Abstract
KillerRed is the only known fluorescent protein that demonstrates notable phototoxicity, exceeding that of the other green and red fluorescent proteins by at least 1,000-fold. KillerRed could serve as an instrument to inactivate target proteins or to kill cell populations in photodynamic therapy. However, the nature of KillerRed phototoxicity has remained unclear, impeding the development of more phototoxic variants. Here we present the results of a high resolution crystallographic study of KillerRed in the active fluorescent and in the photobleached non-fluorescent states. A unique and striking feature of the structure is a water-filled channel reaching the chromophore area from the end cap of the β-barrel that is probably one of the key structural features responsible for phototoxicity. A study of the structure-function relationship of KillerRed, supported by structure-based, site-directed mutagenesis, has also revealed the key residues most likely responsible for the phototoxic effect. In particular, Glu68 and Ser119, located adjacent to the chromophore, have been assigned as the primary trigger of the reaction chain.
Introduction
The green fluorescent protein (GFP)2 and related proteins have become efficient noninvasive tools in cell biology and biomedicine for visualizing and monitoring the internal processes within cells or whole organisms (1–8). The multicolor labeling technologies, based on fluorescent proteins (FPs), have found important biomedical applications in the studies of various aspects of cancer, including primary tumor growth, tumor cell motility and invasion, metastatic seeding, colonization, and angiogenesis (9–11).
The recent development of the first genetically encoded photosensitizer, KillerRed (SWISS-PROT/TrEMBL data base sequence ID Q2TCH5), a highly phototoxic red fluorescent protein (12, 13), opened a new area of FP application. KillerRed is a red fluorescent protein characterized by excitation and emission maxima at 585 and 610 nm, respectively. This genetic variant was engineered from non-fluorescent and non-phototoxic chromoprotein anm2CP from Hydrozoa jellyfish (sequence ID Q6RYS4). Upon irradiation by green light at the wavelength of 520–590 nm, KillerRed generates the reactive oxygen species (ROS), accompanied by profound self photobleaching. The ROS-induced phototoxicity of KillerRed is at least 3 orders of magnitude higher than that of other fluorescent proteins exhibiting low background phototoxicity (12). Such a unique property of KillerRed could find use in light-induced inactivation of target proteins and in precise cell killing. Unlike chemical photosensitizers, KillerRed can be directly expressed by a target cell, both individually and in fusion with a target protein. The most exciting future application of KillerRed may be in photodynamic therapy of cancer. This phototoxic agent, precisely delivered to solid tumors by a viral vector, could serve as an intrinsically generated photosensitizer, causing light-induced tumor destruction. Therefore, understanding the relationship between the structure and properties of KillerRed would be crucially important for further improvement of this unique protein.
We present here the three-dimensional structure of KillerRed in an active fluorescent state at 1.75 Å resolution as well as in photobleached non-fluorescent states at 2.15 Å resolution. We have focused our attention on the distinctive structural features that might be responsible for the observed photophysical properties of the protein. The presented crystallographic data are supported by structure-based, site-directed mutagenesis.
EXPERIMENTAL PROCEDURES
Expression, Purification, and Crystallization
The plasmid pQE30-KillerRed was transformed into Escherichia coli JM109 (DE3) cells. The protein was expressed in 3 liters of LB supplemented with 100 mg/liter ampicillin by overnight incubation at 37 °C. No induction by isopropyl 1-thio-β-d-galactopyranoside was applied, because promoter leakage was sufficient for effective expression. Cells were resuspended in 20 mm Tris, pH 8.0, 100 mm NaCl and lysed by sonication. Supernatant clarified by centrifugation was applied to a metal affinity column (Ni2+-nitrilotriacetic acid-agarose resin, Qiagen) and washed with 10 column volumes of 20 mm Tris, pH 8.0, 100 mm NaCl. The target protein was eluted with 20 mm Tris, pH 8.0, 100 mm NaCl, 50 mm EDTA. Final purification was achieved by size exclusion chromatography on a Superdex 200 HiLoad (16/60) column (Amersham Biosciences) in 20 mm Tris, pH 7.5, 200 mm NaCl, 5 mm EDTA. Protein concentration and buffer exchange were performed in 10,000 molecular weight cut-off concentration units (VivaScience).
Crystals of KillerRed were grown by the hanging drop vapor diffusion method at 20 °C. Each drop consisted of 1 μl of the protein solution at a concentration of 11 mg/ml in 10 mm Tris, pH 7.5, 100 mm NaCl, 2.5 mm EDTA mixed with an equal amount of reservoir solution: 2 m (NH4)2SO4, 100 mm cacodylate, pH 6.5, 200 mm NaCl. The crystals reached their final size in 1 week.
Data Collection, Structure Solution, and Crystallographic Refinement
Diffraction data were collected from both the native (red-colored) and photobleached (colorless) crystals of KillerRed. To prepare photobleached crystals, a hanging drop with crystallized protein was irradiated with 5 milliwatts of 532-nm laser light for 5 h. Each data set was collected from a single crystal flash-cooled in a 100 K nitrogen stream. Prior to cooling, the crystal was transferred to a cryoprotective solution containing 20% glycerol and 80% reservoir solution. Data were collected at a wavelength of 1 Å with a MAR300 CCD detector at the SER-CAT beamline 22ID (Advanced Photon Source, Argonne National Laboratory, Argonne, IL) and were processed with HKL2000 (14).
The crystal structure of KillerRed was solved by the molecular replacement method with MOLREP (15, 16), using the coordinates of the GFP (Aequorea victoria) monomer (25% sequence identity, Protein Data Bank code 1GFL (17)) as a search model. Structural refinement of both the native and photobleached proteins was performed with REFMAC5 (18), alternating with manual revision of the model using COOT (19). Water molecules were located with ARP/wARP (20). Structure validation was performed with PROCHECK (21) and revealed that in the final model, all of the non-Gly/Pro residues resided in allowed regions on the Ramachandran conformational map. Crystallographic data and refinement statistics are presented in Table 1.
TABLE 1.
