Abstract
Lipolysis is an important metabolic pathway controlling energy homeostasis through degradation of triglycerides stored in lipid droplets and release of fatty acids. Lipid droplets of mammalian cells are coated with one or more members of the PAT protein family, which serve important functions in regulating lipolysis. In this study, we investigate the mechanisms by which PAT family members, perilipin A, adipose differentiation-related protein (ADFP), and LSDP5, control lipolysis catalyzed by hormone-sensitive lipase (HSL), a major lipase in adipocytes and several non-adipose cells. We applied fluorescence microscopic tools to analyze proteins in situ in cultured Chinese hamster ovary cells using fluorescence recovery after photobleaching and anisotropy Forster resonance energy transfer. Fluorescence recovery after photobleaching data show that ADFP and LSDP5 exchange between lipid droplet and cytoplasmic pools, whereas perilipin A does not. Differences in protein mobility do not correlate with PAT protein-mediated control of lipolysis catalyzed by HSL or endogenous lipases. Forster resonance energy transfer and co-immunoprecipitation experiments reveal that each of the three PAT proteins bind HSL through interaction of the lipase with amino acids within the highly conserved amino-terminal PAT-1 domain. ADFP and LSDP5 bind HSL under basal conditions, whereas phosphorylation of serine residues within three amino-terminal protein kinase A consensus sequences of perilipin A is required for HSL binding and maximal lipolysis. Finally, protein kinase A-mediated phosphorylation of HSL increases lipolysis in cells expressing ADFP or LSDP5; in contrast, phosphorylation of perilipin A exerts the major control over HSL-mediated lipolysis when perilipin is the main lipid droplet protein.
Introduction
Lipid droplets are cellular organelles, structurally similar to lipoprotein particles. Lipid droplets include a neutral lipid core composed largely of triglycerides, surrounded by a phospholipid monolayer and coated with surface proteins that provide an interface for various aspects of lipid metabolism, including lipid transport, lipogenesis, and lipolysis (1–5). Lipolysis is an important mechanism by which cells release energy stored in lipid droplets; its impairment has been linked to cellular lipotoxicity and insulin resistance (6). Studies are needed to gain an understanding of the underlying molecular mechanisms regulating lipolysis. Although all cells are equipped to perform lipolysis, the extent of lipid accumulation and specific components of the lipolytic pathway are variable, depending on the type of cell.
Numerous recent studies have led to consensus that members of the PAT family of proteins, originally named for Perilipin, Adipose differentiation-related protein (ADFP)4 and Tail Interacting Protein 47 (TIP47), play conserved structural and functional roles on lipid droplets (6–9). Proteomic studies have identified a “signature” composition for lipid droplets from a variety of types of cells that includes at least one PAT family member. In mammalian cells, the PAT family includes perilipin (peri), ADFP, and TIP47, as well as S3-12 and LSDP5 (also known as PAT-1, AMLPAT, MLDP, and OXPAT). Perilipin is the major PAT protein associated with the lipid droplet surface in mature adipocytes and is a major regulatory factor for lipolysis (10, 11). The roles of other PAT proteins in lipolysis are less well defined.
Perilipin facilitates triglyceride storage under basal (fed) conditions in adipocytes by reducing the access of endogenous lipases to stored lipids. Additionally, studies with perilipin null mice have shown that perilipin is required for maximal lipolysis in response to stimulation of the β-adrenergic receptor signaling pathway (10, 11). A key event is the phosphorylation of perilipin A by protein kinase A (PKA), which presumably alters the conformation of perilipin at the lipid droplet surface to facilitate lipolysis. Studies in cultured cells have begun to elucidate the complex mechanisms by which phosphorylated perilipin A promotes lipolysis (12–14). Perilipin is unique among PAT proteins in having multiple PKA consensus sites; to date, none of the other PAT proteins has been demonstrated to be phosphorylated by PKA.
Three lipases are responsible for complete lipolysis of triglycerides in adipocytes (15, 16). Adipose triglyceride lipase (ATGL) catalyzes the initial hydrolysis of triglycerides into diglycerides. Subsequently, hormone-sensitive lipase (HSL) acts as a diglyceride lipase (17, 18). Finally, monoglyceride lipase cleaves the third fatty acid to release glycerol. Although mechanisms regulating the association of ATGL with the lipid droplet surface remain to be elucidated, HSL binding to lipid droplets requires perilipin (19). For most other types of cells, cytosolic lipases have not been extensively characterized.
Significant perilipin expression is limited to adipose tissue and a few other types of cells; however, HSL expression is more widespread, including liver, muscle, and heart. These tissues lack perilipin under normal physiological conditions, suggesting that other PAT proteins likely control HSL-mediated lipolysis. A prior study has demonstrated that HSL binds to the surfaces of lipid droplets in skeletal muscle (20), a tissue that expresses ADFP, TIP47, and LSDP5. Other studies have shown that the amount of ATGL or HSL that binds to the surfaces of lipid droplets is controlled by the PAT protein composition of the lipid droplet (6, 9, 19). This raises the question of whether the association of HSL with lipid droplets is regulated by a common mechanism in adipose and non-adipose tissues. Moreover, although most PAT proteins play a role in shielding stored lipids from cytosolic lipases under basal conditions, they are not equally effective. Under these conditions, perilipin and LSDP5 exhibit equivalent strong repression of lipolysis, whereas ADFP and TIP47 are comparably less protective (7, 8, 19). The goal of this study is to examine the mechanisms by which PAT proteins regulate HSL-mediated lipolysis.
In this study, we have explored the hypothesis that lipolysis is regulated either by the distinct binding affinity of each PAT protein for the lipid droplet or by differences in binding interactions between HSL and the various PAT proteins. In the former case, lipase association with lipid droplets would be inversely proportional to the ease of displacement of a PAT protein from the lipid droplet surface. Alternatively, HSL may interact with amino acid sequences that are conserved between PAT family members.
We have investigated the binding affinity of PAT proteins for lipid droplets using fluorescence recovery after photobleaching (FRAP) to study the movement of PAT proteins within and into the phospholipid monolayer of lipid droplets. Additionally, we have examined interactions of HSL with perilipin A, ADFP, TIP47, and LSDP5 in adipose and non-adipose cells using the following three approaches: measurement of anisotropy Forster resonance energy transfer (AFRET) between fluorescently tagged HSL and PAT proteins; co-immunoprecipitation (co-IP) of HSL with PAT proteins; and assessment of lipolysis. The results show that perilipin A, ADFP, and LSDP5 diffuse rapidly within the phospholipid monolayer of an individual lipid droplet, yet only ADFP and LSDP5 exchange between lipid droplet and cytoplasmic pools. Therefore, displacement of PAT proteins from the surfaces of lipid droplets is not the major determinant that controls lipolysis. Localization of HSL to lipid droplets requires binding of HSL to highly conserved sequences within the amino termini (PAT-1 domains) of PAT proteins, whereas increased lipolysis requires both PAT protein binding and activation of HSL by PKA. Thus, our studies provide evidence that protein/protein interactions between HSL and PAT proteins regulate lipolysis in both adipose and non-adipose cells.
EXPERIMENTAL PROCEDURES
Antibodies
Rabbit anti-GFP antibody was from Clontech, and mouse anti-GFP antibody was from Covance (Princeton, NJ). Antibodies raised in rabbits against HSL, perilipin, ADFP, LSDP5, and CGI-58 were previously characterized (12, 21, 22). Rabbit anti-ATGL antibody was from Cell Signaling Technology (Danvers, MA). Horseradish peroxidase-conjugated goat anti-rabbit IgG and donkey anti-mouse IgG were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit serum was from Rockland Immunochemicals (Gilbertsville, PA). Mouse anti-rabbit IgG was from Jackson ImmunoResearch (West Grove, PA).
