies were obtained for the partial mixed-type inhibition model with values for β > 0 (see Table 1 and supplemental Fig. S4C), indicating that both enzyme-substrate-inhibitor complexes can still release product. This mixed-type inhibition pattern for both products unequivocally confirms the proposed sequential random Bi Bi mechanism (43). For TaPEAMT2 the same pattern of intersecting lines in the double reciprocal plots was observed for both product inhibitors, again confirming a sequential random Bi Bi mechanism. Values for β were very close to 0 in both cases, however, indicating a full mixed-type inhibition that leads to dead-end enzyme-substrate-inhibitor complexes (see Table 1 and supplemental Fig. S4C). This should allow for a very tight control of TaPEAMT2 activity when either of the two products accumulates. Vmax and the apparent Km for P-EA are similar to those found in the initial velocity analysis, but the apparent Km for SAM is significantly lower. Binding of SAH might therefore be the cause of the negative cooperativity observed for TaPEAMT2 in the initial velocity study, leading to an increase in the apparent Km for SAM. It is quite surprising that the Ki for binding of SAH to the free enzyme is 2-fold higher for TaPEAMT2 than for TaPEAMT1, whereas Ki values for P-Cho binding to the free enzyme were almost 3-fold lower for TaPEAMT2 than for TaPEAMT1. Given the more non-competitive nature of TaPEAMT2 inhibition by SAH and the observed negative cooperativity at higher SAM concentrations, it would appear that TaPEAMT2 activity, which is very high at physiologically relevant substrate concentrations, can at the same time be down-regulated very quickly when products accumulate. At substrate saturation TaPEAMT2 is much more sensitive to P-Cho inhibition than TaPEAMT1 (21 ± 3% versus 58 ± 2% of control activity in the presence of 2 mm P-Cho), as a consequence of its higher affinity for P-Cho and the strong non-competitive component of this inhibition. Due to the different inhibition mechanisms both isoforms show a similar relative inhibition by SAH despite their contrasting Ki values (56 ± 4% versus 53 ± 6% of control activity in the presence of 200 μm SAH). For the spinach enzyme an IC50 for P-Cho of 250–490 μm was reported with a mixed competitive behavior toward P-EA, whereas its sensitivity to inhibition by SAH was high with an IC50 of ∼10 μm (6, 33). Although both wheat enzymes show an IC50 of ∼200 μm for P-Cho at 200 μm P-EA and saturating SAM concentrations similar to the spinach enzyme, their IC50 for SAH was significantly higher with 70 μm for TaPEAMT1 and 130 μm for TaPEAMT2 at 300 μm SAM and saturating P-EA concentrations. It therefore seems likely that the wheat enzymes are more robust toward accumulation of SAH in the cell, with TaPEAMT2 being particularly suited to operate at low substrate concentrations.
FIGURE 3.
Product inhibition studies of TaPEAMT1 and TaPEAMT2. A, double reciprocal plot of 1/v versus 1/[SAM] for TaPEAMT1 using four fixed SAH concentrations of 0 (●), 50 (○), 100 (▼), 200 (□), and 500 μm (■), and 1 μg of recombinant protein (mean ± S.E., n = 2). B, double reciprocal plot of 1/v versus 1/[SAM] for TaPEAMT2 using five fixed SAH concentrations of 0 (●), 25 (○), 50 (▼), 100 (s), 200 (□), and 500 (■) μm, and 1 μg of recombinant protein (mean ± S.E., n = 2). C, double reciprocal plot of 1/v versus 1/[P-EA] for TaPEAMT1 using four fixed P-Cho concentrations of 0 (●), 1,000 (○), 5,000 (□), and 10,000 (■) μm, and 1 μg of recombinant protein (n = 2). D, double reciprocal plot of 1/v versus 1/[P-EA] for TaPEAMT2 using four fixed P-Cho concentrations of 0 (●), 50 (○), 200 (▼), 500 (□), and 1000 (■) μm, and 1 μg of recombinant protein (mean ± S.E., n = 2), with some data points only measured once. No forcing or weighting of data points was applied.
