Abstract
A major limitation in biopharmaceutical development is selectively targeting drugs to diseased tissues. Growth factors and viruses have solved this problem by targeting tissue-specific cell-surface heparan sulfates. Neuregulin (NRG), a growth factor important in both nervous system development and cancer, has a unique heparin-binding domain (HBD) that targets to cell surfaces expressing its HER2/3/4 receptors (Esper, R. M., Pankonin, M. S., and Loeb, J. A. (2006) Brain Res. Rev. 51, 161–175). We have harnessed this natural targeting ability of NRG by fusing the HBD of NRG to soluble HER4. This fusion protein retains high affinity heparin binding to heparin and to cells that express heparan sulfates resulting in a more potent NRG antagonist. In vivo, it is targeted to peripheral nerve segments where it blocks the activity of NRG as a Schwann cell survival factor. The fusion protein also efficiently blocks autocrine and paracrine signaling and reduces the proliferation of MCF10CA1 breast cancer cells. These findings demonstrate the utility of the HBD of NRG in biopharmaceutical targeting and provide a new way to block HER signaling in cancer cells.
Introduction
Neuregulin (NRG)2 has been shown to have a wide array of functions with important roles in nervous system and heart development as well as in diseases ranging from breast cancer to schizophrenia (1–4). It mediates its diverse effects through binding homo- and heterodimeric cell-surface epidermal growth factor receptors, including HER2(erbB2), HER3(erbB3), and HER4(erbB4), that leads to rapid receptor-tyrosine phosphorylation and activation of downstream signaling pathways, including the mitogen-activated protein kinase and phosphatidylinositol 3-kinase (5). Many alternatively spliced forms of NRG are secreted after proteolytic cleavage from their transmembrane precursors (6). All of these forms have a receptor-binding epidermal growth factor-like domain together with a distinct C2 immunoglobulin-like domain that functions as a unique heparin-binding domain (HBD) with an alternating array of positively charged amino acids held together in a disulfide-bonded loop. This HBD binds specific sulfate groups on negatively charged heparan sulfate proteoglycans that leads to a highly specific tissue distribution of NRG (7–10).
Because of its strong mitogenic effect in some breast cancers, blocking NRG signaling has become an attractive therapeutic target (11). Although a currently approved humanized monoclonal antibody against HER2 called trastuzumab (12) has become a clinically effective adjuvant therapy for a subgroup of breast cancer patients (13, 14), it does not effectively block HER2 activation induced by NRG autocrine and paracrine signaling (15–17). A major obstacle in blocking NRG signaling both in cancer and in developmental studies comes from its ability to become highly concentrated in the extracellular matrix where it can produce sustained HER activation required for downstream gene activations (7, 15). To overcome this obstacle, we have fused the human the HBD of NRG to the soluble ectodomain of HER4 (H4) with high affinity for epidermal growth factor-like domain as a means to target this NRG antagonist to the same heparan sulfate-rich cell surfaces that bind NRG. In this way, the antagonist could effectively compete with NRG on an even footing and would enable more effective studies of the role of NRG in development and in cancer.
EXPERIMENTAL PROCEDURES
Construction of Fusion Proteins
All fusion proteins were derived entirely from human sequences. The extracellular domain of HER4 receptor (H4) corresponds to 99–2042 bp of the human HER4 NM_005235 mRNA. The sequence from 99–173 bp encodes a 25-amino acid signal sequence that was incorporated onto the N terminus of all constructs for secretion and protein expression. H4 was amplified by PCR and then inserted into pMH vector (Roche Applied Science, Indianapolis IN) between KpnI and EcoRI to generate the H4-HA construct. The HBD (532–849 bp) and HBD-S (532–1023 bp) domains of NRG β1 form (NM_013964) were amplified from the plasmid HARIA PATH2 (gift of Dr. Tejvir Khurana, University of Pennsylvania). Either HBD-S or the HBD domain alone was inserted into pMH-H4-HA in the KpnI site to generate HBD-S-H4-HA and HBD-H4-HA, respectively. H4-HBD-HA was subcloned by inserting the HBD domain into pMH-H4-HA between the EcoRI and BamHI sites. For HBD-S-H4 (GenBankTM accession number GQ912308) and H4 fusion proteins, the HA tag in both constructs was replaced by a His tag using PCR, and they were subcloned into pMH between the Ndel and BamHI sites.