Cell Culture
CHO-K1 and CHO Flp-In cells were obtained from ATCC (Manassas, VA) and Invitrogen, respectively. Both types of cells were grown in Ham's F-12 medium supplemented with 10% fetal calf serum, 2 mmol/liter l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin at 37 °C with 5% CO2 and 95% humidity. Perilipin A overexpressing CHO-K1 cells, as well as CHO-K1 clonal cell lines overexpressing HSL-GFP and TIP47-GFP fusion proteins, have been previously characterized (7, 19).
The constructs peri-YFP, LSDP5-YFP, and ADFP-YFP were introduced into the CHO Flp-In cells according to the manufacturer's instructions (information for all constructs is provided in supplemental Table 1). Stably transfected CHO Flp-In cells were selected 24 h after transfection by adding hygromycin (500 μg/ml) to the growth medium. Wild-type CHO Flp-In cells were grown in the presence of Zeocin (100 μg/ml).
For immunofluorescence studies, CHO-K1 and CHO Flp-In cells were seeded in 35-mm dishes with glass bottoms (MatTek Corp., Ashland, MA) at a density of 2 × 105 cells. The following day, cells were transfected with 1 μg of DNA plasmid containing cDNA coding for the fusion protein of interest per well (co-transfections received 0.5 μg of each of two plasmids per well) using Lipofectamine Plus reagent (Invitrogen), according to the manufacturer's instructions. The following day, cells were incubated for 12 h in growth medium supplemented with 400 μm oleic acid complexed to fatty acid-free bovine serum albumin (FA-free BSA), as described previously (19). To measure lipolysis, cells were incubated with F-12 medium containing 1% FA-free BSA for 2 h. To stimulate PKA, cAMP levels in cells were elevated by the addition of 1 mm isobutylmethylxanthine (IBMX) and 10 μm forskolin. Re-esterification of fatty acids was prevented with addition of 5 μm triacsin C.
For immunoprecipitation studies, cells were plated at 2 × 105 cells per well in 6-well plates (Corning, Corning, NY). For lipolysis experiments, cells were plated at 0.5 × 10 5 cells per well in 24-well plates (Corning, Corning, NY).
Molecular Cloning
Cerulean and Venus mammalian expression vectors to drive expression of monomeric variants of fluorescent proteins, pECFP-C1, pEYFP-C1, and pEYFP-N1, were used to reduce intrinsic dimerization of fluorescent proteins and artifacts during Forster resonance energy transfer experiments (23). In addition, the enhanced CFP coding sequence has been mutated to minimize bleaching that occurs during two-photon laser scanning and to improve detection (23, 24). cDNAs of HSL, perilipin A, ADFP, LSDP5, TIP47, or CGI-58 were cloned in-frame with coding sequences for fluorescent peptides in pECFP-C1, pEYFP-C1, and pEYFP-N1. Names and sequences of subcloning oligonucleotide primers for all fluorescent fusion protein constructs are presented in supplemental Table 1. Perilipin A phosphorylation site point mutations were generated, as described previously (26), and were then subcloned into pEYFP-N1. The 11-mer repeat region from LSDP5 (exon 6) was amplified by PCR using the following primers: 5-LSDP5-E6-SacI, 5′-TAGAGCTCAAGTGGTGACATCAGCCAAGGATAC-3′, and 3-LSDP5-E6-SacI, 5′-TAGAGCTCTGCTAGCTCAGCCTCAGTCATGGGC-3′, and inserted into the SacI site of linearized pEYFP-c1 (Clontech). All constructs were verified by sequencing analysis.
Confocal Imaging and AFRET
Confocal imaging of live cells was performed at 37 °C and 5% CO2 using a Zeiss LSM510 microscope equipped with an S-M incubator (Carl Zeiss MicroImaging, Inc.), and controlled by a CTI temperature regulator along with humidification and an objective heater. Emitted light was passed through bandpass filters for collection of CFP (470–510 nm) and YFP (530–550 nm). For AFRET measurements, images were collected using a ×40, 1.3NA F-FLUAR oil immersion objective lens and vertically polarized 800 nm two-photon excitation. Detection was with photomultiplier tubes in the non-descanned configuration. Emitted fluorescence was filtered using either a HQ480/40m filter (Chroma, Rockingham, VT) for collection of CFP fluorescence or HQ535/30m-2p (Chroma, Rockingham, VT) for detection of YFP fluorescence. Separate photomultiplier tubes collected vertically and horizontally polarized fluorescence by passing the emitted light through a broadband polarizing beam splitter. Anisotropies were calculated, as described previously (27). To examine the movement of HSL to cellular lipid droplets, the subcellular region containing the largest area of lipid droplets was outlined, and average fluorescence intensity values of individual pixels in the enhanced CFP and enhanced YFP channels were calculated and normalized to surface area (LSM510 software).
Co-immunoprecipitations and Immunoblot Analysis
Antibodies used for immunoprecipitations were purified with the Melon gel IgG spin purification kit (Pierce). Rabbit serum or mouse anti-rabbit IgG was used as the negative control. Protein/protein interactions were investigated using extracts from cells co-transfected with the appropriate pair of fusion protein constructs. The list for all combinations of constructs is provided in supplemental Table 2. Cells were lysed by incubation in a buffer containing 150 mm NaCl, 1% Triton X-100, 60 mm n-octyl glucoside (Sigma), 10 mm Tris, pH 8.0, supplemented with protease inhibitors, for 15 min on ice, followed by extrusion through a syringe (28). Cell lysates were pre-cleared with 30 μl of protein A/G-agarose (Pierce) for 15 min on a rotator at room temperature and then centrifuged at 10,000 × g at 4 °C for 30 min. 250 μl of the pre-cleared supernatant was incubated with the precipitating antibodies, as noted in supplemental Table 2, at room temperature for 2 h on a rotator. 50 μl of protein A/G-agarose was then added, and suspensions were incubated at room temperature for 2 h on a rotator. Complexes were precipitated by centrifugation at 2300 × g at 4 °C for 5 min, washed three times with cold phosphate-buffered saline buffer, and finally mixed with 60 μl of Laemmli sample buffer (Bio-Rad) containing β-mercaptoethanol. 20 μl of the pre-cleared supernatant and 30 μl of the immunoprecipitated proteins were separated on 4–12% polyacrylamide NuPAGE gels (Invitrogen), transferred to nitrocellulose membranes, and probed with the specific antibodies indicated, followed by a corresponding horseradish peroxidase-conjugated secondary antibody. The immunoblot signals were detected with Supersignal chemiluminescence reagents (Pierce).
Adenovirus
To generate adenovirus for the expression of HSL-CFP, the HSL-CFP cDNA fragment was excised from HSL-CFP-N1 (see above) with BglII and NotI and was subsequently subcloned into a modified pAdTrack-CMV shuttle vector (29) lacking the GFP cassette. The PmeI-digested vector was used for transformation into AdEasy BJ5183 cells. Correct recombination of the resulting viral vector was confirmed by restriction enzyme digestions. Finally, the PacI-digested viral DNA was transfected into human embryonic kidney 293 cells for virus production and amplification (29). Adenovirus expressing untagged HSL was also generated (30).