Data collection and refinement statistics
| KillerRed (native) | KillerRed (photobleached) | |
|---|---|---|
| Protein Data Bank code | 3GB3 | 3GL4 |
| Data collection | ||
| Space group | P212121 | P212121 |
| Cell dimensions: a, b, c (Å) | 72.0, 73.7, 78.7 | 72.2, 73.8, 76.2 |
| Resolution (Å) | 20.0-1.75 (1.83-1.75)a> | 30.0-2.15 (2.23-2.15) |
| Rmerge | 0.042 (0.546) | 0.052 (0.561) |
| I/σI | 35.5 (2.9) | 29.5 (2.5) |
| Completeness (%) | 92.9 (76.3) | 97.3 (96.2) |
| CompletenessRedundancy | 6.4 (5.5) | 6.0 (5.5) |
| Refinement | ||
| Resolution (Å) | 20.0-1.75 | 30.0-2.15 |
| No. of reflections | 38,123 | 21,447 |
| Rwork/Rfree | 0.186/0.244 | 0.229/0.309 |
| No. of atoms | ||
| Protein | 3,624 | 3,624 |
| SO42− | 20 | |
| Water | 246 | 128 |
| B-factors (Å2) | ||
| Protein | 35.0 | 61.1 |
| Chromophore | 33.5 | 54.8 |
| SO4 | 73.1 | |
| Water | 43.7 | 59.8 |
| r.m.s. deviations | ||
| Bond lengths (Å) | 0.017 | 0.015 |
| Bond angles (degrees) | 1.81 | 1.77 |
a Values in parentheses are for the highest resolution shell.
Mutagenesis and Photophysical Characterization
Preparation of mutant variants by site-directed mutagenesis was performed by PCR using the overlap extension method with primers containing appropriate target substitutions (22). Mutants were cloned into a pQE30 vector. Small scale expression was performed in E. coli XL1 Blue strain (Invitrogen) grown at 37 °C on Petri dishes with LB agar (100 mg/ml ampicillin), with no isopropyl 1-thio-β-d-galactopyranoside induction. After cell sonication, mutant proteins were purified using TALON metal affinity resin (Clontech). Absorption and excitation-emission spectra of the purified proteins were recorded with a Varian Cary 100 UV-visible spectrophotometer and a Varian Cary Eclipse fluorescence spectrophotometer, respectively.
To compare the phototoxicity of KillerRed variants, we used the following bacterial cell killing test. E. coli (XL1-Blue strain) was transformed with pQE30 vector encoding parental KillerRed or its mutants; plates were incubated at 37 °C overnight and then for 3 days at 4 °C to assure complete maturation of the mutant proteins. Fluorescence brightness and absorption efficiency of the colonies taken for the phototoxicity test were measured using fluorescence stereomicroscope SZX-12 (Olympus) equipped with a CCD camera and spectrophotometer SMS 2 VIS (Pannhoff Optische Messtechnik), enabling recording of emission and absorption spectra for individual colonies. Protein-expressing cells were obtained from a single E. coli colony, diluted into 200 μl of phosphate-buffered saline buffer, and divided into two equal portions. One of them was irradiated with white light (about 1 watt/cm2, light source Fiber-Light from Dolan-Jenner Industries, Inc.) for 1 h; the other was kept in the dark. Subsequently, both sample aliquots were plated to Petri dishes at different dilutions. The number of growing colonies corresponded to the number of bacterial cells surviving after irradiation (i.e. colony-forming units). The colony-forming unit number for the irradiated E. coli portion was compared with the non-irradiated one, thus allowing estimation of the relative phototoxic effect for every protein tested.
RESULTS
Monomer Structure
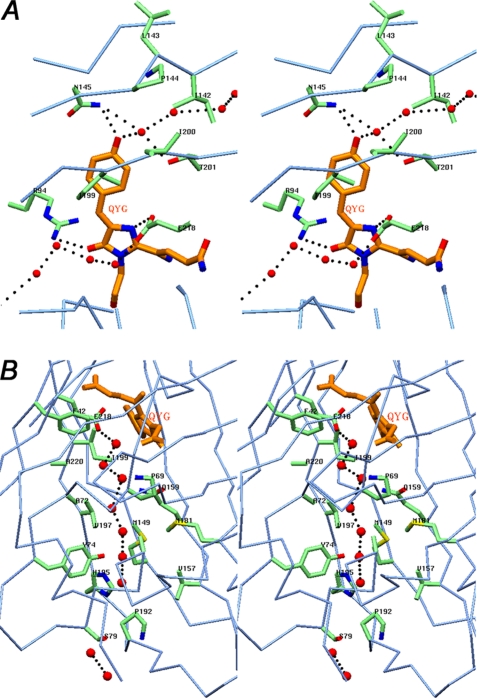
The principal structural fold of KillerRed, shared with all members of the GFP family, is an 11-stranded β-barrel having loop caps from both sides and a chromophore (matured from the sequence Gln65-Tyr66-Gly67) embedded in the middle of an internal α-helix located on the β-barrel axis. The C-terminal tail 224–233, having an irregular conformation, goes away from the β-barrel side surface. Two cis peptide bonds, preceding Pro37 and Pro87, are located at the β-barrel cap loops. The side chains of 11 residues adopt two alternative orientations. The KillerRed structure shows the existence of a pore at the cylindrical β-barrel surface, formed by the backbone of Ile142, Leu143, Pro144, Ile199, Ile200, and Thr201 (Fig. 1A). A chain of three hydrogen-bonded water molecules passes through this pore from the outside to the hydroxyphenyl moiety of the chromophore. The presence of an analogous pore has been reported for several non-phototoxic fluorescent proteins, such as TurboGFP (variant of ppluGFP2, Pontellina plumata) (23); wild type zGFP506, zYFP538, and zRFP574 (Zoanthus) (24, 25); and mKate (variant of eqFP578, Entacmaea quadricolor) (26). Based on mutagenesis experiments, the pore was suggested to be essential for the chromophore maturation rate by providing additional access for molecular oxygen (23).
FIGURE 1.
The environment of the chromophore in KillerRed. A, stereoview of a chain of hydrogen-bonded waters (red spheres in the upper part of the drawing) flowing from the outside to the phenolic ring hydroxyl of the QYG chromophore (shown in orange) through a pore located on the cylindrical β-barrel surface composed of the backbone of residues 142–144 and 199–201. A fraction of the other water chain (lower part of the drawing; see the legend for B) and the key residues Asn145 and Thr201 and the catalytic Arg94 and Glu218, interacting with chromophore, are also shown. B, stereoview of the water-filled channel with a chain of seven consecutive hydrogen-bonded water molecules (red spheres) traversing along the β-barrel from Pro192 at the cap loop to the catalytic Glu218 in the chromophore area. Two other water molecules are shown at the assumed entry point outside of the β-barrel (figure created with SETOR (55)).