Expression and Purification of Fusion Proteins
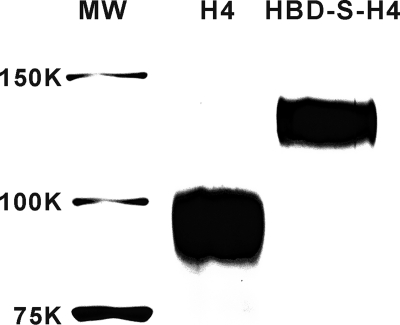
HEK293 cells were transfected with the four recombinant constructs using Lipofectamine 2000 according to the manufacturer (Invitrogen). The G418 Geneticin (Invitrogen) was then added at a concentration of 400 μg/ml to the HEK293 cells to select the positive transfected cells. Stable cell lines were selected following 3 weeks of G418 selection. Stably transfected HEK293 cells were diluted and plated in 96-well plates to yield single positive clones. The single clones that expressed the highest level of each fusion proteins (confirmed by Western blot using hemagglutinin antibody) were then maintained in culture media with 200 μg/ml Geneticin. To express and purify His-tagged HBD-S-H4 and H4 proteins, HEK293 freestyle suspension cell line was transfected with both constructs by 293fectin reagent (Invitrogen) following the manufacturer's instruction. Media from 7- to 8-day-old transfected cell culture was then collected by centrifugation, and fusion proteins were purified using a nickel column (Qiagen, Valencia, CA) for His-tagged proteins, followed by a heparin-Sepharose column (Sigma) for HBD-S-H4 or a second nickel column for H4. Purity was assessed by silver-stained gels (Fig. 2), and protein quantitation was determined using the Bradford Assay (Pierce).
FIGURE 2.
Silver-stained gel of purified HBD-S-H4 and H4 fusion proteins. The predicted molecular masses of HBD-S-H4 and H4 are about 88 and 70 kDa, respectively. The higher apparent molecular mass on the gel reflects that both fusion proteins are glycosylated during expression in mammalian cells.
Heparin Binding Assay
Opti-Mem I (Invitrogen) was incubated with transfected HEK293 cell lines for 2 days before the conditioned media containing each of four recombinant HA-tagged constructs were collected and passed through a heparin-Sepharose column (Sigma). The flow-through was collected, and the column was washed with PBS. The binding proteins were then eluted with increasing concentrations of NaCl (0.25, 0.3, 0.4, 0.5, 0.6, and 1 m). The flow-through and elution fractions were analyzed by Western blotting with an anti-HA antibody (Covance, Princeton, NJ) to determine the extent of heparin binding for each protein.
L6 Assay
Fusion proteins were pre-mixed with 75 pm NRG at room temperature for 30 min and then applied to L6 myotubes in 48-well plates for 45 min at 37 °C. In some cases, HBD-S-H4 or H4 was added to L6 myotubes first, incubated for 45 min, then washed with medium three times, and challenged with 75 pm NRG for 45 min. The erbB receptor phosphorylation (p185) assay was performed as described previously (18) using the phosphotyrosine antibody (pY, 4G10, Upstate Biotechnology, Billerica, MA). The blot was stripped and reprobed with erbB2 and erbB3 (Neomarkers, Fremont, CA) antibodies for the quantitation of total erbB proteins present in the lysate. Band intensity was quantified with Metamorph Image analysis software (Universal Imaging, Sunnyvale, CA) (15). Recombinant NRG protein, corresponding to amino acids 14–276, was purchased from R&D Systems.
Silver Staining
Purity of recombinant proteins was assessed by resolving 250 ng of HBD-S-H4 and H4 fusion proteins on a 7.5% reducing SDS-polyacrylamide gel followed by silver staining using the SilverSNAP Stain Kit (Pierce) following the manufacturer's instruction.
CHO Cell Binding Assay and Immunofluorescent Staining
HBD-S-H4 and H4 were conjugated with biotin by a biotin protein labeling kit (Solulink, San Diego, CA) following the manufacturer's instructions. 1 × 104 wild-type CHO or CHO-pgsD677 cells that lack heparan sulfate (ATTC#: CRL-2244) were plated in each well of a 96-well plate or chamber-slide and cultured for 3 days. Some cells were treated with 2 units/ml heparinase (Sigma) or media for 3 h and then incubated with 50 nm biotin-conjugated HBD-S-H4 or H4 at room temperature for 2 h. Cells were washed with PBS for three times. For the cell binding assay, streptavidin-horseradish peroxidase was added to live cells in a 96-well plate and incubated for 30 min. Signals were measured by addition of chemiluminescence reagents (PerkinElmer Life Sciences) and using a microplate luminometer. For staining, cells were fixed in 4% paraformaldehyde for 30 min, and streptavidin-horseradish peroxidase and tyramide-Alexa fluor 647 (Invitrogen) were used to visualize the signal.
Heparin Binding Assay
A heparin-binding plate (BD Bioscience) was coated with 150 ml/well of 100 mg/ml heparin (Sigma) overnight at room temperature (19). Wells were blocked with 0.2% gelatin/PBS for 1 h at 37 °C. Biotin-conjugated HBD-S-H4, H4, or IgG was added to each well at the concentration from 0 to 150 nm and incubated for 2 h at 37 °C. Wells were then washed three times with PBS and incubated with avidin-horseradish peroxidase (Sigma) for 30 min. Binding was detected by adding substrate p-nitrophenyl phosphate, and the absorbance was measured at 405 nm. The dissociation constant (Kd) of the saturation curve was calculated by a online Kd calculator (Invitrogen).