Lipolysis Measurements
Lipolysis was assessed in the absence or presence of IBMX and forskolin according to Tansey et al. (12) and Sztalryd et al. (19). CHO-FlpIn cells stably expressing perilipin-YFP, LSDP5-YFP, and ADFP-YFP were seeded into a 24-well plate at a density of 0.5 × 105 cells per well. The following day, cells were infected with recombinant adenovirus for the expression of HSL-CFP-N1 (Ad-HSL-CN1) or green fluorescent protein (Ad-GFP). 24 h after adenovirus transduction, the medium was replaced with growth medium supplemented with 400 μm oleic acid plus 1 μCi/well [3H]oleic acid complexed to 0.4% FA-free BSA overnight to promote triacylglycerol deposition. Cells were then incubated in the absence of supplemental fatty acids with or without 1 mm IBMX and 10 μm forskolin, with 5 μm triacsin and 1% FA-free BSA for 2 or 3 h (as indicated in the figure legends). Radioactivity released into the culture medium was quantified. Efficiency of transduction with Ad-HSL-CN1 was determined by immunoblotting.
Assay for Triglyceride Hydrolase Activities of Cell Extracts
Cells were harvested by scraping with a rubber policeman, and homogenization was performed on ice in lysis buffer A (0.25 m sucrose, 1 mm EDTA, 1 mm dithiothreitol, 20 μg/ml leupeptin, 2 μg/ml antipain, 1 μg/ml pepstatin, pH 7.0). The substrate for the measurement of triglyceride hydrolase activity, containing triolein and [9,10-3H]triolein (PerkinElmer Life Sciences), was emulsified with phosphatidylcholine/phosphatidylinositol (3:1) using a probe sonicator (Virsonic 475, Virtis, Gardiner, NJ), as described previously (31). Cell lysates (0.1 ml) from cells expressing the constructs described in supplemental Table 1 were added to 0.1 ml of the substrate and then incubated in a water bath at 37 °C for 30 min. The reaction was terminated by adding 3.25 ml of methanol/chloroform/heptane (10:9:7) and 1 ml of 0.1 m potassium carbonate, 0.1 m boric acid, pH 10.5. After centrifugation at 800 × g for 20 min, the radioactivity in 0.5 ml of the upper phase was determined by liquid scintillation counting, as described previously (31).
Statistical Analysis
Statistical significance was tested using either one-way analysis of variance or a two-tailed Student's t test (GraphPad Software, Inc).
RESULTS
Lipolysis Is Not Controlled by PAT Protein Displacement from Lipid Droplets
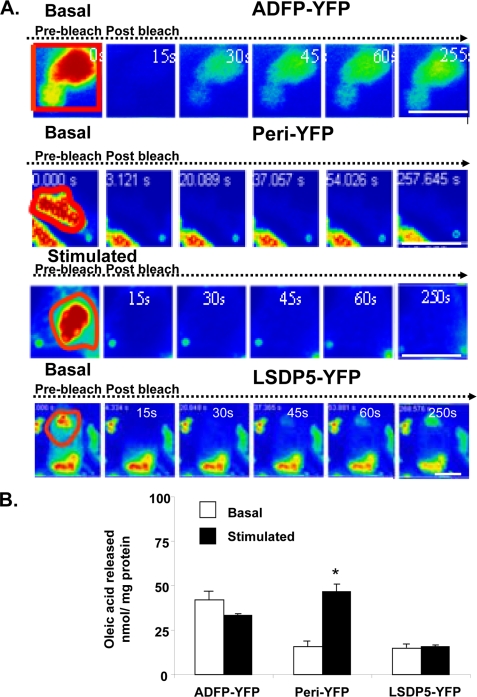
We tested the hypothesis that lipolysis is regulated by the displacement of PAT proteins from lipid droplets, thus allowing cytosolic lipases to gain access to stored triglyceride. ADFP-YFP, LSDP5-YFP, and perilipin-YFP fusion proteins were stably expressed in CHO-K1 cells, which lack HSL. ATGL is present in these cells (2), although the cytosolic lipases have not been fully characterized. Lipolysis by endogenous lipases was measured under both basal conditions and following addition of forskolin and IBMX to increase levels of cAMP and activate PKA. The expression of perilipin-YFP resulted in low basal (unstimulated) lipolysis, with increased triglyceride hydrolysis upon PKA activation (Fig. 1B). Thus, as reported previously, perilipin shields stored triglycerides from cytosolic lipases under basal conditions but becomes permissive to lipolysis upon phosphorylation by PKA. Comparable low basal release of fatty acids was observed when LSDP5-YFP was expressed; however, activation of PKA failed to increase lipolysis. Thus, LSDP5 protects against basal lipolysis as effectively as perilipin but remains protective when PKA is activated. Cells expressing ADFP-YFP showed elevated lipolysis under both basal and PKA-activated conditions; thus, ADFP is permissive to lipolysis. Because similar findings have been reported for untagged PAT proteins (8, 12, 33), addition of the YFP tag does not alter the function of PAT proteins in regulation of lipolysis.
FIGURE 1.
PAT protein association with lipid droplets and effects on lipolysis. A, FRAP analysis of ADFP-YFP, perilipin-YFP (under basal and stimulated conditions), and LDSP5-YFP in CHO-K1 cells following overnight incubation with 400 μm oleic acid. The cells were incubated with 5 μm triacsin C in the absence of supplemental fatty acids, and perilipin-YFP cells were stimulated for 15 min with 10 μm forskolin, 1 mm IBMX, and 5 μm triacsin C. Live cells were examined with a confocal laser microscope using a ×40 oil immersion objective. Red boxed regions were bleached at time 0, and fluorescence within these regions was monitored at 15-s intervals. Bar, 10 μm. B, wild-type or CHO-K1 cells stably expressing ADFP-YFP, perilipin-YFP, or LSDP5-YFP were incubated overnight with 400 μm [3H]oleic acid. Supplemental fatty acids were withdrawn, and cells were incubated with 5 μm triacsin C with (stimulated) or without (basal) 1 mm IBMX and 10 μm forskolin; the amount of [3H]oleic acid released into the medium was measured after 3 h. Data represent means ± S.E. (n = 8) (*, p < 0.05, for PKA-stimulated value compared with basal value).
We next used FRAP in live cells to measure the movement of YFP-tagged PAT proteins both laterally within the surface phospholipid monolayer and to the lipid droplet from another pool, presumably cytoplasm. For the latter, 100% of lipid droplet-associated fluorescence was photobleached, and recovery of fluorescence was measured over time. When perilipin-YFP on lipid droplets was 100% photobleached, fluorescence failed to recover over a period of more than 4 min, under both basal and PKA-stimulated conditions (Fig. 1A). Thus, perilipin A is not rapidly recycled between lipid droplets and another cellular pool. In contrast, ADFP and LSDP5 exhibit similar partial recovery from 100% photobleaching (40% for ADFP and 35% for LSDP5) over 250 s (Fig. 1A and supplemental Fig. 1). Thus, both ADFP and LSDP5 exchange between lipid droplets and the cytoplasm.
Next, 50% of the fluorescence associated with a single lipid droplet was photobleached, and recovery of fluorescence was measured over time. All three PAT proteins recovered equally well from 50% photobleaching (supplemental Fig. 2). Thus, these PAT proteins are not anchored rigidly at the lipid droplet surface but are capable of lateral movement within the phospholipid monolayer.
Differences in control of basal lipolysis were not correlated to the ease of exchange of PAT proteins between lipid droplets and the cytoplasm. ADFP is more permissive to lipolysis than LSDP5, yet LSDP5 and ADFP show equivalent exchange between lipid droplets and the cytoplasm. Furthermore, activation of PKA increased lipolysis in cells expressing perilipin A yet did not promote egress of perilipin from the lipid droplet. Therefore, we evaluated differences in protein/protein interactions between HSL and PAT proteins as an alternative mechanism to control lipolysis.