Effect of Ions and Downstream Metabolites on Wheat PEAMT Activity
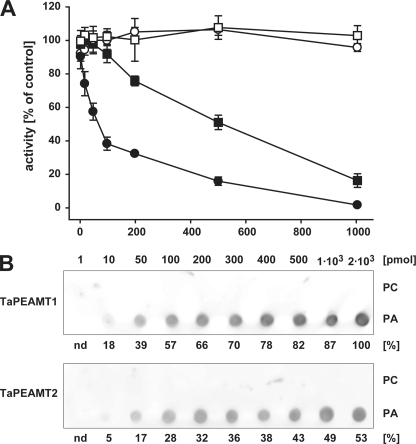
The native spinach PEAMT has previously been shown to be sensitive to inhibition not only by P-Cho and SAH, but also by phosphate, calcium, and manganese ions (6). We therefore tested these compounds as well as downstream metabolites choline, GlyBet, PC, and PA at substrate saturation (2 mm P-EA and 4 mm SAM, n = 2). 10 mm choline or 50 mm GlyBet had no effect on the two enzymes, with TaPEAMT1/TaPEAMT2 retaining 96 ± 2%/93 ± 3% and 97 ± 2%/102 ± 3% of control activities, respectively. The activity of both isoforms was mildly repressed by 10 mm potassium (76 ± 9%/86 ± 3%), magnesium (79 ± 9%/84 ± 3%), or calcium phosphate (77 ± 9%/84 ± 3% of TaPEAMT1/TaPEAMT2 control activity). Manganese ions, however, had a much more drastic effect on the two enzymes: both showed 86% inhibition at 10 mm MnCl2 (14 ± 5%/14 ± 2% of TaPEAMT1/TaPEAMT2 control activity), much higher than the 43% inhibition reported for the spinach enzyme (6). Apart from manganese all the other inhibitors tested were less effective than in the spinach PEAMT assay. By far the most effective inhibitor tested in this experiment was PA (Fig. 4A). The response of the two isoforms was remarkably different. At PA concentrations as low as 100 μm TaPEAMT1 activity was already reduced to 33 ± 4%, whereas TaPEAMT2 activity was affected only at much higher concentrations. In contrast, even concentrations as high as 1 mm PC did not inhibit either enzyme (see Fig. 4A), indicating a high degree of specificity and potency of the PA effect on TaPEAMT1 activity.
FIGURE 4.
PA inhibition of TaPEAMT activity and PA binding to TaPEAMT isoforms. A, in vitro activity of recombinant TaPEAMT1 (circles) and TaPEAMT2 (squares) in the presence of increasing PA (solid symbols) or PC (open symbols) concentrations, expressed as percentage of activity in control reactions in the absence of lipids. Reactions contained 1 μg of recombinant protein and saturating substrate concentrations (2 mm SAM and 4 mm P-EA; mean ± S.E., n ≥ 2). Although the presence of PC vesicles does not alter the activity of either enzyme, addition of PA vesicles lead to a rapid decrease in TaPEAMT1 activity with an IC50 of ∼70 μm PA, while TaPEAMT2 is only inhibited by very high PA concentrations with an IC50 of ∼470 μm PA. B, specificity of PA binding. 80 nm recombinant TaPEAMT protein was incubated with membranes spotted with concentration series for PC and PA, followed by decoration with primary and secondary antibodies and chemiluminescence detection. Bound TaPEAMT protein can be detected with as little as 10 pmol of PA spotted, whereas PC does not attract any protein to the membrane, even with as much as 2 nmol being spotted. A representative blot selected out of three membranes for each PEAMT isoform in two independent experiments is shown. Numbers below chemiluminescence signals represent the average spot intensity across replicated membranes expressed as the percentage of signal intensity obtained for TaPEAMT1 at the maximal PA concentration spotted.