A striking feature of the structure of the phototoxic KillerRed is the presence of a long water-filled channel extending along the β-barrel axis (Fig. 1B). The continuous chain of hydrogen-bonded waters in the channel covers the distance from Pro192 at the flanking cap to the catalytic Glu218 in the chromophore area. The stereochemical composition of the channel perfectly defines the route of the water stream. Each water molecule makes hydrogen bonds with the preceding and following waters and with the proximal amino acids lining the channel. This feature is unique to KillerRed and was not observed in the structures of other FPs. No evident entry point for these waters was found in the β-barrel cap. Only the enhanced thermal motion of the extreme loop segment 184–190, exposed to bulk solvent and with the atomic B factors 10–15 Å2 higher than the mean value, might indirectly indicate a possibility of leakage in this area. It should be noted that another red fluorescent protein, DsRed, despite possessing the same chromophore (27, 28), does not show any evidence of phototoxicity (12). An important difference between KillerRed and DsRed (Protein Data Bank code 1GGX) is that, in the latter protein, a water network originating from the end cap does not reach the chromophore. The side chain of the proximal Lys residue at position 69, hydrogen-bonded to the Ser side chain at position 199, blocks the flow and protects the chromophore from direct water access. On the contrary, the respective Pro and Ile residues, present at these positions in KillerRed, leave the gate open.
Oligomer Structure
Remarkably, the tetrameric arrangement of the protein subunits, typical for all red fluorescent and non-fluorescent Anthozoa-derived GFP-like proteins, has not been observed in the crystal structure of KillerRed. Both in solution (12, 13) and in the crystalline state, KillerRed exists as a dimer of two monomers (A and B in Fig. 2A) related by a non-crystallographic 2-fold symmetry axis with side-to-side packing at ∼90°. The buried area between the monomers (∼1,700 Å2/monomer) is stabilized by dispersed van der Waals contacts, 10 hydrogen bonds (≤3.3 Å), and two symmetry-related strong salt bridges between the side chains of Glu99 (A/B) and Arg158 (B/A) (Fig. 2A). Two aromatic side chains of Phe162 (A/B) make a stabilizing stack (at ∼3.5 Å) shielded by the side chains of the nearest four His and two Leu residues at the interface center near the symmetry axis. The irregular C-terminal tail (residues 224–233) extends away from the β-barrel and sticks to the cylindrical surface of the interacting counterpart, contributing to interface stabilization. Removal of the observed salt bridges and stacking interactions, accompanied by deletion of a portion of the C-terminal tail, might favor the monomeric state of the protein, highly desirable for the chromophore-assisted, light-induced inactivation techniques.
FIGURE 2.
Oligomerization of KillerRed. A, stereoview of the interface between the A and B subunits in the KillerRed dimer that are related by a 2-fold non-crystallographic symmetry axis. B, KillerRed oligomeric assembly in the crystal lattice showing three consecutive dimers composed from the β-barrel monomeric units (figure created with SETOR (55)).
In the crystal, the dimers are packed at a 45° angle, forming an oligomeric macrohelix along the crystallographic 2-fold screw axis (Fig. 2B). The contact surface between the monomers from the adjacent dimers is less extensive (∼620 Å2/monomer) than within a dimer and is stabilized by a single salt bridge, three hydrogen bonds, and a few hydrophobic contacts.
Native Chromophore Structure
The internal chromophore in KillerRed exhibits the fluorescent properties in a red spectral region with maxima λex = 585 nm and λem = 610 nm. The high quality weighted 2mFo − DFc and mFo − DFc electron density maps (18) are consistent with the modeled structure of the chromophore, with no indication of any disorder in the relatively tightly packed chromophore environment. The atomic temperature factors of the chromophore are below the average for the protein (Table 1).
The post-translational modification of the chromophore-forming sequence Gln65-Tyr66-Gly67 results in a two-ring fluorophore structure typical for the family of FPs, consisting of a five-membered imidazolone heterocycle with a p-hydroxybenzylidene substituent (Fig. 3A). The phenolic ring of Tyr66 is in cis orientation relative to the Cα–N bond and is almost coplanar with the imidazolone ring (the torsion angles around the corresponding Cα–Cβ and Cβ–Cγ bonds are ∼10.5 and ∼−1.5°, respectively). The geometry at the Cα carbon of the first chromophore residue Gln65 in KillerRed is consistent with the Anthozoa-derived red and far red fluorescent proteins and involves an sp2 hybridized center accompanied by formation of an N-acylimine double bond N=Cα. The latter bond extends the chromophore-conjugated π-electronic system, apparently resulting in a bathochromic shift in spectra. In all reported structures of the red FPs, the peptide linkage between the chromophore and the preceding residue has a cis configuration (24, 27–31). Different geometrical restraint schemes were tested in the refinement of KillerRed in order to determine optimal geometry of the link bridging the first chromophore residue Gln65 and the preceding Ile64. Weak restraints in crystallographic refinement resulted in an Ile64-Gln65 peptide link with strongly linearized bond angle (O)C-N-Cα (∼165°), showing an inclination to trans configuration.
FIGURE 3.
Stereoviews of the QYG chromophore with a preceding Ile residue in the weighted omit mFo − DFc electron density map (18) (cut-off ρ = 3.0σ). A, active fluorescent state; B, photobleached non-fluorescent state after 5 h of green laser light irradiation at λ = 532 nm. The latter density indicates considerable damage to the Gln65 and Tyr66 side chains of the chromophore (figure created with PyMOL (56)).
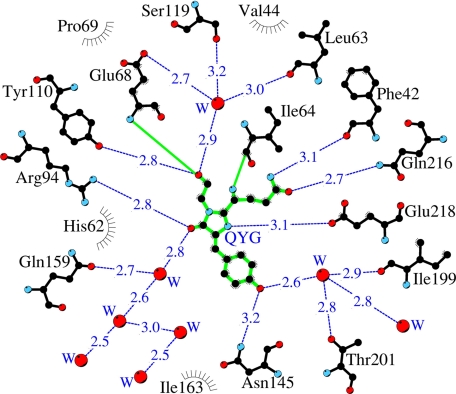
A shell close to the chromophore in the KillerRed structure is composed of 17 residues (within 3.9 Å), including the catalytic Arg94 and Glu218 (corresponding to Arg96 and Glu222 in GFP). Most of these residues are involved in an extensive network of hydrogen bonds formed by the side chain and backbone interactions and with active participation of mediating waters. Some of those waters are presumed to be reaction products of the chromophore maturation process. Several residues from the network, including Glu218 and Arg94, make direct and water-mediated hydrogen bonds with the chromophore (Fig. 4). This network is apparently functionally important, creating a potential proton wire in the maturation or degradation processes.