Chicken Eggs and in Ovo Treatment
Fertilized chicken eggs were obtained from Michigan State University Poultry Farms (East Lansing, MI) and incubated at 37 °C in a Kuhl rocking humidified incubator (Flemington, NJ) at 50% humidity. Chicken embryo experiments were performed with approval of Institutional Animal Care and Use Committee at Wayne State University. Either HBD-S-H4 or H4 proteins were dissolved in sterile saline containing 0.2% BSA to prepare 600 nm concentration in total volume of 200 μl of solution, and applied daily to embryos at E5 and E6 through a small hole on a choroallantoic membrane (20). Treatment with the identical volume of saline without recombinant proteins was used as a control. The eggs were sealed after each treatment with clear packing tape and embryos were sacrificed at E7 for immunostaining as described below.
Immunostaining
Chicken embryos were fixed with 4% paraformaldehyde overnight. After washing briefly with PBS, embryos were placed in 30% sucrose and cut transversely at 12 μm on a cryostat. Immunofluorescence was performed as described previously (9). Sections were incubated with antibodies to chicken NRG 183N (9) (1:100) or human NRG AD03 (10) (21) (1:300, Assay Designs Inc., Ann Arbor, MI) and Schwann cells 1E8 (1:10, Developmental Studies Hybridoma Bank, UIowa) or His6 tag (1:100, Abcam, Cambridge, MA) in blocking solution (10% normal goat serum, 0.5% Triton X-100 in PBS) for overnight at 4 °C, followed by incubation with goat anti-mouse or anti-rabbit Alexa fluor 546 (1:250, Invitrogen). For AD03 or His tag immunostaining, biotin-conjugated goat anti-rabbit (1:500,PerkinElmer Life Sciences) or horseradish peroxidase-conjugated goat anti-mouse (1:100, Invitrogen) was used as secondary antibody, and signal was detected using a tyramide signal amplification kit (Invitrogen) following the manufacturer's instructions. Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assays were performed with in situ cell death kit (Roche Applied Science) after the incubation of secondary antibodies for AD03 and 1E8 double labeling. Some sections were treated with 1 m NaCl/PBS at 37 °C for 90 min as described previously to disrupt ionic interactions (9) before the immunostaining procedure.
Quantitative Analysis
Digital images were obtained with a Nikon Eclipse 600 epifluorescence microscope with a Princeton Instruments Micromax 5-MHz cooled charge-coupled device camera. Metamorph Image analysis software (Universal Imaging) was used to quantify the percentage of apoptotic Schwann cells in the area of ventral nerve root at the lumbar level of spinal cord. Regions of interests were first defined using a Schwann cell marker staining for a non-biased selection of ventral nerve regions. These nerve segments were further divided in half at the midpoint between the spinal cord and the union of the motor and sensory axons. The total number of Schwann cells in each nerve segment was quantified by counting the number of full nuclei in each of these regions of interest using the nuclear dye 4′,6-diamidino-2-phenylindole. This was achieved by dividing total pixel area of signal thresholded for the nuclear signal by the average pixel area for an individual Schwann cell nucleus. Manual counts were used to validate this method. The number of TUNEL-positive Schwann cell nuclei were then counted in both proximal and distal nerve areas. 12–20 sections for each animal, and at least 5 animals for each condition were quantified. Statistical significance was defined as p < 0.01 using a two-tailed Student's t test.
Receptor Phosphorylation and Proliferation Assay of MCF10CA1 Cells
Cell proliferation and phosphorylation assays were performed as described previously (15). Briefly, HBD-S-H4 (0, 1, 5, and 10 nm) was applied to 3-day-old MCF10CA1 cells in culture with or without recombinant NRG. Phosphotyrosine Western blots were used as described above. 5000 MCF10CA1 cells were plated in each well of a 48-well plate for the proliferation assay. After the first 2 days in culture, cells were treated with 1 nm HBD-S-H4, H4, or trastuzumab (Gift from Wei-Zen Wei, Karmanos Cancer Institute). Then cell numbers were counted in quadruplicate wells by using a hemocytometer from the 3rd to 8th day.
RESULTS
Optimal Fusion of the NRG HBD to the HER4 Receptor
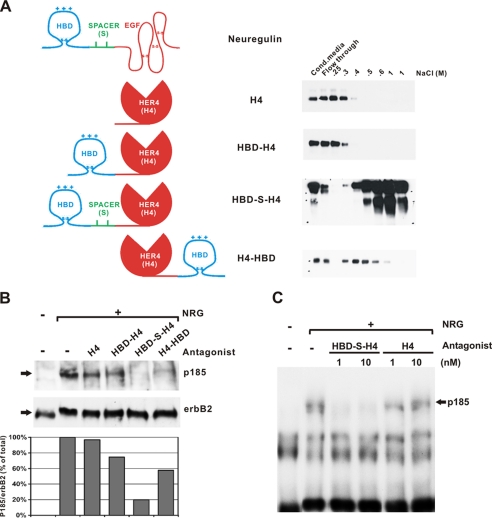
As a first step, fusion proteins were generated to determine the optimal arrangement of the HBD to retain high affinity heparin binding (Fig. 1A). The HBD was inserted either C-terminal (H4-HBD) or N-terminal (HBD-H4 and HBD-S-H4) to H4. The HBD-S-H4 contains the natural occurring glycosylated spacer domain (S) from NRG placed between the HBD and H4 domains (22). Heparin-Sepharose chromatography was used to compare the heparin-binding ability of each relative to the H4 protein alone by determining the salt concentration required to elute it from the column. Although the HBD-S-H4 and H4-HBD proteins bind the heparin column, the H4 and HBD-H4 constructs did not (Fig. 1A). Interestingly, the construct with the spacer, that most closely resembles native NRG (HBD-S-H4), had the highest heparin binding affinity requiring 1 m NaCl for elution. This may be due to the physical separation of the HBD from H4 that could either allow an optimal protein conformation necessary for heparin binding or prevent steric inhibition between the two domains. The ability of each construct to block NRG-induced receptor activation was determined based on its ability to block NRG-induced erbB receptor tyrosine phosphorylation (p185) in rat L6 cells (Fig. 1B). Consistent with the heparin binding results, antagonist potency paralleled the heparin-binding activities. Whereas H4 and HBD-H4 had little effect on reducing receptor phosphorylation, HBD-S-H4 and H4-HBD significantly inhibited erbB receptor activation with HBD-S-H4, reducing the p185 signal by 80%. These results demonstrate that the isolated HBD retains high affinity heparin-binding activity, even when fused to another protein. This results in an NRG antagonist with significantly improved potency.