Binding of PA to Recombinant TaPEAMT Proteins
A number of cytosolic enzymes have been identified as PA-binding proteins in eukaryotes (1, 44). The binding of PA can result in either inhibition or activation of enzyme activity. Because PA was shown to strongly inhibit the PEAMT1 isoform at low concentration, we wanted to look more closely at the specificity of this interaction using lipid protein overlay assays (41). Fig. 4B shows that binding of PA to both isoforms could be detected with as little as 10 pmol of PA spotted onto the nitrocellulose membrane and incubation with 40–80 nm recombinant protein, whereas no binding to PC was detected even when spotting as much as 2 nmol of PC. In these assays PA binding to TaPEAMT1 appeared to be stronger than for TaPEAMT2 (see Fig. 4B). The specificity of this interaction was examined by analyzing the binding of TaPEAMT isoforms to commercially available membrane lipid strips. For both proteins PA was indeed the strongest interacting lipid, but there was also some affinity for phosphatidylinositol (PtdIns)(4)P and to some extent to PtdIns(4,5)P2 and cardiolipin (Fig. 5A). To see whether the position of the phosphate group within the hexose moiety affects the interaction, commercial PIP strips were incubated with the two recombinant proteins (Fig. 5B). Again, the strongest interaction by far was observed with PA, followed by PtdIns monophosphates (PtdIns(5)P, -(4)P, and -(3)P) and two of the PtdIns diphosphates (PtdIns(3,4)P2 and PtdIns(4,5)P2). Interestingly, no binding could be detected for PtdIns(3,5)P2 or PtdIns(3,4,5)P3 suggesting specificity of the recognition. The binding to cardiolipin, but not to PC or other phospholipids, indicates that the spacing of phosphate groups might be important for binding specificity and that it is not merely determined by the size of the ligand. The same holds true for the strong interaction with PA, but not lyso-PA or sphingosine 1-phosphate that merely lack one acyl side chain. To date no lipid binding motif has been identified for PEAMT proteins, and there is not enough sequence similarity between our two PEAMT proteins and published motifs to predict PA or PtdIns monophosphate binding sites. Therefore it has to be assumed that there are yet other binding motifs to be discovered in this protein family.
FIGURE 5.
Screen of putative lipid ligands for the two TaPEAMT isoforms. A, membrane lipid strips spotted with 100 pmol of each lipid were incubated with 80 nm recombinant TaPEAMT protein as before. As expected both isoforms strongly interact with PA, followed by some binding to PtdIns(4)p > PtdIns(4,5)P2/cardiolipin. Numbers next to each chemiluminescent signal are given as percent spot intensity of PA binding to TaPEAMT1. B, to test the specificity of the interaction with phosphoinositides PIP strips (spots contain 100 pmol of lipid each) were incubated with 80 nm recombinant TaPEAMT protein as before. The strongest interaction again is observed with PA, followed by interactions with the three PtdIns monophosphates ((5)P > (4)P > (3)P) and two of the PtdIns diphosphates ((3,4)P2 ≥ (4,5)P2) consistent with earlier findings, except that TaPEAMT2 binding to PtdIns(4,5)P2 was not detected in A most likely due to a more stringent wash. The recognition appears to be specific, because no binding to PtdIns, PtdIns(3,5)P2, or PtdIns(3,4,5)P3 was observed. Numbers next to each chemiluminescent signal describe spot intensities expressed as percentage of spot intensity for PA binding to TaPEAMT1.
DISCUSSION
PEAMT catalyzes a rate-limiting step in plant choline and PC biosynthesis (10, 30). Wheat and barley appear to have evolved an alternative pathway for GlyBet accumulation compared with chenopods (5), because they synthesize choline via PC hydrolysis just like GlyBet non-accumulators tobacco and Arabidopsis (10, 30, 44). In the light of this fundamental difference it is important to biochemically characterize PEAMTs from different plant species. The presence of more than one isoform of this enzyme in many species adds a level of complexity to possible regulatory mechanisms that has so far not been explored. Here we report the identification of a second PEAMT isoform in wheat and the comparative biochemical characterization of both TaPEAMT1 and TaPEAMT2 enzymes. We found that the two genes differ in their expression response to cold exposure in the spring wheat cultivar Egret. An increase in TaPEAMT2 transcript levels was detected in both leaves and roots after 6 days in the cold. TaPEAMT1 gene expression showed no significant variation. In both roots and leaves of unstressed plants TaPEAMT1 transcripts were much more abundant than TaPEAMT2, suggesting that this isoform is likely to be responsible for the bulk turnover of P-Cho. An increase in total PEAMT protein and activity levels could already be detected within 24–48 h of transfer to cold, thus preceding TaPEAMT2 gene induction by several days. In Plasmodium the rapid reduction in PEAMT protein upon choline addition can be inhibited by bortezombid, a proteasome inhibitor (5, 45). It will therefore be interesting to see whether targeted proteasome-mediated protein degradation or preferential loading of PEAMT mRNA onto the ribosome could explain our observation of a rapid increase in PEAMT protein upon cold treatment ahead of the transcriptional induction of TaPEAMT2.