FIGURE 4.
A schematic diagram illustrating the environment of the chromophore in the KillerRed structure. Hydrogen bonds (≤3.3 Å) are shown as blue dashed lines, water molecules (W) are shown as red spheres, and van der Waals contacts (≤3.9 Å) are shown as black “eyelashes” (figure prepared with LIGPLOT/HBPLUS (57)).
Photobleached Chromophore Structure
The crystal structure of KillerRed, irradiated with green laser light at λ = 532 nm, presents the first example of a photobleached chromophore in the FP family. The resulting weighted omit mFo − DFc electron density reflects a mixture of the chromophore states at different stages of degradation (Fig. 3B), which is probably the main reason for the relatively large difference between the R and Rfree (Table 1). The most visible damage caused by 5 h of light exposure is to the side chains of Tyr66 and Gln65. On the other hand, the five-membered imidazolone ring and the protein backbone remain mostly intact. The weighted omit mFo − DFc map for the peptide linker preceding the chromophore (the common place for backbone fragmentation in FPs) as well as the proper geometry of the refined peptide fragment unambiguously confirm the integrity of the backbone (Fig. 3B).
Spectral Transitions of KillerRed in Solution
The long water channel and the pore extending from the protein surface to the chromophore in KillerRed as well as the high phototoxicity of this protein suggest that the chromophore might be accessible to external agents. Taking into account that photoreduction is characteristic for many photosensitizers, we tested, using fluorescence microscopy, several commonly used reductants on KillerRed immobilized on beads. The red fluorescence was found to be quenched by β-mercaptoethanol (βME), dithiothreitol, and dithionite (in concentrations of 10–100 mm), whereas the reduced glutathione and cysteine had no effect (at concentrations of up to 300–500 mm). The most prominent quenching was observed for βME, and this reaction was studied in detail. βME-induced KillerRed quenching was reversible, with red fluorescence largely recovering after washing out βME (Fig. 5A). The recovery of the red signal was fast and occurred with a half-life of about 3 min. Importantly, KillerRed photostability was found to be noticeably lower in the presence of βME (Fig. 5B), suggesting possible photoreduction. Measurement of KillerRed absorption spectra in the presence of different concentrations of βME showed a gradual decrease of the “red” peak at 585 nm and a corresponding increase of a 410 nm peak, with the βME EC50 (half-maximal effective concentration) of about 10 mm (Fig. 5C). Interestingly, βME-induced KillerRed spectral changes are similar to those observed during the initial stages of KillerRed photobleaching (Fig. 5D).
FIGURE 5.
Spectral transitions of KillerRed in solution. A, reduction of red fluorescence intensity of KillerRed immobilized on beads upon the addition of 100 mm βME and its recovery after βME has been washed out. B, normalized photobleaching curves for KillerRed immobilized on beads in the absence and presence of βME upon excitation by mercury arc lamp (540–580 nm, 0.2 watt/cm2). C, absorbance spectrum changes in the presence of βME at different concentrations (shown in mm above each curve). An inset shows the dependence of absorbance at 585 nm on βME concentration. D, absorbance spectrum changes during photobleaching with a green laser at 532 nm.
We also tested the influence of βME on other red fluorescent proteins and chromoproteins of different origin and oligomeric states. These FPs included the tetrameric DsRed2 (32), zFP574 (33), and asFP595 (34); dimeric TurboRFP (35); and monomeric TagRFP (35) and mCherry (36). None of these proteins was sensitive to βME in concentrations of up to 250 mm. The KillerRed progenitor chromoprotein anm2CP showed some sensitivity to βME; however, the effect was much lower compared with KillerRed. Even in 250 mm βME, anm2CP saved more than 60% of red absorption peak intensity. Thus, KillerRed sensitivity to βME appears to be an exceptional case among GFP-like proteins that correlates well with its high phototoxicity.
Structure-based, Site-directed Mutagenesis
The identity of residues potentially important for KillerRed phototoxic properties has been tested by site-directed mutagenesis. In the KillerRed structure, the most critical site in the vicinity of the chromophore is occupied by Glu68. Its negatively charged side chain resides in a mostly hydrophobic pocket formed (within 3.5–4 Å) by the side chains of five aromatic residues (Phe14, Phe42, Phe70, Phe71, and the hydrophobic part of Tyr110), by Ile16, and by Ser119 (Fig. 6). The negative charge of Glu68 is partially compensated by hydrogen bonds with two water molecules, mediating interactions with the carbonyl of the chromophore Gly67 and the side chain of Ser119. The latter residue adopts two alternative orientations and is involved in a hydrogen bond network that interacts with the chromophore. A buried negative charge in an essentially hydrophobic shell apparently gives rise to local energy instability, making Glu68 potentially highly reactive.
FIGURE 6.
The key residues adjacent to the chromophore. Glu68 is hydrogen-bonded via two waters to Ser119, in a mostly hydrophobic pocket. The side chain of Ser119 shows two alternative orientations (created with SETOR (55)).
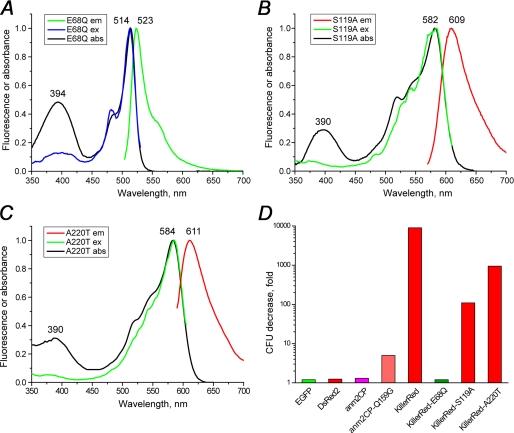
Based on the structural observations, the roles of both Glu68 and Ser119 were studied by site-directed mutagenesis. The replacement E68Q resulted in complete inhibition of red chromophore formation. The KillerRed-E68Q mutant exhibited absorption maxima at 394 and 514 nm (protonated and deprotonated GFP-like chromophore) and a weak green fluorescence with λem = 523 nm (Fig. 7A). In contrast, the substitution S119A had only a negligible effect on the spectral properties; absorption, excitation, and emission spectra, overall brightness, maturation efficiency, and photostability of KillerRed-S119A were all very similar to those of the unmodified KillerRed (Fig. 7B).