FIGURE 1.
The HBD of NRG retains heparin-binding ability and potentiates an HER4 antagonist. A, schematic diagram of secreted NRG and four antagonist constructs. The heparin-binding domain (HBD) was fused N-terminal or C-terminal to the extracellular binding domain of HER4 receptor (H4) with or without a spacer domain (S). H4 alone was made as a control. Each was applied to a heparin column. The flow-through and successive elutions with increasing concentrations of NaCl were measured by immunoblotting with an anti-HA antibody. The HBD-S-H4 protein had the highest affinity for heparin. B, comparable amounts of each fusion protein were premixed with 50 pm NRG and applied to L6 muscle cells to determine the effect of each protein on the phosphorylation of erbB receptors (p185) normalized to erbB2 levels (bottom gel). Quantitation of phosphorylated erbB protein to total erbB2 protein reveals that HBD-S-H4 is the most potent antagonist to block NRG-induced erbB activation in L6 cells. C, L6 cells were treated with either purified HBD-S-H4 or H4 as a control at 1 and 10 nm for 1 h. The cells were washed to remove unbound fusion protein and then challenged with 75 pm NRG. Although H4 alone had no residual effects, HBD-S-H4 had sustained effects completely blocking NRG-induced erbB receptor phosphorylation. This experiment was repeated three times.
HBD-S-H4 Is Targeted to Cell Surface Heparan Sulfates
Based on the above results, highly purified HBD-S-H4 and native H4 His-tagged fusion proteins were produced in HEK293 cells for direct comparisons (Fig. 2). To demonstrate the enhanced blocking activity of HBD-S-H4 is due to its ability to concentrate on cell-surfaces expressing heparan sulfates, L6 cells were pre-treated with either HBD-S-H4 or H4 and then vigorously washed to remove any antagonist not bound to the cell surface. They were then challenged with soluble NRG as shown in Fig. 1C. Cells pre-treated with HBD-S-H4 were resistant to NRG activation, whereas no effect was observed with H4 treatment.
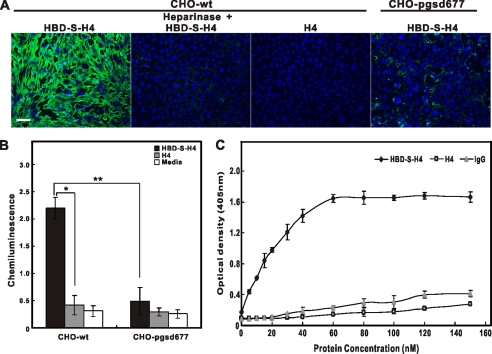
Using CHO cells that do and do not express heparan sulfates on their surfaces, we compared the binding of HBD-S-H4 onto wild-type CHO cells without and with heparinase treatment and to pgsd677 mutant CHO cells that lack the ability to synthesize heparan sulfate (23). While the HBD-S-H4 construct adhered to the wild type CHO cells, the H4 construct did not. Similarly, the absence of heparan sulfate in mutant cells or heparinase treatment of wild type cells significantly decreased HBD-S-H4 binding (Fig. 3A, B). Finally, a binding assay was used to determine the affinity of HBD-S-H4 to heparin. While both H4 and IgG had minimal binding, HBD-S-H4, bound to heparin-coated plates saturating at 60 nm and a calculated dissociation constant (Kd) of 14.7 nm (Fig. 3C). All these results demonstrate that the increased potency of HBD-S-H4 fusion is in part due to its ability to specifically interact with heparan sulfate and concentrate on cell surfaces where it exhibits sustained activity.
FIGURE 3.