The present study indicates that on top of this tight control of the absolute protein amount within a given tissue the two differentially expressed wheat PEAMT isoforms also have unique biochemical properties that may be important for plant adaptation to changing environmental conditions: although TaPEAMT1 is less active than TaPEAMT2 and has a lower affinity for one of its substrates, P-EA, it has a higher affinity for the second substrate, SAM, and is less sensitive to feedback inhibition by P-Cho, especially due to the partial nature of that inhibition. This probably makes this enzyme more suitable to maintaining a moderate flux through the pathway under conditions where the methylation index, that is the cellular SAM:SAH ratio (32, 46) is high and the cytosolic P-Cho pool builds up to some extent as has been observed in unstressed tobacco leaves with subsequent salinization rapidly depleting this pool (30). The sensitivity of the TaPEAMT1 protein to SAH inhibition means, however, that any disturbance in the activated methyl cycle will quickly down-regulate this enzyme. In this context it is of note that both PEAMTs have unusually low Ki values for SAH compared with very high Km values for SAM (20- and 17-fold differences in Km (SAM) over Ki (SAH) for TaPEAMT1 and TaPEAMT2, respectively) making them very susceptible to inhibition by decreases in the methylation index compared with other methyltransferases (32). It is therefore likely that any increase in PEAMT activity must be accompanied by concomitant increases in activated methyl cycle activities. This coordinated up-regulation has been observed under salt stress for SAM synthetase and PEAMT in the halophyte Atriplex nummularia (29) and for SAH hydrolase, adenosine kinase, and PEAMT in GlyBet accumulators spinach and sugar beet while there was no evidence for it in the non-accumulators tobacco or canola (32). In the light of the kinetic differences we observed between the two wheat enzymes, it is interesting to look at the kinetic mechanisms that have evolved in PEAMT enzymes of other organisms (33, 47–49) (see also supplemental Table S2). Compared with the wheat enzymes the single domain Plasmodium enzyme has a lower specific activity, but higher substrate affinities and is highly sensitive to feedback inhibition by P-Cho. The two separate methyltransferases of C. elegans exhibit a random sequential Bi Bi mechanism as the wheat (and spinach) enzymes but are distinctive by their insensitivity to P-Cho inhibition. It may be that the evolution of the two-domain structure of higher plant PEAMTs has facilitated the selection of a moderate sensitivity to P-Cho inhibition that sits in between that of the highly sensitive Plasmodium and the virtually insensitive C. elegans proteins.
Given the tight control of PEAMT activity by its products but also the post-transcriptional control most likely exerted by choline (45, 50, 51), we were interested to see whether metabolites further downstream would also act as regulators. It has recently been shown that knockout of Arabidopsis NMT1 leads to cell death in the root elongation and differentiation zones that can be reverted by exogenous application of PA (44) and that cyanobacterial SAM synthetase activity can be stimulated in the presence of PC (52). We therefore tested the effects of these two phospholipids on PEAMT activity. Although PC did not affect the in vitro activity of the two wheat enzymes, PA led to a very rapid repression of both catalytic activities, with TaPEAMT1 being much more sensitive to PA inhibition than TaPEAMT2. The IC50 values obtained for PA inhibition of both enzymes make it likely that repression of TaPEAMT1 activity can occur at physiologically relevant PA concentrations (50–150 μm in Arabidopsis leaves (2, 53)), whereas TaPEAMT2 inhibition most likely only occurs under circumstances where endogenous PA levels rise dramatically, as has been demonstrated under hyperosmotic stress and dehydration conditions (54, 55). Lipid-protein-overlay studies confirmed the strong specific binding of both PEAMT isoforms to PA but not to glycerolipids, lysolipids, diacylglycerol, or most other phospholipids. PA signaling has been implicated in numerous plant stress responses (for review see Ref. 1). The identification of PA as a negative regulator of choline and phospholipid biosynthesis therefore provides a potential mechanism for how plants amplify or attenuate the production of lipid signals. This discovery adds a layer of complexity to the regulation of this biosynthetic pathway, because PA and choline are the products of PC hydrolysis by phospholipase D, and both lead to a rapid down-regulation of PEAMT. Our results indicate that there is a very strong metabolic feedback loop tightly controlling the level of PC synthesis under conditions where rapid turnover of this phospholipid is occurring. In this context it is interesting that wheat PEAMTs also bind to the substrate for PLC hydrolysis, PtdIns(4,5)P2 in vitro. In Arabidopsis this phospholipid is a potent activator of phospholipase D activity (56), whereas phospholipase D derived PA is an activator of some PtdIns(4)P 5-kinase isoforms, which synthesize PtdIns(4,5)P2 (57). PEAMT binding to PtdIns(4,5)P2 could therefore constitute a regulatory circuit that may in itself be required as an overriding safety switch that cuts off further substrate/PC supply.