FIGURE 7.
Absorption, excitation, and emission spectra of the mutants of KillerRed. A, KillerRed-E68Q; B, KillerRed-S119A; C, KillerRed-A220T. D, decrease of viable bacterial counts (in colony-forming units (CFU)) after white light illumination of E. coli expressing the designated fluorescent proteins.
To check the influence of these substitutions on the phototoxic properties, we compared the phototoxicity of KillerRed and its mutants in bacterial cells. The E. coli cells expressing these proteins were irradiated with intense white light, and their survival was compared with a corresponding control sample kept in the dark. These tests showed that KillerRed-E68Q is practically non-phototoxic, whereas KillerRed-S119A possesses about 100-fold reduced ability to kill bacterial cells compared with KillerRed (Fig. 7D).
A total of 20 amino acids differ between the fluorescent, highly phototoxic KillerRed and its non-fluorescent, non-phototoxic progenitor, chromoprotein anm2CP (Figs. 8 and 9A). Most of these differences are found at the periphery of the β-barrel. Three of these substitutions, N145T, T201I, and I199V, reside in the immediate vicinity of the chromophore phenolic ring (Fig. 4). The nature of Thr at position 201 in KillerRed is distinguished from those of Ile and Leu in the respective anm2CP progenitor and DsRed, characterized by the chromophore-forming sequence identical to that in KillerRed (Fig. 9B). The first two replacements are the most essential, because they provide a significant contribution to stabilization of the fluorescent cis form of the chromophore by hydrogen-bonding the hydroxyl of Tyr66 to the side chains of Asn145 and via a water molecule to Thr201. Mutagenesis experiments demonstrated that, among several important substitutions, the combination of Asn145 and Ala161, replacing Thr145 and Cys161 in anm2CP, was mandatory for the KillerRed phototoxic effect (12). Our structure-based analysis has shown that the side chain of Cys161 in one of the optimal orientations is too close to the key residue Asn145 (d < 2.2 Å), which apparently would prevent the latter from optimal hydrogen bonding to the chromophore hydroxyl. The role of Ala161, having a smaller side chain, is expected to be the elimination of this negative effect.
FIGURE 8.
Structure-based sequence alignment of KillerRed, its wild-type progenitor anm2CP, DsRed, and avGFP. The numbering of residues in KillerRed and avGFP is given above and below the corresponding sequences. Buried residues are highlighted in gray. The chromophore-forming sequence is underlined. In red are shown residues residing in the nearest environment of the chromophore (≤3.9 Å); among them are the catalytic residues, Arg94 and Glu218 (highlighted in yellow), whereas the KillerRed/anm2CP differences are highlighted in blue.
FIGURE 9.
A comparison of KillerRed with other FPs. A, stereoview showing the distribution of the amino acid differences (see Fig. 8) between the phototoxic, fluorescent KillerRed (element C in green) and the non-toxic, non-fluorescent anm2CP (element C in orange). B, the neighborhood of the chromophore in the superimposed three-dimensional structures of KillerRed (element C in green) and DsRed (element C in orange) (created with SETOR (55)).
Analogously to non-fluorescent chromoproteins characterized by the trans state of the chromophore, the non-fluorescent progenitor anm2CP has also been proposed to have its chromophore in the trans state. Based on the KillerRed structure, that state has been modeled for anm2CP with 180° rotation of the Tyr66 side chain around the Cα–Cβ bond and by the introduction of all replacements corresponding to amino acid differences. The resulting model has demonstrated that this proposed non-fluorescent trans state is most likely stabilized by a hydrogen bond between the Tyr66 hydroxyl and the Gln159 side chain. This hydrogen bond is apparently energetically preferable to the assumed competing hydrogen bond with Thr145, designated to stabilize the cis form. DsRed has a Gly residue at position 159, which provides an opportunity for Ser145 to stabilize the cis form of the chromophore through a similar hydrogen bond with the hydroxyl of Tyr66. We suggest that a substitution of Gln159 by a Gly in anm2CP has to destabilize the non-fluorescent trans form of the chromophore and shift the equilibrium in favor of the fluorescent cis form, stabilized by a hydrogen bond with Thr145. Indeed, a new mutant variant anm2CP-Q159G has acquired evident red fluorescence (about 4-fold brighter compared with anm2CP) and noticeable phototoxicity (Fig. 7D). The same Q159G mutation in KillerRed has not resulted in any evident effect on its spectral properties and phototoxicity (data not shown), which most likely indicates the predominance of the cis chromophore form over the potential trans form.
Our results show that the residues at positions 145 and 159 compete in the ability to stabilize the cis and trans states of the chromophore. The choice between these two states depends significantly on the nature of the corresponding residues. Asn145 (supported by the water-mediated Thr201) in KillerRed and Ser145 in DsRed, both stabilizing the cis form, evidently have more influence than Gln and Gly at position 159, designated to stabilize the trans form. On the contrary, Gln159, presumably stabilizing the trans form in anm2CP, seemingly predominates over Thr145, designated to stabilize the cis form.
We have also carried out a number of mutagenesis experiments that were expected to modify the water channel extending along the β-barrel axis. In particular, we constructed mutants I78A, I78T, I78V, P192A, H195G, H195A, H195S, and H195N with the aim of increasing the access of external water to the channel. We also created mutants A72L, A82K, P69K/I199S, and A82K/Y74H/H195Y, with the aim of blocking the water flow in the channel. The barrier produced by the hydrogen bond between Lys69 and Ser199 was found to protect the chromophore from direct water access in DsRed. Unfortunately, all of these mutants exhibited very poor maturation rates, thus preventing direct comparison of their phototoxicity with that of the parental KillerRed. However, more encouraging results have been obtained for the variant A220T. The larger Thr220 apparently partially blocks the channel, resulting in about 10-fold decreased phototoxicity, while retaining the spectral properties and maturation efficiency close to those in KillerRed (Fig. 7, C and D).