Intracellular Localization of HSL Is Dependent on the Type of PAT Protein Present at the Lipid Droplet Surface
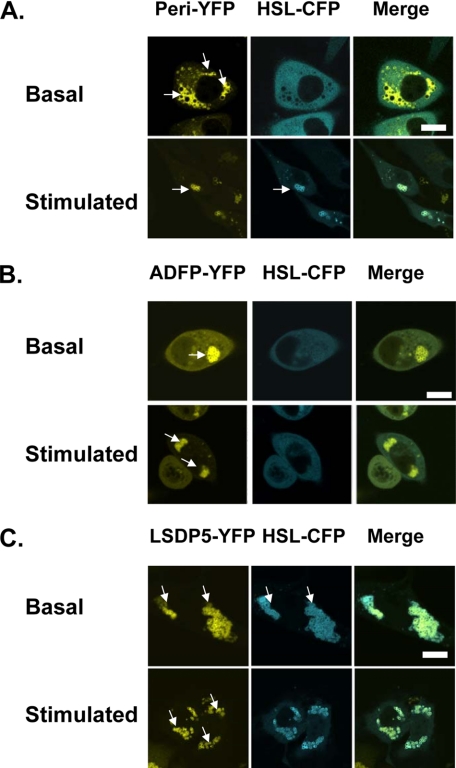
We used fluorescence microscopy to study localization of HSL and PAT proteins under basal and PKA-stimulated conditions. HSL-CFP was expressed in cells with stably expressed PAT-YFP fusion proteins. HSL-CFP was observed in the cytoplasm of cells expressing perilipin-YFP under basal conditions and on perilipin-coated lipid droplets upon activation of PKA (Fig. 2A), consistent with the localization of endogenous HSL in 3T3-L1 adipocytes (34). In contrast, HSL-CFP remains mainly in the cytoplasm of cells expressing ADFP-YFP, even after PKA stimulation (Fig. 2B). Unexpectedly, the majority of HSL-CFP fluorescence was observed on lipid droplets in cells expressing LSDP5-YFP, whether or not PKA was activated (Fig. 2C). Thus, localization of HSL to lipid droplets is dependent on the PAT protein present and, for perilipin, PKA stimulation. We next used AFRET and immunoprecipitations to assess binding interactions between HSL and PAT proteins.
FIGURE 2.
Recruitment of HSL to lipid droplets is dependent on the PAT protein composition of the droplet. Recruitment of HSL to perilipin-covered lipid droplets requires activation of PKA. CHO-K1 cells were co-transfected with HSL-CFP and a PAT fusion protein as follows: perilipin-YFP (A), ADFP-YFP (B), or LDSP-5-YFP (C). Cells were incubated overnight with 400 μm oleic acid. The following day, supplemental fatty acids were removed, and the cells were incubated with 5 μm triacsin C and no further additions (basal) or stimulated for 30 min with 10 μm forskolin, 1 mm IBMX, and 5 μm triacsin C (stimulated). Live cells were examined with a confocal microscope as in Fig. 1. Bar, 10 μm. Top, middle, and bottom rows show representative cells from 6 to 12 separate experiments.
HSL Interacts Directly with Most PAT Proteins
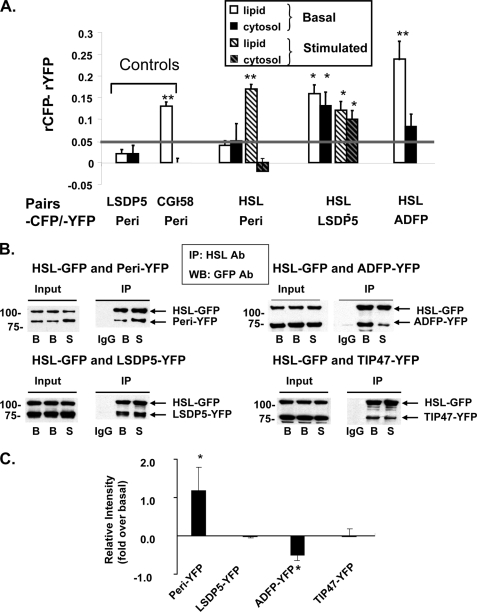
AFRET occurs when fluorescent probes are no more than 8–10 nm apart; consequently, it detects proteins that are in close proximity and likely interact. To validate our measurements, we selected two pairs of fusion proteins that target to lipid droplets for use as controls. LSDP5-CFP and perilipin-YFP (peri-YFP) were selected as negative controls; LSDP5 and perilipin are unlikely to interact because their tissue expression profiles are different (muscle and liver cells for LSDP5 and adipocytes for perilipin). CGI-58-CFP and perilipin-YFP were used as positive controls, because these proteins have been demonstrated previously to interact on lipid droplets (28, 35, 36). We observed that both pairs of proteins co-localize at the surfaces of lipid droplets (supplemental Fig. 3A), yet only CGI-58-CFP and perilipin-YFP display the expected interactions in AFRET and IP experiments (Fig. 3A and supplemental Fig. 3, B–D).
FIGURE 3.
HSL binds to PAT family proteins, as determined by in situ AFRET and co-immunoprecipitation (IP). A, images were collected by confocal microscopy of live cells expressing pairs of fluorescent fusion proteins shown along the x axis and calculations for AFRET were performed (29). The gray line indicates the threshold of significance. Data are means ± S.E. from 6 to 12 experiments. *, p < 0.05; **, p < 0.01. B, CHO-K1 cells expressing HSL-CFP were transfected with a PAT fusion protein as follows: perilipin-YFP, ADFP-YFP, or LDSP-5-YFP. The cells were then incubated with 400 μm oleic acid overnight. The following day, cells were incubated with 5 μm triacsin C (B, basal conditions) or with 10 μm forskolin, 1 mm IBMX, and 5 μm triacsin C for 30 min (S, stimulated conditions). Cell lysates were incubated with rabbit anti-HSL IgG (basal or stimulated) or rabbit pre-immune control IgG. Immunoprecipitates were analyzed by Western blot for PAT family proteins and HSL using a commercial GFP antibody that cross-reacts with YFP. One of three similar experiments is shown. IP, immunoprecipitation; WB, Western blot; Ab, antibody. C, Western blot ECL signals were quantified by densitometry using ImageJ software. Each data point represents the average of the calculated ratio of the amount of each PAT protein recovered in stimulated conditions relative to the amount recovered in basal conditions. Data represent means ± S.E. of three experiments (*, p < 0.05, basal versus stimulated conditions).
We collected AFRET data in cells expressing three experimental fusion pairs, perilipin-YFP/HSL-CFP, LSDP5-YFP/HSL-CFP, and ADFP-YFP/HSL-CFP, under basal conditions and following stimulation of PKA. Interactions were detected between perilipin-YFP and HSL-CFP on lipid droplets in stimulated cells but not in basal conditions (Fig. 3A). No interaction was detected between these proteins in the cytoplasm under either condition, consistent with previous observations that perilipin does not localize to the cytoplasm (26, 37–39). Interactions were detected between LSDP5-YFP and HSL-CFP both in the cytoplasm and on lipid droplets under both basal and stimulated conditions (Fig. 3A). Interactions were detected between ADFP-YFP and HSL-CFP on lipid droplets under basal conditions but not in the cytoplasm (Fig. 3A); measurements were not taken for stimulated cells.