NRG1's HBD targets HBD-S-H4 to cell surfaces through heparan sulfate interactions. A, CHO-wt and CHO-pgsd677 cells were incubated with biotinylated HBD-S-H4 or H4 (50 nm) with or without heparinase treatment. Compare with other conditions, HBD-S-H4-treated CHO-WT had the brightest green fluorescence intensity that appeared to concentrate in extracellular matrix between cells. Scale bar is 50 μm. B, a binding assay of protein constructs to CHO cells shows the enhanced binding ability of HBD-S-H4 to wild-type CHO cells (CHO-wt) but not to mutant cells with deficient heparan sulfate synthesis (CHO-pgsd677). The H4 construct did not bind to these cells. Error bars represent standard error of four experiments. * and ** indicate significant differences, and p values are 0.0006 and 0.0017, respectively, using a Student's t test comparing the indicated conditions. C, increasing concentrations of biotin-conjugated HBD-S-H4, H4, or IgG were added to heparin-coated 96-well plates to produce a saturation binding curve. The amount of HBD-S-H4 bound to heparin increased until saturation of binding sites. Data points are shown as mean ± S.E. of quadruplicate wells.
In Vivo Targeting to Peripheral Nerve Segments Induces Schwann Cell Apoptosis
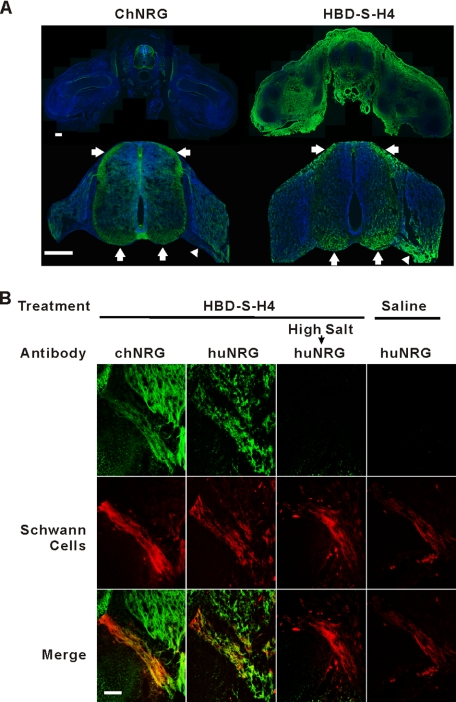
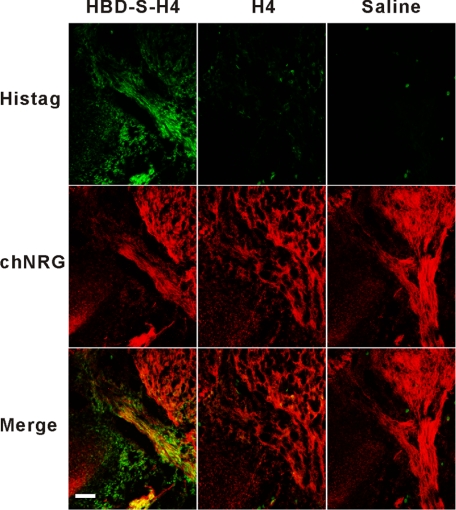
Within the developing spinal cord, NRGs are highly expressed in motor and sensory neurons and have critical functions in peripheral nerve development, including Schwann cell proliferation, survival, and myelination (9, 24, 25). To determine if HBD-S-H4 targets the same peripheral nerve regions as endogenous NRG, we compared the tissue distribution of exogenously added HBD-S-H4 in chicken embryos and compare this to H4 and saline treatment (Fig. 4A). HBD-S-H4 was localized on tissue sections of these embryos using an antibody (AD03) that only recognizes the human extracellular domain of NRG (huNRG) and does not cross-react with endogenous chicken NRG (chNRG) (21). Within the spinal cord, HBD-S-H4 became concentrated in the same region of the spinal cord and peripheral nerve as endogenous chNRG. The fusion protein also accumulated outside the nervous system in regions that do not normally express NRG. This distribution outside the nervous system is likely due to its adherence to developmentally expressed heparan sulfate proteoglycans. No huNRG immunoreactivity was present in the saline- or H4-treated animals. Double labeling the ventral nerve root shows that the same regions of peripheral nerve where chNRG was concentrated, bound high levels of HBD-S-H4 (Fig. 4B). Slight differences in patterns may reflect mild protein sequence differences between the human and chicken HBD. High salt treatment, which interrupts NRG-heparan sulfate proteoglycan ionic interactions, removed the HBD-S-H4 immunoreactivity (Fig. 4B). Immunostaining with a His tag antibody shows that only HBD-S-H4, but not H4, became concentrated in this nerve area (Fig. 5). These results demonstrate that systemically administered HBD-S-H4 is targeted to and concentrated on the same regions of the developing nervous system as endogenous NRG thereby making it a useful tool to study the role of matrix-bound NRG in peripheral nerve development.
FIGURE 4.