DISCUSSION
The phototoxicity of the GFP-like proteins is usually very low, perhaps because of screening of the internal chromophore from water media by the protein shell. KillerRed is the only currently known fluorescent protein exhibiting prominent phototoxicity. The observed water-filled channel reaching the chromophore area from the end cap of the β-barrel is one of the important distinctive features of KillerRed responsible for its unique phototoxic properties (Fig. 1B). Indeed, increased access to the chromophore might facilitate its degradation and/or promote the accompanying toxic effect by conveying O2 and the photoinduced ROS species, respectively, inside and outside of the β-barrel. Alternatively, the water channel could be a “proton wire,” providing proton transfer that stabilizes a negatively charged chromophore radical in the course of primary electron transfer from an external electron donor (37, 38). The light-driven proton-pump function was recently proposed for avGFP (39). Notably, a similar water channel containing five ordered water molecules reaching from the protein surface to the heme was revealed in cytochrome f and showed to be crucial for its in vivo electron transfer function (40, 41). In some models, it was demonstrated that ordered water molecules can directly facilitate electron transfer, working as an “electron wire” (42, 43). Therefore, the highly ordered water molecules in KillerRed could possibly play a role of electron wire, conducting electrons from the attacked external molecule.
There is currently no direct evidence that the pore at the cylindrical surface of the β-barrel of KillerRed, with a string of water molecules reaching the chromophore hydroxyl, is critical for its toxic properties (Fig. 1A). A similar pore was observed in several non-phototoxic FPs and was shown to be essential for the rate of the chromophore maturation, presumably by providing additional access for molecular oxygen (23–26). Nevertheless, we believe that the supply of extra oxygen may provide additional support for the ROS generation.
We propose that Glu68 and Ser119 are among the key functional residues defining the unique features of KillerRed. The reactive role of a buried charge in a hydrophobic environment was previously demonstrated for the yellow fluorescent protein zYFP538 (Zoanthus sp.) (25). In addition, Asp68 in zYFP538 was shown to be one of the key reactive residues promoting the posttranslational modification of the chromophore beyond the green emitting form (44). The results of KillerRed mutagenesis showed that Glu68 is actively involved in posttranslational modification of the chromophore beyond the green emitting form and, in combination with Ser119, is the most probable photoinduced trigger for the reaction chain causing ROS generation and the observed phototoxic effect. Replacement of the hydrophilic Ser119 by the hydrophobic Ala may result in removal of water molecules connecting Glu68 with Ser119 and the chromophore from the hydrophobic pocket (Fig. 6). The proposed hydrophobic isolation of the negatively charged Glu68 from the network might play a critical role in the ROS-generating reaction process. The adjacent, potentially reactive catalytic Glu218 and Arg94 residues (45–48), both making direct hydrogen bonds with the chromophore, might contribute to the reaction chain triggered by Glu68.
It was demonstrated that non-fluorescent chromoproteins, including anm2CP, are characterized by a very short lifetime of the excited state (less than 1 ps), whereas the lifetime of the fluorescent proteins lies in the nanosecond time scale (49). A much longer lifetime of the excited cis state of the chromophore in KillerRed (by 3 orders of magnitude) most likely increases the probability of photodynamic reactions.
The proposed structural differences between the parental wild type anm2CP and its variant KillerRed are of primary importance for the evolution of the KillerRed properties. In contrast to the trans conformation of the chromophore that is most likely present in anm2CP, the cis conformation in KillerRed, stabilized by hydrogen bonds with Asn145 and Thr201, the latter water-mediated, brings the Tyr66 phenolic ring close to the key residues Glu68 and Ser119 and points this ring toward the water pore at the cylindrical β-barrel surface (Fig. 1A). The improved access of oxygen to the chromophore through the pore is expected to enhance the photodynamic response. In the Q159G variant of the non-toxic and non-fluorescent progenitor anm2CP, the proposed transition of the chromophore from the non-fluorescent trans to the fluorescent cis state resulted in the appearance of red fluorescence (data not shown) and toxicity (Fig. 7D). This remarkable effect is obviously due to creating a stereochemical environment of chromophore similar to that found in KillerRed.
Better access of water to the chromophore area through the long channel in KillerRed is another proposed key feature responsible for its phototoxicity. The replacement of Ala220 by the larger Thr interferes with the passage of water molecules through the channel, resulting in lower phototoxicity (Fig. 7D).
The biochemical data presented here demonstrate an exceptional accessibility of the KillerRed chromophore to external reductants. Spectral changes upon treatment with βME may be attributed to reduction of the acylimine group of the “red” chromophore, resulting in a “green” protonated state, which absorbs at ∼400 nm (Fig. 5C). The observed reversibility of the red fluorescence after washing out βME suggests that the chromophore environment in KillerRed ensures a dynamic equilibrium between the reduced and oxidized states of the chromophore. Although the molecular mechanism of the phototoxic effect of KillerRed remains largely unclear, we have shown here that this effect is accompanied by extensive chromophore degradation (Fig. 3B). Significant degradation was observed for the side chains of Gln65 and Tyr66 of the chromophore. Under most physiological conditions, the aromatic amino acids were reported to be among the primary targets for ROS-mediated oxidation (50). Surprisingly, the five-membered imidazolone ring of the chromophore remained relatively stable to ROS attacks, whereas a closely related imidazole ring of a histidine was shown to be susceptible to ROS-induced degradation (50). We hypothesize that the degradation of Gln65 and Tyr66 side chain is a side effect of the repeated cycles of photoreduction and the following oxidative restoration of the chromophore acylimine group during photodynamic reactions. Indeed, spectral changes during the early stages of the light-induced KillerRed bleaching most likely correspond to conversion of a “red” chromophore (absorption peak at 585 nm) into a protonated “green” chromophore (absorption peak at 400 nm) (Fig. 5D).
Generally, photosensitizers can act via photodynamic reactions of type I or type II (51–53). In type I reactions, a photoinduced electron transfer from an appropriate molecule to the photosensitizer occurs as a primary step, followed by interaction of the reduced photosensitizer with molecular oxygen and formation of the superoxide anion radical O2˙̄. Type II reactions perform direct energy transfer from an excited photosensitizer to oxygen, resulting in generation of singlet oxygen.