We confirmed the AFRET results by co-IP and also tested interactions of TIP47-YFP with HSL-CFP (Fig. 3, B and C). For perilipin A, ADFP, and LSDP5, the amount of PAT proteins that co-immunoprecipitated with HSL correlated well with the detection of protein/protein interactions by AFRET; co-IP of perilipin-YFP and HSL-CFP increased following PKA stimulation of cells, whereas co-IP of ADFP-YFP and HSL-CFP was reduced by PKA stimulation, and co-IP of LSDP5-YFP and HSL-CFP was unchanged. TIP47-YFP and HSL-CFP were co-immunoprecipitated under both basal and stimulated conditions. AFRET measurements for this protein pair were inconclusive due to diffuse distribution of TIP47-YFP fluorescence throughout the cell that is unaltered by stimulation of cells (data not shown and see Ref. 40).
The possible contribution of YFP and CFP tags to artifactual interactions was tested by co-IP of untagged perilipin with untagged HSL. As observed with tagged proteins, perilipin interacted more highly with HSL in PKA-stimulated conditions than in basal conditions (supplemental Fig. 4). Moreover, CGI-58-CFP binds untagged perilipin A (supplemental Fig. 3D), suggesting that the interaction does not require YFP. Finally, a control IP using lysates from cells expressing perilipin-YFP and LSDP5-YFP showed that these two proteins, which do not interact in AFRET experiments, do not co-immunoprecipitate (supplemental Fig. 3C). Hence, the interactions between protein pairs that are depicted in Fig. 3B do not require fluorescent peptide tags.
Interaction of HSL with PAT Proteins Is Not Sufficient to Induce Lipolysis; Phosphorylation of HSL Plays an Important Role in Lipolysis
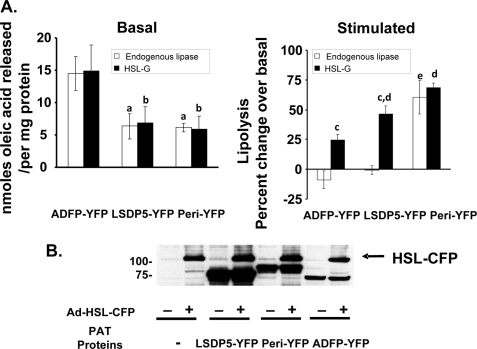
To investigate the consequences of HSL interaction with PAT proteins, we assessed lipolysis under both basal and PKA-activated conditions in cells expressing fluorescent fusion proteins of HSL and each of the three major PAT proteins (Fig. 4B). Cells expressing a PAT protein without ectopic HSL were also studied to assess lipolysis catalyzed by endogenous lipases. Interestingly, expression of HSL did not significantly increase basal lipolysis in any of the cell lines, although cells expressing ADFP-YFP (with or without ectopic HSL) released twice the level of fatty acids over 2 h relative to cells expressing either perilipin-YFP or LSDP5-YFP (Fig. 4A).
FIGURE 4.
Activation of PKA is required for increased lipolysis. CHO-Flip-In cells expressing ADFP-YFP, LSDP5-YFP, or perilipin-YFP were transduced with an adenoviral HSL-CFP construct for 48 h prior to lipolysis measurements. Control cells (without HSL-CFP) were infected with an adenoviral construct for lacZ. A, cells were loaded overnight with 400 μm [3H]oleic acid and then incubated in the absence of supplemental fatty acids with 5 μm triacsin C for 2 h. Basal activities are shown in the left graph, and activities from cells stimulated with IBMX and forskolin are expressed as a percentage over basal and shown in the right graph. Data are means ± S.E. from three separate experiments (a, p < 0.01 comparing lipolysis of cells lacking HSL but containing peri-YFP or LSDP5-YFP-coated droplets to lipolysis of cells containing ADFP-YFP-coated lipid droplets; b, p < 0.01 comparing lipolysis of HSL-expressing cells containing peri-YFP or LSDP5-coated droplets to lipolysis of HSL-expressing cells containing ADFP-YFP-coated droplets; c, p < 0.05 comparing % change over basal from cells lacking HSL to % change over basal in HSL-expressing cells; d, p < 0.05 comparing % change over basal in HSL-expressing cells containing peri-YFP or LSDP5-YFP-coated droplets to HSL-expressing cells containing ADFP-YFP-coated droplets; e, p < 0.01 comparing % change over basal in cells lacking HSL but containing peri-YFP-coated lipid droplets to % change over basal in cells lacking HSL but containing ADFP-YFP- or LSDP5-YFP-coated lipid droplets. B, immunoblot of cellular proteins extracted from CHO-Flip-In cells expressing ADFP-YFP, LSDP5-YFP, or perilipin-YFP and transduced with an adenoviral HSL-CFP (or lacZ) construct. Immunoblot shown is representative of three separate experiments.
After PKA stimulation, lipolysis increased for all cell lines expressing HSL. In cells expressing ADFP-YFP or LSDP5-YFP (but not HSL), activation of PKA did not increase lipolysis catalyzed by endogenous lipases (Fig. 4A). In contrast, in cells expressing perilipin-YFP, similar increases in lipolysis were observed in the presence and absence of ectopic HSL. Thus, phosphorylation of HSL is required to substantially increase lipolysis in cells expressing PAT proteins, which lack sites for phosphorylation by PKA, such as ADFP and LSDP5. Moreover, only perilipin modulates lipolysis catalyzed by endogenous lipases through a PKA-mediated mechanism.
PAT-1 Domains of Perilipin and LSDP5 Bind HSL
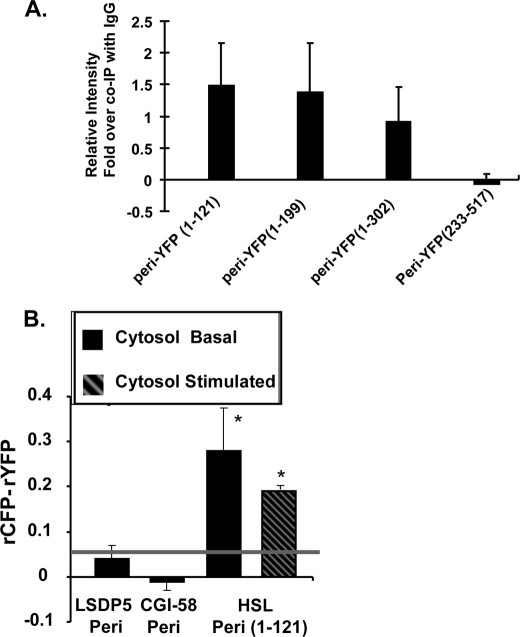
To identify the sequence of PAT proteins that binds HSL, we expressed a series of truncated fluorescent perilipin fusion proteins in cells, which also express HSL-CFP, and used cell lysates in co-IP experiments. The minimum sequence of perilipin that binds HSL is the PAT-1 domain (amino acids 1–121) (Fig. 5A); this region shares the highest homology among all PAT proteins except S3-12. HSL co-immunoprecipitated with all longer truncated variants of perilipin that included this region.
FIGURE 5.
AT-1 domain is the site of HSL binding. A, Western blot ECL signals were quantified by densitometry using ImageJ software. Each data point represents the average of the calculated ratio of the amount of each truncated form of perilipin-YFP recovered in IP performed with HSL IgGs relative to the amount recovered in IP performed with control rabbit IgGs. Plasmids for fusion constructs of perilipin-(1–121), perilipin-(1–199), perilipin-(1–302), and perilipin-(233–517) with YFP were transfected into CHO-K1 cells stably expressing HSL-GFP. Cells were incubated under stimulated conditions, extracts prepared for co-IP with rabbit anti-HSL antibodies, and blots probed with a GFP antibody. Data represent means ± S.E. of three experiments. B, in situ AFRET measurements; data were collected in live cells incubated in basal conditions and expressing pairs of fluorescent fusion proteins, as described in Fig. 3. Measurements were acquired solely in the cytoplasm. The gray line indicates the threshold of significance. Data are means ± S.E. from three experiments for each condition. *, p < 0.05, for the comparison of HSL-CFP/Peri-(1–121)-YFP to LSDP-5-CFP/Peri-YFP.