HBD-S-H4 targets to the same regions of the developing nervous system where NRG accumulates. A, 20 μg of HBD-S-H4 was added to the chorioallantoic membrane of embryonic chicken embryos. Tissue sections through the spinal cord 2 days later at E7 show a comparison between endogenous chicken NRG expression (green, left) and HBD-S-H4 distribution (green, right) at low power (top). Higher power images (bottom panel) focusing on the spinal cord show that HBD-S-H4 adhered to the same regions as endogenous NRG along axonal tracts in the spinal cord (arrows) and along the ventral root (arrowhead). Sections were counterstained for nuclei with 4′,6-diamidino-2-phenylindole (blue). Scale bars are 200 μm. B, both endogenous chicken NRG (chNRG, green) and HBD-S-H4 (huNRG, green) were concentrated in the ventral nerve root identified by a Schwann cell marker (red). High salt (1 m NaCl for 1.5 h) treatment on sections of HBD-S-H4 removed the signal in the ventral root. Control treatments with saline did not reveal any immunoreactivity with the same antibody in the ventral nerve root. Scale bar is 50 μm.
FIGURE 5.
Immunostaining with an antibody against His tag present in both HBD-S-H4 and H4 constructs. It shows that His tag immunoreactivity (His tag, green) was only present in the HBD-S-H4-treated embryo and concentrated in the same ventral nerve region as endogenous chicken NRG (chNRG, red). Scale bar is 50 μm.
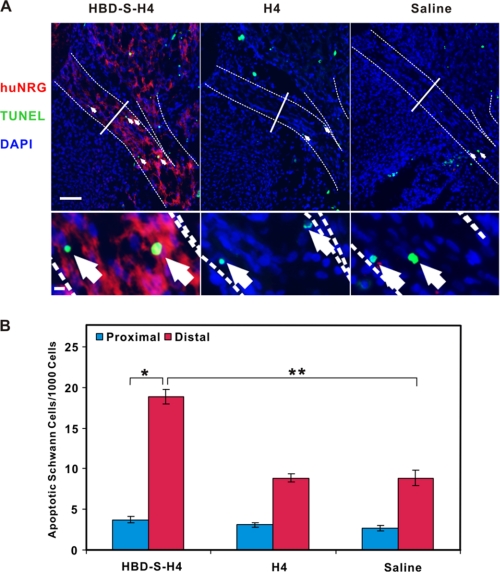
NRG signaling is critical for Schwann cell survival during early embryonic development (20, 25). Given its ability to concentrate in these developing nerves, the in vivo efficacy of HBD-S-H4 was determined by counting apoptotic Schwann cells in the ventral nerve root. HBD-S-H4 treatment significantly increased the number of apoptotic Schwann cells, while H4 had no effect (Fig. 6). Schwann cell apoptosis was maximal in nerve regions with maximal HBD-S-H4 staining, corresponding to regions rich in endogenous chNRG. Taken together, these findings demonstrate that HBD-S-H4 is specifically targeted in vivo to the same nerve segments as endogenous NRG where it effectively antagonizes NRG's actions as a Schwann cell survival factor.
FIGURE 6.
HBD-S-H4 induces Schwann cell apoptosis in the ventral nerve root. A, chicken embryos were treated with HBD-S-H4, H4, or saline from E5-E7. Increased numbers of TUNEL-positive Schwann cells (green, arrows) were seen in ventral root regions (highlighted with dashed lines) that were stained positively for HBD-S-H4 (red) with the human NRG antibody (huNRG). Each section was also stained with 4′,6-diamidino-2-phenylindole (DAPI) to show cell nuclei (blue). The ventral root was divided equally into proximal and distal regions (solid line) showing increased HBD-S-H4 accumulation in the distal versus proximal nerve segments. Scale bar is 50 μm. The bottom panel shows high power images of TUNEL-positive Schwann cells in each condition. Scale bar is 5 μm. B, quantitation of apoptotic Schwann cells shows significantly more apoptotic Schwann cells in the distal nerve root regions with higher levels of HBD-S-H4. Results are expressed as mean ± S.E. with n = 5, 7, and 6 for each, respectively. * and ** indicate significant differences and p values are 0.00009 and 0.00004, respectively.
Inhibition of HER Phosphorylation and Proliferation of Breast Cancer Cells
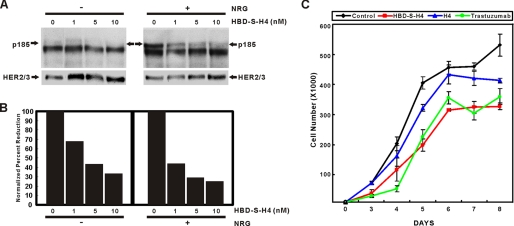
A potent NRG antagonist that targets the same cell surfaces as NRG has the potential to be an adjunctive treatment for human breast cancers that proliferate in response to HER2/3/4 activation by NRG (3, 26–28). We have found that as some human breast epithelial cells become more malignant; they develop autocrine NRG signaling that promotes proliferation (15, 29). We therefore compared the ability of HBD-S-H4 versus H4 to block HER phosphorylation and cell proliferation in MCF10CA1 breast cancer cells. HBD-S-H4 effectively blocked both autocrine and paracrine NRG-induced receptor phosphorylation in a dose-dependent manner (Fig. 7A). At the same concentration, HBD-S-H4 significantly reduced the proliferation of these cells compared with either no treatment or H4 alone (Fig. 7B). The level of growth inhibition achieved was comparable to the currently approved therapeutic antibody, trastuzumab. Further in vivo efficacy studies will be needed to determine the therapeutic potential of HBD-S-H4 in human breast cancers.