The combination of available data allows us to propose that KillerRed is, most likely, a type I photosensitizer. A recent study demonstrated that its phototoxicity for mammalian cells is lower in D2O compared with control samples in H2O (54). For type II photosensitizer, we would expect an opposite result, because the corresponding ROS agent, singlet oxygen, exhibits an extended lifetime in D2O. The decreased phototoxicity of KillerRed in D2O also indicates that proton transfer is an important step in the photodynamic reaction, supporting the proposed role of a proton wire in the long water channel. The spectral changes at the initial stages of KillerRed photobleaching (Fig. 5D) have been found to be similar to those observed for βME-induced KillerRed. This suggests that the primary step in photodynamic action of KillerRed is a photoinduced reduction of acylimine group of the red chromophore by an electron from an external moiety. Subsequently, the molecular oxygen may abstract this electron with formation of O2˙̄ and restoration of the original chromophore structure. This process may occur many times before resulting ultimately in chromophore degradation.
To summarize, we emphasize the key combination of the structural features as proposed primary prerequisites for the observed unique properties of KillerRed: (i) the water-filled channel and the pore that increase accessibility to the chromophore; (ii) the presence of Asn145 and Thr201 residues, stabilizing the cis form of the chromophore; and (iii) Glu68 and Ser119, adjacent to the chromophore, becoming the key reactive residues. We believe that structural and mutagenesis data presented here provide a solid base for a further interdisciplinary study of KillerRed and for development of more effective, genetically encoded photosensitizers, which could be used in a variety of applications.
A short publication describing the structure of KillerRed was published on-line when this paper was undergoing review (58). The main conclusions reached by its authors are in agreement with the results presented here.
Acknowledgment
We acknowledge the use of beamline 22-ID of the Southeast Regional Collaborative Access Team (SER-CAT), located at the Advanced Photon Source, Argonne National Laboratory.
This work was supported, in whole or in part, by the National Institutes of Health Intramural Research Program, NCI, Center for Cancer Research and by National Institutes of Health, NCI, Contract HHSN2612008000001E. This project was also supported in part by Russian Foundation for Basic Research Grants 07-04-00054 and 08-04-01702-a; by Russian Federal Agency for Science and Innovations Grant 02.513.12.3013; by the Molecular and Cell Biology Program RAS; and by grants from the President of the Russian Federation (MD-2780.2009.4). Use of the Advanced Photon Source was supported by the United States Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract W-31-109-Eng-38.
The atomic coordinates and structure factors (codes 3GB3 and 3GL4) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- GFP
- green fluorescent protein
- ROS
- reactive oxygen species
- βME
- β-mercaptoethanol.
REFERENCES
- 1.Verkhusha V. V., Lukyanov K. A. (2004) Nat. Biotechnol. 22, 289–296 [DOI] [PubMed] [Google Scholar]
- 2.Chudakov D. M., Lukyanov S., Lukyanov K. A. (2005) Trends Biotechnol. 23, 605–613 [DOI] [PubMed] [Google Scholar]
- 3.Zubova N. N., Savitsky A. P. (2005) Usp. Biol. Khim. 45, 1–66 [Google Scholar]
- 4.Seitz G., Warmann S. W., Fuchs J., Mau-Holzmann U. A., Ruck P., Heitmann H., Hoffman R. M., Mahrt J., Müller G. A., Wessels J. T. (2006) J. Pediatr. Surg. 41, 1369–1376 [DOI] [PubMed] [Google Scholar]
- 5.Remington S. J. (2006) Curr. Opin. Struct. Biol. 16, 714–721 [DOI] [PubMed] [Google Scholar]
- 6.Shcherbo D., Merzlyak E. M., Chepurnykh T. V., Fradkov A. F., Ermakova G. V., Solovieva E. A., Lukyanov K. A., Bogdanova E. A., Zaraisky A. G., Lukyanov S., Chudakov D. M. (2007) Nat. Methods 4, 741–746 [DOI] [PubMed] [Google Scholar]
- 7.Shaner N. C., Patterson G. H., Davidson M. W. (2007) J. Cell Sci. 120, 4247–4260 [DOI] [PubMed] [Google Scholar]
- 8.Wang Y., Shyy J. Y., Chien S. (2008) Annu. Rev. Biomed. Eng 10, 1–38 [DOI] [PubMed] [Google Scholar]
- 9.Hoffman R. M. (2005) J. Biomed. Opt. 10, 41202. [DOI] [PubMed] [Google Scholar]
- 10.Hoffman R. M. (2005) Curr. Top. Dev. Biol. 70, 121–144 [DOI] [PubMed] [Google Scholar]
- 11.Hoffman R. M. (2005) Nat. Rev. Cancer 5, 796–806 [DOI] [PubMed] [Google Scholar]
- 12.Bulina M. E., Chudakov D. M., Britanova O. V., Yanushevich Y. G., Staroverov D. B., Chepurnykh T. V., Merzlyak E. M., Shkrob M. A., Lukyanov S., Lukyanov K. A. (2006) Nat. Biotechnol. 24, 95–99 [DOI] [PubMed] [Google Scholar]
- 13.Bulina M. E., Lukyanov K. A., Britanova O. V., Onichtchouk D., Lukyanov S., Chudakov D. M. (2006) Nat. Protoc. 1, 947–953 [DOI] [PubMed] [Google Scholar]
- 14.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 15.Collaborative Computational Project 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 16.Vagin A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 17.Yang F., Moss L. G., Phillips G. N., Jr. (1996) Nat. Biotechnol. 14, 1246–1251 [DOI] [PubMed] [Google Scholar]
- 18.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 19.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 20.Perrakis A., Morris R., Lamzin V. S. (1999) Nat. Struct. Biol. 6, 458–463 [DOI] [PubMed] [Google Scholar]
- 21.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 22.Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. (1989) Gene 77, 51–59 [DOI] [PubMed] [Google Scholar]