The PAT-1 domain lacks sequences required to target perilipin to lipid droplets (41). Consistent with these earlier findings, we observed diffuse localization of a fusion protein of YFP with perilipin amino acids 1–121 throughout the cytoplasm (supplemental Fig. 5). Cells expressing this construct and HSL-CFP were used for AFRET measurements (Fig. 5B); to confirm the co-IP results, truncated perilipin (Peri-(1–121)-YFP) interacted with HSL-CFP in the cytoplasm under both basal and stimulated conditions. Furthermore, HSL was retained in the cytoplasm of PKA-stimulated cells expressing the perilipin PAT-1 domain, even when full-length untagged perilipin was present on lipid droplets (supplemental Fig. 5). Addition of the PAT-1 domain of perilipin to cell lysates containing HSL did not reduce triglyceride hydrolase activity measured with an exogenous substrate (data not shown). Thus, the PAT-1 peptide of perilipin serves a dominant negative function in preventing HSL from binding to lipid droplets, yet it does not act as an inhibitor of lipase enzymatic activity.
Because the PAT-1 domain is highly conserved between members of the PAT family, we tested the comparable PAT-1 sequence of LSDP5 for HSL binding. A truncated variant of LSDP5 (amino acids 1–123 fused to YFP) co-immunoprecipitated with HSL (supplemental Fig. 6A). Another sequence that is conserved among PAT proteins has been termed the PAT-2 domain and includes a region of 11-mer repeats that are predicted to form amphipathic α-helices (42, 43). This region is in the amino terminus of PAT proteins proximal to the PAT-1 sequence. We used co-IP to test for a potential interaction between the 11-mer repeat region of LSDP5 and HSL; we were unable to detect a significant interaction (supplemental Fig. 6B).
Phosphorylation of Serine Residues in Amino-terminal PKA Sites of Perilipin A Facilitates HSL Binding and Recruitment to Lipid Droplets
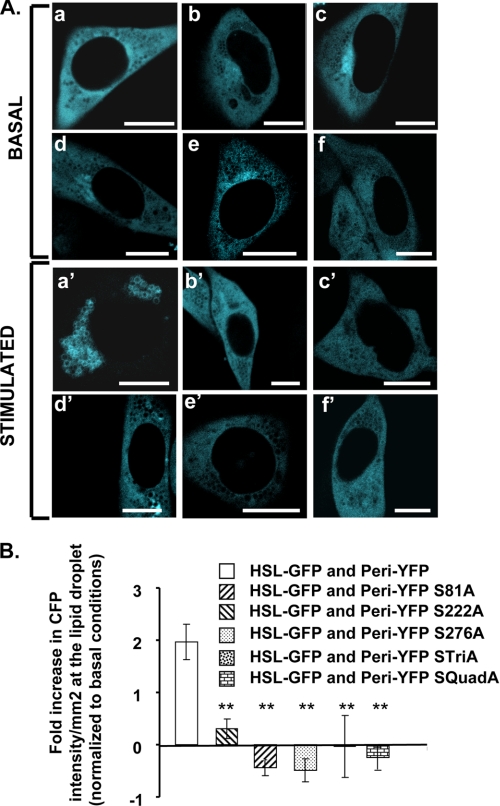
Mouse perilipin A has 6 serine residues in PKA consensus sequences. We generated five mutated variants of perilipin harboring single or multiple replacements of PKA-site serine residues with alanine residues; these mutated variants of perilipin were expressed in cells as fusion proteins with YFP. All variants of perilipin A with PKA-site mutations localize exclusively to lipid droplets (supplemental Fig. 7, A and B). A single mutation altering serine 81 in the first phosphorylation site is sufficient to reduce HSL translocation to lipid droplets following activation of PKA (Fig. 6). Interestingly, single mutations of sites 2 (serine 222) and 3 (serine 276) also reduce HSL translocation to perilipin-coated lipid droplets in stimulated cells. As expected, perilipin A with mutations in all three of these sites failed to recruit HSL to lipid droplets. These results were confirmed by lack of co-IP of HSL with mutated forms of perilipin in PKA-stimulated conditions (supplemental Fig. 8). Thus, PKA-mediated phosphorylation of serines 81, 222, and 276 of perilipin A is critical for recruitment of HSL to perilipin-coated lipid droplets.
FIGURE 6.
HSL recruitment to lipid droplets requires the phosphorylation of serines 81, 222, and 276 of perilipin A. A, CHO-K1 cells were co-transfected with HSL-CFP and one of the following mutated phosphorylation site perilipin fusion proteins: perilipin-YFP (panels a and a′); perilipin S81A-YFP (panels b and b′); perilipin S222A-YFP (panels c and c′); perilipin S276A-YFP (panels d and d′); perilipin STriA-YFP (Ser-81, Ser-222, and Ser-276 mutated to Ala) (panels e and e′); perilipin SQuadA-YFP (Ser-81, Ser-222, Ser-276, and Ser-433 mutated to Ala) (panels f and f′). Cells were incubated overnight with 400 μm oleic acid. The following day, the cells were incubated with 5 μm triacsin C with no further additions (basal) or were stimulated for 10 min with 10 μm forskolin, 1 mm IBMX, and 5 μm triacsin C; live cells were examined with a confocal microscope as in Fig. 1. Bar, 10 μm. B, quantitative analysis for HSL binding to lipid droplets covered with perilipin A or mutated variants of perilipin. Data are means ± S.E. from 2 to 14 experiments. HSL binding to lipid droplets coated with each of the mutated variants of perilipin was significantly different from the binding of HSL to unmodified perilipin A (**, p < 0.001).
DISCUSSION
The major objective of these studies was to determine whether a common mechanism is responsible for the recruitment of HSL to lipid droplets coated with different members of the PAT protein family. A second objective was to determine whether binding of HSL to lipid droplets is sufficient for HSL to gain access to its substrate lipids and increase lipolysis.
We first tested the hypothesis that cytosolic lipases gain access to stored lipids when PAT proteins are displaced from lipid droplets; ease of release of PAT proteins from lipid droplets would be inversely proportional to the binding affinity of the protein for the droplet surface. FRAP studies revealed that perilipin, ADFP, and LSDP5 have similar lateral mobility in the phospholipid monolayer coating lipid droplets, but only ADFP and LSDP5 exchange between the cytoplasm and lipid droplets. Because LSDP5 and perilipin restrict lipolysis equally under basal conditions, these data suggest that lipolysis is not controlled by replacement of PAT proteins with lipases at the surfaces of lipid droplets. Furthermore, phosphorylation of perilipin by PKA did not promote movement of perilipin between lipid droplets and the cytoplasm, consistent with a prior observation that perilipin A always fractionates with lipid droplets in lysates from both basal and PKA-stimulated adipocytes (26). Overall, the data suggest that perilipin has the highest affinity for lipid droplets, whereas LSDP5 and ADFP have comparable lower binding affinity. Moreover, PAT proteins are not rigidly anchored into the lipid droplet. Thus, the structure of the lipid droplet is fluid, allowing lateral mobility of associated proteins; however, the mechanism by which perilipin A anchors into the lipid droplet is distinctly different from that of LSDP5 and ADFP and prevents rapid exchange with a cytoplasmic pool of proteins.