The strongest PIP interaction was observed with PtdIns monophosphates, particularly PtdIns(5)P. This is the most recent member of phosphoinositides detected in plants, and it was shown to rapidly accumulate in Chlamydomonas cells subjected to hyperosmotic stress alongside its precursor PtdIns(3,5)P2 (58). Interestingly, no binding of the latter to wheat PEAMTs was observed. There was no binding either to PtdIns(3,4,5)P3 (the only phosphoinositide isomer that has not yet been detected in plants (1)). Phosphoinositide binding could allow for the recruitment of PEAMT to specific membrane domains (59, 60) as has been observed for the association of Plasmodium PEAMT to the Golgi apparatus (61).
The relative contribution of the free phosphobase methylation pathway versus the CDP-choline pathway to de novo PC biosynthesis in plants is still a standing question (13). The work in Plasmodium suggests that these pathways are not necessarily interchangeable and might lead to structurally different PC pools important for different cellular processes (8). Beyond the present in vitro work it will therefore be essential to examine the regulatory differences between the two wheat PEAMT enzymes in vivo and to gain insights into their contributions to the regulation of PC synthesis and ultimately plant growth and adaptation to stress. Our data open the way for genetic engineering approaches to these questions. Overexpression of a recombinant version of PEAMT that is less sensitive to feedback inhibition by P-Cho has already been discussed as one important tool to increase pool sizes of choline and possibly GlyBet in plants (33). The recent work on two virtually P-Cho-insensitive enzymes from C. elegans might further assist future engineering efforts (47, 48). A challenge will clearly be to overcome the observed strong repression of the PEAMT enzymes by both choline and PA; the influence of the proteasome on PEAMT stability in plants must be addressed and PA binding motifs identified.
Supplementary Material
Acknowledgments
We thank Jan Elliot for antibody production and Charles Hocart for helpful discussion.
This work was supported by the Australian National University and the Grains Research and Development Corporation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Tables S1 and S2.
- PC
- phosphatidylcholine
- P-Cho
- phosphocholine
- GlyBet
- glycine betaine
- PA
- phosphatidic acid
- P-EA
- phosphoethanolamine
- (Ta)PEAMT
- (wheat) phosphoethanolamine N-methyltransferase
- PIP
- phosphoinositide
- SAH
- S-adenosyl-l-homocysteine
- SAM
- S-adenosylmethionine
- PtdIns
- phosphatidylinositol (PtdIns(3)P, PtdIns 3- phosphate
- PtdIns(4)P
- PtdIns 4-phosphate
- PtdIns(5)P
- PtdIns 5-phosphate
- PtdIns(3,4)P2
- PtdIns 3,4-diphosphate
- PtdIns(4,5)P2
- PtdIns 4,5-diphosphate
- PtdIns(3,5)P2
- PtdIns 3,5-diphosphate
- PtdIns(3,4,5)P3
- PtdIns 3,4,5-triphosphate).