- 23.Evdokimov A. G., Pokross M. E., Egorov N. S., Zaraisky A. G., Yampolsky I. V., Merzlyak E. M., Shkoporov A. N., Sander I., Lukyanov K. A., Chudakov D. M. (2006) EMBO Rep. 7, 1006–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pletneva N., Pletnev V., Tikhonova T., Pakhomov A. A., Popov V., Martynov V. I., Wlodawer A., Dauter Z., Pletnev S. (2007) Acta Crystallogr. D Biol. Crystallogr. 63, 1082–1093 [DOI] [PubMed] [Google Scholar]
- 25.Pletneva N. V., Pletnev S. V., Chudakov D. M., Tikhonova T. V., Popov V. O., Martynov V. I., Wlodawer A., Dauter Z., Pletnev V. Z. (2007) Bioorg. Khim. 33, 421–430 [DOI] [PubMed] [Google Scholar]
- 26.Pletnev S., Shcherbo D., Chudakov D. M., Pletneva N., Merzlyak E. M., Wlodawer A., Dauter Z., Pletnev V. (2008) J. Biol. Chem. 283, 28980–28987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yarbrough D., Wachter R. M., Kallio K., Matz M. V., Remington S. J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 462–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tubbs J. L., Tainer J. A., Getzoff E. D. (2005) Biochemistry 44, 9833–9840 [DOI] [PubMed] [Google Scholar]
- 29.Wall M. A., Socolich M., Ranganathan R. (2000) Nat. Struct. Biol. 7, 1133–1138 [DOI] [PubMed] [Google Scholar]
- 30.Petersen J., Wilmann P. G., Beddoe T., Oakley A. J., Devenish R. J., Prescott M., Rossjohn J. (2003) J. Biol. Chem. 278, 44626–44631 [DOI] [PubMed] [Google Scholar]
- 31.Wilmann P. G., Petersen J., Pettikiriarachchi A., Buckle A. M., Smith S. C., Olsen S., Perugini M. A., Devenish R. J., Prescott M., Rossjohn J. (2005) J. Mol. Biol. 349, 223–237 [DOI] [PubMed] [Google Scholar]
- 32.Yanushevich Y. G., Staroverov D. B., Savitsky A. P., Fradkov A. F., Gurskaya N. G., Bulina M. E., Lukyanov K. A., Lukyanov S. A. (2002) FEBS Lett. 511, 11–14 [DOI] [PubMed] [Google Scholar]
- 33.Labas Y. A., Gurskaya N. G., Yanushevich Y. G., Fradkov A. F., Lukyanov K. A., Lukyanov S. A., Matz M. V. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 4256–4261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lukyanov K. A., Fradkov A. F., Gurskaya N. G., Matz M. V., Labas Y. A., Savitsky A. P., Markelov M. L., Zaraisky A. G., Zhao X., Fang Y., Tan W., Lukyanov S. A. (2000) J. Biol. Chem. 275, 25879–25882 [DOI] [PubMed] [Google Scholar]
- 35.Merzlyak E. M., Goedhart J., Shcherbo D., Bulina M. E., Shcheglov A. S., Fradkov A. F., Gaintzeva A., Lukyanov K. A., Lukyanov S., Gadella T. W., Chudakov D. M. (2007) Nat. Methods 4, 555–557 [DOI] [PubMed] [Google Scholar]
- 36.Shaner N. C., Campbell R. E., Steinbach P. A., Giepmans B. N., Palmer A. E., Tsien R. Y. (2004) Nat. Biotechnol. 22, 1567–1572 [DOI] [PubMed] [Google Scholar]
- 37.Paschenko V. Z., Knox P. P., Chamorovsky S. K., Krasilnikov P. M., Mamedov M. D., Semenov A. Y., Zakharova N. I., Renger G., Rubin A. B. (2001) Bioelectrochemistry 53, 233–241 [DOI] [PubMed] [Google Scholar]
- 38.Bisby R. H., Parker A. W. (1998) Biochem. Biophys. Res. Commun. 244, 263–267 [DOI] [PubMed] [Google Scholar]
- 39.Agmon N. (2005) Biophys. J. 88, 2452–2461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ponamarev M. V., Cramer W. A. (1998) Biochemistry 37, 17199–17208 [DOI] [PubMed] [Google Scholar]
- 41.Sainz G., Carrell C. J., Ponamarev M. V., Soriano G. M., Cramer W. A., Smith J. L. (2000) Biochemistry 39, 9164–9173 [DOI] [PubMed] [Google Scholar]
- 42.Lin J., Balabin I. A., Beratan D. N. (2005) Science 310, 1311–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miyashita O., Okamura M. Y., Onuchic J. N. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 3558–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gurskaya N. G., Savitsky A. P., Yanushevich Y. G., Lukyanov S. A., Lukyanov K. A. (2001) BMC Biochem. 2, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barondeau D. P., Putnam C. D., Kassmann C. J., Tainer J. A., Getzoff E. D. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 12111–12116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sniegowski J. A., Lappe J. W., Patel H. N., Huffman H. A., Wachter R. M. (2005) J. Biol. Chem. 280, 26248–26255 [DOI] [PubMed] [Google Scholar]
- 47.Barondeau D. P., Kassmann C. J., Tainer J. A., Getzoff E. D. (2006) J. Am. Chem. Soc. 128, 4685–4693 [DOI] [PubMed] [Google Scholar]
- 48.Pouwels L. J., Zhang L., Chan N. H., Dorrestein P. C., Wachter R. M. (2008) Biochemistry 47, 10111–10122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schuttrigkeit T. A., von Feilitzsch T., Kompa C. K., Lukyanov K. A., Savitsky A. P., Voityuk A. A., Michel-Beyerle M. E. (2006) Chem. Phys. 323, 149–160 [Google Scholar]
- 50.Gilbert D. L., Colton C. A. (1999) Reactive Oxygen Species in Biological Systems: An Interdisciplinary Approach, 1st Ed., Springer-Verlag New York Inc., New York [Google Scholar]
- 51.Rosenthal I., Ben-Hur E. (1995) Int. J. Radiat. Biol. 67, 85–91 [DOI] [PubMed] [Google Scholar]
- 52.Krasnovskiı̆ A. A. (2004) Biofizika 49, 305–321 [PubMed] [Google Scholar]
- 53.Buytaert E., Dewaele M., Agostinis P. (2007) Biochim. Biophys. Acta 1776, 86–107 [DOI] [PubMed] [Google Scholar]
- 54.Serebrovskaya E. O., Edelweiss E. F., Stremovskiy O. A., Lukyanov K. A., Chudakov D. M., Deyev S. M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 9221–9225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Evans S. V. (1993) J. Mol. Graph. 11, 134–138 [DOI] [PubMed] [Google Scholar]
- 56.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, Palo Alto, CA [Google Scholar]
- 57.Wallace A. C., Laskowski R. A., Thornton J. M. (1995) Protein Eng. 8, 127–134 [DOI] [PubMed] [Google Scholar]
- 58.Carpentier P., Violot S., Blanchoin L, Bourgeois D. (2009) FEBS Lett. 583, 2839–2842 [DOI] [PubMed] [Google Scholar]