FIGURE 7.
HBD-S-H4 blocks the growth of malignant human breast cancer MCF10CA1 cells through disruption of autocrine and paracrine NRG signaling. A, increasing concentrations of HBD-S-H4 were added to MCF-10CA1 cells for 45 min without or with 75 pm recombinant NRG. The top panel shows that HBD-S-H4 blocked both autocrine (without added NRG) and paracrine (with NRG) HER2/3 receptor phosphorylation (p185). The lower panel shows the same Western blot reprobed with HER2 and HER3 antibodies. B, quantitation of the p185 signal normalized to HER2/3 levels is shown. The experiment was repeated three times. C, growth curves of MCF10CA1 cells were compared without or with HBD-S-H4, H4, or trastuzumab treatment. HBD-S-H4 (1 nm) markedly reduced the proliferation rate of these cancer cells compared with an equal molar amount of H4 and was comparable to treatment with trastuzumab.
DISCUSSION
These studies provide evidence that the HBD of NRG can retain heparan-binding specificity when fused to other polypeptides and demonstrate the efficacy of a novel and highly effective way to block NRG/HER signaling. The fusion protein we generated not only targets the same regions of the developing nervous system where NRG binds but also effectively blocks endogenous NRG activity leading to excessive Schwann cell apoptosis. This result affirms NRG's in vivo role as a Schwann cell survival factor and provides a new reagent to examine the role of matrix-bound forms of NRG in development and disease. This new reagent could have therapeutic potential, because it is derived entirely from natural human polypeptide sequences. It Its in vitro efficacy in breast cancer cells, suggests that this protein may also be useful therapeutically in breast and other cancer cells in vivo that proliferate in response to autocrine and paracrine HER signaling.
A key feature of the HBD of NRG that sets it apart from other growth factors and viral heparin-binding proteins, is its use of a unique disulfide-linked C2 immunoglobulin domain capable of maintaining a structure necessary for high affinity and tissue-specific heparin binding. When fused to HER4, a spacer peptide appears to be needed to keep the HBD away from the HER4 domain and maintain maximal biological activity of both domains. Fusion of the HBD of NRG to a soluble HER4 receptor not only converts it into a heparin-binding protein, but also dramatically potentiates its ability to block NRG signaling through cell-surface concentration. This concentration not only provides sustained antagonist activity, but also targets the antagonist to the same tissues where the agonist is expressed, thus placing it on a more equal footing.
One of the most important obstacles in the development of biological therapeutics is getting a therapeutic agent to diseased tissues without causing undue toxicity in normal tissues. The natural ability of the HBD of NRG to concentrate on cell surfaces in a tissue-specific manner may help overcome some of these obstacles for biopharmaceuticals that might otherwise fail due to poor efficacy and excessive toxicity. There is a growing literature that points to a tremendous diversity, yet specificity of glycosaminoglycan structures on different tissues critical for normal development and affected by human disease (30–32). These glycosaminoglycans maintain tissue-specific heterogeneity through highly regulated enzymes that modify the sugar and sulfation patterns needed for the selective targeting of a number of normal growth factors and viruses. Thus, not only could NRG's native HBD be a useful targeting vector for tissues that express and/or bind NRG, but modifications in this HBD sequence could modulate glycosaminoglycan and tissue-specific targeting specificity in many diseases, including neurodegenerative diseases and cancers.
Acknowledgment
We thank Dr. Fei Song for discussions and critical reading of the manuscript.
This work was supported by the National Multiple Sclerosis Society (Grant RG 3410B3) and the Michigan Universities Commercialization Initiative.
The nucleotide sequence(s) reported in this paper has been submitted to GenBankTM/EBI Data Bank with the accession number(s) GQ912308.
- NRG
- neuregulin
- HER
- human epidermal growth factor receptor
- HBD
- heparin-binding domain
- PBS
- phosphate-buffered saline
- HA
- hemagglutinin
- CHO
- Chinese hamster ovary
- TUNEL
- terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling
- huNRG
- human NRG
- chNRG
- chicken NRG.