REFERENCES
- 1.Meijer H. J., Munnik T. (2003) Annu. Rev. Plant Biol. 54, 265–306 [DOI] [PubMed] [Google Scholar]
- 2.Wang X., Devaiah S. P., Zhang W., Welti R. (2006) Prog. Lipid Res. 45, 250–278 [DOI] [PubMed] [Google Scholar]
- 3.Michel V., Yuan Z. F., Ramsubir S., Bakovic M. (2006) Exp. Biol. Med. 231, 490–504 [DOI] [PubMed] [Google Scholar]
- 4.Rontein D., Nishida I., Tashiro G., Yoshioka K., Wu W. I., Voelker D. R., Basset G., Hanson A. D. (2001) J. Biol. Chem. 276, 35523–35529 [DOI] [PubMed] [Google Scholar]
- 5.Hanson A. D., Rhodes D. (1983) Plant Physiol. 71, 692–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith D. D., Summers P. S., Weretilnyk E. A. (2000) Physiol. Plant. 108, 286–294 [Google Scholar]
- 7.Kent C. (1995) Annu. Rev. Biochem. 64, 315–343 [DOI] [PubMed] [Google Scholar]
- 8.Witola W. H., El Bissati K., Pessi G., Xie C., Roepe P. D., Mamoun C. B. (2008) J. Biol. Chem. 283, 27636–27643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lorenzin D., Webb C., Summers P. S., Weretilnyk E. A. (2001) Can. J. Bot. 79, 897–904 [Google Scholar]
- 10.Hitz W. D., Rhodes D., Hanson A. D. (1981) Plant Physiol. 68, 814–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mou Z., Wang X., Fu Z., Dai Y., Han C., Ouyang J., Bao F., Hu Y., Li J. (2002) Plant Cell 14, 2031–2043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanaka K., Tolbert N. E., Gohlke A. F. (1966) Plant Physiol. 41, 307–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tasseva G., Richard L., Zachowski A. (2004) FEBS Lett. 566, 115–120 [DOI] [PubMed] [Google Scholar]
- 14.Summers P. S., Weretilnyk E. A. (1993) Plant Physiol. 103, 1269–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McDonnell E., Jones R. G. (1988) J. Exp. Bot. 39, 421–430 [Google Scholar]
- 16.Wyn Jones R. G., Storey R. (1981) in The Physiology and Biochemistry of Drought Resistance in Plants (Paleg L. G., Aspinall D. eds) pp. 172–204, Academic Press, Sydney [Google Scholar]
- 17.Rhodes D., Hanson A. D. (1993) Annu. Rev. Plant Phys. 44, 357–384 [Google Scholar]
- 18.Ashraf M., Foolad M. R. (2007) Environ. Exp. Bot. 59, 206–216 [Google Scholar]
- 19.Allard F., Houde M., Krol M., Ivanov A., Huner N. P., Sarhan F. (1998) Plant Cell Physiol. 39, 1194–1202 [Google Scholar]
- 20.Wyn Jones R. G., Storey R. (1978) Aust. J. Plant Physiol. 5, 817–829 [Google Scholar]
- 21.Yang W. J., Nadolska-Orczyk A., Wood K. V., Hahn D. T., Rich P. J., Wood A. J., Saneoka H., Premachandra G. S., Bonham C. C., Rhodes J. C. (1995) Plant Physiol. 107, 621–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen T. H., Murata N. (2008) Trends Plant Sci. 13, 499–505 [DOI] [PubMed] [Google Scholar]
- 23.Tasseva G., de Virville J. D., Cantrel C., Moreau F., Zachowski A. (2004) Plant Physiol. Biochem. 42, 811–822 [DOI] [PubMed] [Google Scholar]
- 24.Droppa M., Horvath G., Hideg E., Farkas T. (1995) Photosynth. Res. 46, 287–293 [DOI] [PubMed] [Google Scholar]
- 25.Horvath I., Vigh L., Belea A., Farkas T. (1979) Physiol. Plantarum. 45, 57–62 [Google Scholar]
- 26.Park J., Gu Y., Lee Y., Yang Z., Lee Y. (2004) Plant Physiol. 134, 129–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Z., Cuin T. A., Zhou M., Twomey A., Naidu B. P., Shabala S. (2007) J. Exp. Bot. 58, 4245–4255 [DOI] [PubMed] [Google Scholar]
- 28.Charron J. B., Breton G., Danyluk J., Muzac I., Ibrahim R. K., Sarhan F. (2002) Plant Physiol. 129, 363–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tabuchi T., Kawaguchi Y., Azuma T., Nanmori T., Yasuda T. (2005) Plant Cell Physiol. 46, 505–513 [DOI] [PubMed] [Google Scholar]
- 30.McNeil S. D., Nuccio M. L., Rhodes D., Shachar-Hill Y., Hanson A. D. (2000) Plant Physiol. 123, 371–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inatsugi R., Nakamura M., Nishida I. (2002) Plant Cell Physiol. 43, 1342–1350 [DOI] [PubMed] [Google Scholar]