Because a competitive binding model does not describe recruitment of lipases to the lipid droplet, we next asked whether lipases, specifically HSL, bind directly to PAT proteins. Our AFRET and co-IP experiments show that HSL binds to ADFP and LSDP5, but not perilipin, in cells incubated in basal conditions and to PKA-phosphorylated perilipin in stimulated cells. The PAT-1 domain, a region sharing the highest level of sequence similarity among four of five PAT proteins, is the minimal sequence required for HSL binding. Most importantly, although LSDP5 can recruit HSL to the lipid droplet surface in basal conditions, lipolytic activity can only be induced upon HSL phosphorylation. Thus, we propose a model whereby two steps are required for the activation of lipolysis by HSL. The first step is recruitment of HSL to PAT-1 domain sequences of PAT proteins on lipid droplets; the second step is phosphorylation of HSL by PKA to initiate lipolysis.
While this manuscript was undergoing revision, Shen et al. (45) reported co-IP of HSL with a FLAG-tagged (carboxyl-terminal tag) truncated variant of perilipin consisting of amino acids 1–200, but not a perilipin fusion protein with amino acids 1–141, from forskolin-treated CHO cells overexpressing both the lipase and tagged perilipin. Our study differs from the report by Shen et al. (45) in identifying the PAT-1 domain (amino acids 1–121) of perilipin as the most likely HSL-binding site by AFRET experiments, co-IP of HSL with PAT-1 domains of both perilipin and LSDP5, and competition of a soluble PAT-1 domain fusion protein with lipid droplet-bound perilipin for binding of HSL in intact cells. The reason for differences between this recent study (45) and our own are unclear but may be due in part to differences in the fusion constructs employed.
We hypothesize that the PAT-1 domain of perilipin, or a binding sequence within this domain, is inaccessible to HSL in basal conditions and requires phosphorylation of perilipin to trigger a conformational change. Our data suggest that phosphorylation of the three amino-terminal-most PKA sites, serines 81, 222, and 276, promotes this conformational change to reveal the binding site. Alternatively, addition of negative charges to the serine residues at these positions may be critical to provide a surface for HSL binding through electrostatic interactions.
Earlier reports showed ambiguous results concerning the importance of PKA-mediated phosphorylation of perilipin to recruit HSL to lipid droplets. In 3T3-L1 pre-adipocytes transfected with expression constructs for fluorescent fusion proteins, CFP-HSL was observed at the surfaces of lipid droplets coated with perilipin-YFP mutated in all six PKA phosphorylation sites (36); however, PKA-phosphorylated perilipin bound HSL most effectively in co-IP experiments (44). Our data from co-IP experiments and live cell imaging confirm that the phosphorylation of perilipin is necessary to bind HSL for recruitment to lipid droplets, and our data further demonstrate that the first three perilipin phosphorylation sites are critical for this protein/protein interaction.
An important new finding of this study is that phosphorylation of HSL is required to promote lipolysis of triglyceride substrates packaged in lipid droplets; recruitment of HSL to the PAT protein scaffold on lipid droplets is insufficient to increase lipolysis without activation of PKA. Thus, phosphorylation of HSL increases either enzyme activity or lipase access to substrates within lipid droplets. Ex vivo, phosphorylation of HSL increases enzyme activity by ∼2-fold against a traditional exogenous substrate composed of a phospholipid-stabilized emulsion of triolein. A recent study has identified Ser-650 in human HSL (corresponding to Ser-660 in rat HSL) as the major determinant of PKA-mediated activation of HSL (46). These in vitro data provide support for the two-step model of HSL activation proposed in this study. PKA phosphorylates HSL at several sites, some of which are more important for the first step, translocation and binding of HSL to a PAT protein (32), whereas phosphorylation of Ser-650 is critical for the second step of interfacial activation.
PAT family proteins, signature proteins of lipid droplets, are both structural components that stabilize lipid droplets and essential regulators of lipolysis. This study contributes to a developing model whereby PAT proteins are required for the docking of HSL on lipid droplets and activation of lipolysis. PKA-mediated phosphorylation events are required for both docking of HSL on perilipin-coated lipid droplets and activation of HSL catalytic activity.
Supplementary Material
Acknowledgments
We thank Ming Bell for valuable technical advice. We thank Dr. Martin Woodle for careful review and helpful editing of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1 DK075017 (to C. S.) and RO1 DK054797 (to D. L. B.) and NIDDK Intramural Research Programs. This work was also supported by Career Development Award 1-05-CD-17 from the American Diabetes Association (to C. S.), the Geriatric Research, Education, and Clinical Center, Baltimore Veterans Affairs Health Care Center, and the Clinical Nutrition Research Unit of Maryland Grant DK072488.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–8 and Table 1.
- ADFP
- adipose differentiation-related protein
- peri
- perilipin A
- CHO
- Chinese hamster ovary
- co-IP
- co-immunoprecipitation
- AFRET
- anisotropy Forster resonance energy transfer
- FRAP
- fluorescence recovery after bleaching
- YFP
- yellow fluorescent protein
- BSA
- bovine serum albumin
- ATGL
- adipose triglyceride lipase
- HSL
- hormone-sensitive lipase
- PKA
- protein kinase A
- IBMX
- isobutylmethylxanthine
- GFP
- green fluorescent protein
- CFP
- cyan fluorescent protein
- FA-free
- fatty acid-free
- IP
- immunoprecipitation.
REFERENCES
- 1.Brasaemle D. L., Dolios G., Shapiro L., Wang R. (2004) J. Biol. Chem. 279, 46835–46842 [DOI] [PubMed] [Google Scholar]
- 2.Liu P., Ying Y., Zhao Y., Mundy D. I., Zhu M., Anderson R. G. (2004) J. Biol. Chem. 279, 3787–3792 [DOI] [PubMed] [Google Scholar]
- 3.Wu C. C., Howell K. E., Neville M. C., Yates J. R., 3rd, McManaman J. L. (2000) Electrophoresis 21, 3470–3482 [DOI] [PubMed] [Google Scholar]
- 4.Fujimoto Y., Itabe H., Sakai J., Makita M., Noda J., Mori M., Higashi Y., Kojima S., Takano T. (2004) Biochim. Biophys. Acta 1644, 47–59 [DOI] [PubMed] [Google Scholar]
- 5.Ozeki S., Cheng J., Tauchi-Sato K., Hatano N., Taniguchi H., Fujimoto T. (2005) J. Cell Sci. 118, 2601–2611 [DOI] [PubMed] [Google Scholar]
