Abstract
Ubiquitin modification of endosomal membrane proteins is a signal for active inclusion into the Multivesicular Body (MVB) pathway, resulting in lysosomal degradation. However, the endosome represents a dynamic site of protein sorting with a majority of proteins destined for recycling, rather than MVB targeting. Substrate recognition by ubiquitin ligases is therefore highly regulated. We have investigated substrate recognition by the Nedd4 ortholog Rsp5 as a model for understanding ligase-substrate interactions. Rsp5 interacts directly with its substrate Cps1 via a novel interaction mode. Perturbation of this mode of interaction revealed a compensatory role for the Rsp5 adaptor Bsd2. These results highlight the ability of Rsp5 to interact with substrates via multiple modalities, suggesting additional mechanisms of regulating this interaction and relevant outcomes.
Introduction
The post-translational addition of ubiquitin to protein substrates is a regulatory modification of remarkable scope in eukaryotic biology. Cellular processes as diverse as protein degradation, protein trafficking, DNA repair, and nuclear signaling are regulated by ubiquitination, and as a consequence, numerous pathologies and developmental defects have been linked to defects in the ubiquitin system (reviewed in Refs. 1–3). A cascade of reactions culminates in the formation of an isopeptide bond between the C-terminal glycine of ubiquitin and an acceptor amine within the substrate. Ubiquitin modification is tightly regulated, with the third enzyme in the ubiquitination cascade, the ubiquitin ligase, responsible for substrate selection (reviewed in Refs. 4, 5).
Ubiquitin modification of endosomal transmembrane proteins has previously been demonstrated to play a major role in targeting proteins into multivesicular bodies (MVBs)3 en route to lysosomal degradation (6–9). Entry into intralumenal vesicles during MVB sorting is tightly regulated, and Carboxypeptidase S (Cps1) has served as a model MVB cargo in analyses demonstrating the role of ubiquitin modification as a positive cis-acting MVB sorting determinant (6–9). Considerable evidence supports a model wherein the HECT ubiquitin ligase Rsp5 plays the major role in targeting a number of MVB cargoes, including Cps1, into this pathway in Saccharomyces cerevisiae (9–20). Rsp5 is the yeast ortholog of Nedd4 family ligases, all of which contain WW protein interaction domains involved in substrate recognition (21). These WW domains participate in substrate recognition either directly through “PY” motifs within the substrates (9, 18–20, 22–26) or indirectly via adaptors that contain PY motifs (27–29). Bsd2 is one such cofactor that has been implicated in Cps1 ubiquitination and subsequent MVB targeting (17, 30). However, we have previously observed a direct interaction between Rsp5 and Cps1 in vitro (9), suggesting that the interactions leading to Cps1 ubiquitination may be more complicated. Cps1 contains MVB targeting information within the amino acid sequence “PVEKAPR” (6), which does not possess a PY motif capable of WW domain interaction. To resolve the mode of Rsp5 interaction with Cps1, we have utilized a variety of in vitro and in vivo approaches that have uncovered a novel direct interaction between the HECT domain of Rsp5 and the Cps1 sequence PVEKAPR. Mutations within PVEKAPR perturb the functional interaction between Rsp5 and Cps1 in the absence of Bsd2 both in vitro and in vivo. Additionally, we observe distinct Cps1 MVB sorting phenotypes upon loss of Bsd2 as compared with loss of Rsp5 function in vivo. This suggests that although the interaction between Rsp5 and Cps1 is direct, this interaction can be enhanced by Bsd2 to regulate modification in vivo. These results suggest a model wherein ligase-substrate interactions can be subject to multiple levels of modulation to achieve appropriate levels of substrate ubiquitination.