- 32.Weretilnyk E. A., Alexander K. J., Drebenstedt M., Snider J. D., Summers P. S., Moffatt B. A. (2001) Plant Physiol. 125, 856–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nuccio M. L., Ziemak M. J., Henry S. A., Weretilnyk E. A., Hanson A. D. (2000) J. Biol. Chem. 275, 14095–14101 [DOI] [PubMed] [Google Scholar]
- 34.Bolognese C. P., McGraw P. (2000) Plant Physiol. 124, 1800–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu S., Yu Z., Wang F., Li W., Ye C., Li J., Tang J., Ding J., Zhao J., Wang B. (2007) Mol. Biotechnol. 36, 102–112 [DOI] [PubMed] [Google Scholar]
- 36.Hoagland D. R., Arnon D. I. (1941) Soil Science 51, 431–444 [Google Scholar]
- 37.Berkowitz O., Jost R., Pollmann S., Masle J. (2008) Plant Cell 20, 3430–3447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jost R., Berkowitz O., Masle J. (2007) BioTechniques 43, 206–211 [DOI] [PubMed] [Google Scholar]
- 39.Potocký M., Eliás M., Profotová B., Novotná Z., Valentová O., Zarský V. (2003) Planta 217, 122–130 [DOI] [PubMed] [Google Scholar]
- 40.Henderson P. J. F. (1993) in Enzyme assays - A practical approach (Eisenthal R., Danson M. J. eds) pp. 276–316, Oxford University Press, Oxford [Google Scholar]
- 41.Dowler S., Kular G., Alessi D. R. (2002) Sci. STKE 2002, PL6. [DOI] [PubMed] [Google Scholar]
- 42.Arabidopsis Genome Initiative (2000) Nature 408, 796–815 [DOI] [PubMed] [Google Scholar]
- 43.Segel I. H. (1975) Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-state Enzyme Systems, John Wiley & Sons, New York [Google Scholar]
- 44.Cruz-Ramírez A., López-Bucio J., Ramírez-Pimentel G., Zurita-Silva A., Sánchez-Calderon L., Ramírez-Chávez E., González-Ortega E., Herrera-Estrella L. (2004) Plant Cell 16, 2020–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Witola W. H., Ben Mamoun C. (2007) Eukaryot. Cell 6, 1618–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moffatt B. A., Weretilnyk E. A. (2001) Physiol. Plantarum. 113, 435–442 [Google Scholar]
- 47.Brendza K. M., Haakenson W., Cahoon R. E., Hicks L. M., Palavalli L. H., Chiapelli B. J., McLaird M., McCarter J. P., Williams D. J., Hresko M. C., Jez J. M. (2007) Biochem. J. 404, 439–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Palavalli L. H., Brendza K. M., Haakenson W., Cahoon R. E., McLaird M., Hicks L. M., McCarter J. P., Williams D. J., Hresko M. C., Jez J. M. (2006) Biochemistry 45, 6056–6065 [DOI] [PubMed] [Google Scholar]
- 49.Pessi G., Kociubinski G., Mamoun C. B. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 6206–6211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hayden C. A., Jorgensen R. A. (2007) BMC Biol. 5, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tabuchi T., Okada T., Azuma T., Nanmori T., Yasuda T. (2006) Biosci. Biotechnol. Biochem. 70, 2330–2334 [DOI] [PubMed] [Google Scholar]
- 52.Sawicki A., Willows R. D. (2007) Biochem. J. 406, 469–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li M., Welti R., Wang X. (2006) Plant Physiol. 142, 750–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Katagiri T., Takahashi S., Shinozaki K. (2001) Plant J. 26, 595–605 [DOI] [PubMed] [Google Scholar]
- 55.Bargmann B. O., Laxalt A. M., ter Riet B., van Schooten B., Merquiol E., Testerink C., Haring M. A., Bartels D., Munnik T. (2009) Plant Cell Physiol. 50, 78–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qin C., Wang C., Wang X. (2002) J. Biol. Chem. 277, 49685–49690 [DOI] [PubMed] [Google Scholar]
- 57.Perera I. Y., Davis A. J., Galanopoulou D., Im Y. J., Boss W. F. (2005) FEBS Lett. 579, 3427–3432 [DOI] [PubMed] [Google Scholar]
- 58.Meijer H. J., Berrie C. P., Iurisci C., Divecha N., Musgrave A., Munnik T. (2001) Biochem. J. 360, 491–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thole J. M., Nielsen E. (2008) Curr. Opin. Plant Biol. 11, 620–631 [DOI] [PubMed] [Google Scholar]
- 60.Corvera S., D'Arrigo A., Stenmark H. (1999) Curr. Opin. Cell Biol. 11, 460–465 [DOI] [PubMed] [Google Scholar]
- 61.Witola W. H., Pessi G., El Bissati K., Reynolds J. M., Mamoun C. B. (2006) J. Biol. Chem. 281, 21305–21311 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.