- 6.Bell M., Wang H., Chen H., McLenithan J. C., Gong D. W., Yang R. Z., Yu D., Fried S. K., Quon M. J., Londos C., Sztalryd C. (2008) Diabetes 57, 2037–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sztalryd C., Bell M., Lu X., Mertz P., Hickenbottom S., Chang B. H., Chan L., Kimmel A. R., Londos C. (2006) J. Biol. Chem. 281, 34341–34348 [DOI] [PubMed] [Google Scholar]
- 8.Dalen K. T., Dahl T., Holter E., Arntsen B., Londos C., Sztalryd C., Nebb H. I. (2007) Biochim. Biophys. Acta 1771, 210–227 [DOI] [PubMed] [Google Scholar]
- 9.Listenberger L. L., Ostermeyer-Fay A. G., Goldberg E. B., Brown W. J., Brown D. A. (2007) J. Lipid Res. 48, 2751–2761 [DOI] [PubMed] [Google Scholar]
- 10.Tansey J. T., Sztalryd C., Gruia-Gray J., Roush D. L., Zee J. V., Gavrilova O., Reitman M. L., Deng C. X., Li C., Kimmel A. R., Londos C. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 6494–6499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinez-Botas J., Anderson J. B., Tessier D., Lapillonne A., Chang B. H., Quast M. J., Gorenstein D., Chen K. H., Chan L. (2000) Nat. Genet. 26, 474–479 [DOI] [PubMed] [Google Scholar]
- 12.Tansey J. T., Huml A. M., Vogt R., Davis K. E., Jones J. M., Fraser K. A., Brasaemle D. L., Kimmel A. R., Londos C. (2003) J. Biol. Chem. 278, 8401–8406 [DOI] [PubMed] [Google Scholar]
- 13.Souza S. C., Muliro K. V., Liscum L., Lien P., Yamamoto M. T., Schaffer J. E., Dallal G. E., Wang X., Kraemer F. B., Obin M., Greenberg A. S. (2002) J. Biol. Chem. 277, 8267–8272 [DOI] [PubMed] [Google Scholar]
- 14.Miyoshi H., Perfield J. W., 2nd, Souza S. C., Shen W. J., Zhang H. H., Stancheva Z. S., Kraemer F. B., Obin M. S., Greenberg A. S. (2007) J. Biol. Chem. 282, 996–1002 [DOI] [PubMed] [Google Scholar]
- 15.Belfrage P., Jergil B., Strålfors P., Tornqvist H. (1977) FEBS Lett. 75, 259–264 [DOI] [PubMed] [Google Scholar]
- 16.Zimmermann R., Strauss J. G., Haemmerle G., Schoiswohl G., Birner-Gruenberger R., Riederer M., Lass A., Neuberger G., Eisenhaber F., Hermetter A., Zechner R. (2004) Science 306, 1383–1386 [DOI] [PubMed] [Google Scholar]
- 17.Kershaw E. E., Hamm J. K., Verhagen L. A., Peroni O., Katic M., Flier J. S. (2006) Diabetes 55, 148–157 [PMC free article] [PubMed] [Google Scholar]
- 18.Miyoshi H., Perfield J. W., 2nd, Obin M. S., Greenberg A. S. (2008) J. Cell. Biochem. 105, 1430–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sztalryd C., Xu G., Dorward H., Tansey J. T., Contreras J. A., Kimmel A. R., Londos C. (2003) J. Cell Biol. 161, 1093–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prats C., Donsmark M., Qvortrup K., Londos C., Sztalryd C., Holm C., Galbo H., Ploug T. (2006) J. Lipid Res. 47, 2392–2399 [DOI] [PubMed] [Google Scholar]
- 21.Osterlund T., Danielsson B., Degerman E., Contreras J. A., Edgren G., Davis R. C., Schotz M. C., Holm C. (1996) Biochem. J. 319, 411–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamaguchi T., Omatsu N., Matsushita S., Osumi T. J. (2004) J. Biol. Chem. 279, 30490–30497 [DOI] [PubMed] [Google Scholar]
- 23.Rizzo M. A., Springer G. H., Granada B., Piston D. W. (2004) Nat. Biotechnol. 22, 445–449 [DOI] [PubMed] [Google Scholar]
- 24.Rizzo M. A., Piston D. W. (2005) Biophys. J. 88, L14–L16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deleted in proof
- 26.Marcinkiewicz A., Gauthier D., Garcia A., Brasaemle D. L. (2006) J. Biol. Chem. 281, 11901–11909 [DOI] [PubMed] [Google Scholar]
- 27.Piston D. W., Rizzo M. A. (2008) Methods Cell Biol. 85, 415–430 [DOI] [PubMed] [Google Scholar]
- 28.Subramanian V., Rothenberg A., Gomez C., Cohen A. W., Garcia A., Bhattacharyya S., Shapiro L., Dolios G., Wang R., Lisanti M. P., Brasaemle D. L. (2004) J. Biol. Chem. 279, 42062–42071 [DOI] [PubMed] [Google Scholar]
- 29.Luo J., Deng Z. L., Luo X., Tang N., Song W. X., Chen J., Sharff K. A., Luu H. H., Haydon R. C., Kinzler K. W., Vogelstein B., He T. C. (2007) Nat. Protoc. 2, 1236–1247 [DOI] [PubMed] [Google Scholar]
- 30.Becker T. C., Noel R. J., Coats W. S., Gómez-Foix A. M., Alam T., Gerard R. D., Newgard C. B. (1994) Methods Cell Biol. 43, 161–189 [DOI] [PubMed] [Google Scholar]
- 31.Contreras J. A., Danielsson B., Johansson C., Osterlund T., Langin D., Holm C. (1998) Protein Expr. Purif. 12, 93–99 [DOI] [PubMed] [Google Scholar]
- 32.Su C. L., Sztalryd C., Contreras J. A., Holm C., Kimmel A. R., Londos C. (2003) J. Biol. Chem. 278, 43615–43619 [DOI] [PubMed] [Google Scholar]
- 33.Brasaemle D. L., Rubin B., Harten I. A., Gruia-Gray J., Kimmel A. R., Londos C. (2000) J. Biol. Chem. 275, 38486–38493 [DOI] [PubMed] [Google Scholar]
- 34.Egan J. J., Greenberg A. S., Chang M. K., Wek S. A., Moos M. C., Jr., Londos C. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 8537–8541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamaguchi T., Omatsu N., Matsushita S., Osumi T. (2004) J. Biol. Chem. 279, 30490–304907 [DOI] [PubMed] [Google Scholar]
- 36.Granneman J. G., Moore H. P., Granneman R. L., Greenberg A. S., Obin M. S., Zhu Z. J. (2007) J. Biol. Chem. 282, 5726–5735 [DOI] [PubMed] [Google Scholar]
- 37.Greenberg A. S., Egan J. J., Wek S. A., Garty N. B., Blanchette-Mackie E. J., Londos C. (1991) J. Biol. Chem. 266, 11341–11346 [PubMed] [Google Scholar]
- 38.Blanchette-Mackie E. J., Dwyer N. K., Barber T., Coxey R. A., Takeda T., Rondinone C. M., Theodorakis J. L., Greenberg A. S., Londos C. (1995) J. Lipid Res. 36, 1211–1226 [PubMed] [Google Scholar]
- 39.Brasaemle D. L., Barber T., Kimmel A. R., Londos C. (1997) J. Biol. Chem. 272, 9378–9387 [DOI] [PubMed] [Google Scholar]
- 40.Miura S., Gan J. W., Brzostowski J., Parisi M. J., Schultz C. J., Londos C., Oliver B., Kimmel A. R. (2002) J. Biol. Chem. 277, 32253–32257 [DOI] [PubMed] [Google Scholar]
- 41.Garcia A., Sekowski A., Subramanian V., Brasaemle D. L. (2003) J. Biol. Chem. 278, 625–635 [DOI] [PubMed] [Google Scholar]
- 42.Bussell R., Jr., Ramlall T. F., Eliezer D. (2005) Protein Sci. 14, 862–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bussell R., Jr., Eliezer D. (2003) J. Mol. Biol. 329, 763–778 [DOI] [PubMed] [Google Scholar]
- 44.Miyoshi H., Souza S. C., Zhang H. H., Strissel K. J., Christoffolete M. A., Kovsan J., Rudich A., Kraemer F. B., Bianco A. C., Obin M. S., Greenberg A. S. (2006) J. Biol. Chem. 281, 15837–15844 [DOI] [PubMed] [Google Scholar]
- 45.Shen W. J., Patel S., Miyoshi H., Greenberg A. S., Kraemer F. B. (2009) J. Lipid Res., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krintel C., Osmark P., Larsen M. R., Resjö S., Logan D. T., Holm C. (2008) PLoS ONE 3, e3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.