REFERENCES
- 1.Esper R. M., Pankonin M. S., Loeb J. A. (2006) Brain Res. Rev. 51, 161–175 [DOI] [PubMed] [Google Scholar]
- 2.Buonanno A., Fischbach G. D. (2001) Curr. Opin. Neurobiol. 11, 287–296 [DOI] [PubMed] [Google Scholar]
- 3.Lupu R., Cardillo M., Cho C., Harris L., Hijazi M., Perez C., Rosenberg K., Yang D., Tang C. (1996) Breast Cancer Res. Treat 38, 57–66 [DOI] [PubMed] [Google Scholar]
- 4.Mei L., Xiong W. C. (2008) Nat. Rev. Neurosci. 9, 437–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yarden Y., Sliwkowski M. X. (2001) Nat. Rev. Mol. Cell Biol. 2, 127–137 [DOI] [PubMed] [Google Scholar]
- 6.Falls D. L. (2003) Exp. Cell Res. 284, 14–30 [DOI] [PubMed] [Google Scholar]
- 7.Li Q., Loeb J. A. (2001) J. Biol. Chem. 276, 38068–38075 [DOI] [PubMed] [Google Scholar]
- 8.Loeb J. A., Fischbach G. D. (1995) J. Cell Biol. 130, 127–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loeb J. A., Khurana T. S., Robbins J. T., Yee A. G., Fischbach G. D. (1999) Development 126, 781–791 [DOI] [PubMed] [Google Scholar]
- 10.Pankonin M. S., Gallagher J. T., Loeb J. A. (2005) J. Biol. Chem. 280, 383–388 [DOI] [PubMed] [Google Scholar]
- 11.Montero J. C., Rodríguez-Barrueco R., Ocaña A., Díaz-Rodríguez E., Esparís-Ogando A., Pandiella A. (2008) Clin Cancer Res. 14, 3237–3241 [DOI] [PubMed] [Google Scholar]
- 12.Carter P., Presta L., Gorman C. M., Ridgway J. B., Henner D., Wong W. L., Rowland A. M., Kotts C., Carver M. E., Shepard H. M. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 4285–4289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Slamon D. J., Leyland-Jones B., Shak S., Fuchs H., Paton V., Bajamonde A., Fleming T., Eiermann W., Wolter J., Pegram M., Baselga J., Norton L. (2001) N. Engl. J. Med. 344, 783–792 [DOI] [PubMed] [Google Scholar]
- 14.Vogel C. L., Cobleigh M. A., Tripathy D., Gutheil J. C., Harris L. N., Fehrenbacher L., Slamon D. J., Murphy M., Novotny W. F., Burchmore M., Shak S., Stewart S. J., Press M. (2002) J. Clin. Oncol. 20, 719–726 [DOI] [PubMed] [Google Scholar]
- 15.Li Q., Ahmed S., Loeb J. A. (2004) Cancer Res. 64, 7078–7085 [DOI] [PubMed] [Google Scholar]
- 16.Agus D. B., Akita R. W., Fox W. D., Lewis G. D., Higgins B., Pisacane P. I., Lofgren J. A., Tindell C., Evans D. P., Maiese K., Scher H. I., Sliwkowski M. X. (2002) Cancer Cell 2, 127–137 [DOI] [PubMed] [Google Scholar]
- 17.Cho H. S., Mason K., Ramyar K. X., Stanley A. M., Gabelli S. B., Denney D. W., Jr., Leahy D. J. (2003) Nature 421, 756–760 [DOI] [PubMed] [Google Scholar]
- 18.Esper R. M., Loeb J. A. (2004) J. Neurosci. 24, 6218–6227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahoney D. J., Whittle J. D., Milner C. M., Clark S. J., Mulloy B., Buttle D. J., Jones G. C., Day A. J., Short R. D. (2004) Anal. Biochem. 330, 123–129 [DOI] [PubMed] [Google Scholar]
- 20.Winseck A. K., Caldero J., Ciutat D., Prevette D., Scott S. A., Wang G., Esquerda J. E., Oppenheim R. W. (2002) J. Neurosci. 22, 4509–4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pankonin M. S., Sohi J., Kamholz J., Loeb J. A. (2009) Brain Res. 1258, 1–11 [DOI] [PubMed] [Google Scholar]
- 22.Fischbach G. D., Rosen K. M. (1997) Annu. Rev. Neurosci. 20, 429–458 [DOI] [PubMed] [Google Scholar]
- 23.Lidholt K., Weinke J. L., Kiser C. S., Lugemwa F. N., Bame K. J., Cheifetz S., Massagué J., Lindahl U., Esko J. D. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 2267–2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nave K. A., Salzer J. L. (2006) Curr. Opin. Neurobiol. 16, 492–500 [DOI] [PubMed] [Google Scholar]
- 25.Jessen K. R., Mirsky R. (2005) Nat. Rev. Neurosci. 6, 671–682 [DOI] [PubMed] [Google Scholar]
- 26.Tsai M. S., Shamon-Taylor L. A., Mehmi I., Tang C. K., Lupu R. (2003) Oncogene 22, 761–768 [DOI] [PubMed] [Google Scholar]
- 27.Atlas E., Cardillo M., Mehmi I., Zahedkargaran H., Tang C., Lupu R. (2003) Mol. Cancer Res. 1, 165–175 [PubMed] [Google Scholar]
- 28.Krane I. M., Leder P. (1996) Oncogene 12, 1781–1788 [PubMed] [Google Scholar]
- 29.Yao B., Rakhade S. N., Li Q., Ahmed S., Krauss R., Draghici S., Loeb J. A. (2004) BMC Bioinformatics 5, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gorsi B., Stringer S. E. (2007) Trends Cell Biol. 17, 173–177 [DOI] [PubMed] [Google Scholar]
- 31.Fuster M. M., Esko J. D. (2005) Nat. Rev. Cancer 5, 526–542 [DOI] [PubMed] [Google Scholar]
- 32.Bishop J. R., Schuksz M., Esko J. D. (2007) Nature 446, 1030–1037 [DOI] [PubMed] [Google Scholar]