EXPERIMENTAL PROCEDURES
Plasmids and Strains
His-MBP-RSP5WT, His-MBP-RSP5WW1,2,3, DPAP B NTD-GST, Cps1 NTD-GST (pRI1), Sna3 CTD-GST, Sna3 CTD PPAY-AAAA–GST, nTAP416-RSP5WT, nTAP416-RSP5WW1,2,3 have been previously described (9). His-Rsp5 bacterial expression plasmids were generated by subcloning the sequence encoding Rsp5 residues 1–809 amplified from yeast genomic DNA into pQE31 (Qiagen). His-Rsp5 mutants were generated by site-directed mutagenesis at sites indicated in the text. Rsp5 fragments were cloned into pET28-MBP to generate His-MBP epitope-tagged constructs: E2 Binding domain, residues 580–654; C-lobe, residues 695–802; N-lobe, residues 451–809; and MBP HECT+, residues 434–802. Cps1 NTD-GST mutants were generated by site-directed mutagenesis of pRI1 using the GeneTailor kit (Invitrogen) and sequenced to ensure a lack of spurious mutations. PVEKAPR-DPAP B NTD-GST was constructed by ligating duplex oligos encoding the sequence PVEKAPR in-frame with the 26 N-terminal residues of DPAP B with flanking BamHI/NotI restriction sites into pET28 GST (9) digested with BamHI and NotI. PVEKAPR-GST was constructed by the same method, without the DPAP B sequence. PVERAPR-DPAP B NTD-GST was generated by site-directed mutagenesis of PVEKAPR-DPAP B NTD-GST. DsRed415-RSP5WT and DsRed415-rsp5G753I have been previously described (31). A yeast shuttle vector encoding His-Rsp5I603A D604A under control of the RSP5 promoter was constructed as follows: the rsp5-containing EcoRI/HindIII from pQE31-rsp5I603A D604A was ligated with pBC SK (Stratagene) linearized by the same digested to create pBC-rsp5I603A D604A. The RSP5 promoter region consisting of 775 base pairs 5′ of the RSP5 start codon was amplified with primers encoding 5′ SacII and 3′ EcoRI sites from SEY6210 genomic DNA and ligated into pBC-rsp5I603A D604A linearized by SacII/EcoRI digest. pBC-rsp5I603A D604A was digested with SacII, ClaI and NcoI and the rsp5-containing SacII/ClaI fragment was ligated with pRS414 linearized by SacII/ClaI digest to create pRS414-HIS-rsp5I603A D604A. Construction of pGO45 (GFP-Cps1) and pGO89 (GFP-DPAP B) (32), pMB225 (GFP-PVEKAPR-DPAP B) (6), and ubiquitin-GFP-Cps1 (12) have been described previously. GFP-Cps1 mutants were generated by site-directed mutagenesis of pBC CPS1WT and subcloned into pGO35 using Kpn1. Duplex oligos encoding the sequence PVRKAPR or PVERAPR and flanking BglII and HindIII sites were ligated with a BglII/HindIII fragment from pG089 to create GFP-PVrKAPR-DPAP B and GFP-PVErAPR-DPAP B. pRS414-BSD2WT was created by amplifying the Bsd2 coding sequence together with 200 base pairs 5′ of the sequence from SEY6210 genomic DNA and cloned into pRS414 (33). pRS414-BSD2WT was verified by diagnostic digest and expression was confirmed by complementation of Cps1 sorting and ubiquitination defects in the bsd2Δ and bsdΔ pep12Δ backgrounds (data not shown). pRS414-bsd2Y140A and pRS414-bsd2P149A were generated by site-directed mutagenesis.
All strains are based on the wild type genetic backgrounds SEY6210 (MATα leu2–3,112 ura3–52 his3-Δ200 trp1-Δ901 lys2–801 suc2-Δ9) (34) or SEY6210.1 (SEY6210; MATa) (35). SEY6210-based rsp5Δ::HIS3 yeast covered by pRS415-DsRed-RSP5WT and pRS415-DsRed-rsp5WW1,2,3 have been previously described (9). The pep12Δ (CBY31) strain is described in Ref. 36. The SEY6210 BSD2 coding region was interrupted with a HIS3 cassette to create bsd2Δ::HIS3 and confirmed by PCR. To make the bsd2Δ pep12Δ double knock-out strain, bsd2Δ and pep12Δ strains were mated, sporulated, and tetrads were dissected, subjected to nutritional selection, and deletions were verified by PCR. Yeast expressing rsp5I603A D604A were generated by transforming rsp5Δ::HIS3 covered by nTAP416-RSP5WT with pRS414-HIS-rsp5I603A D604A, followed by treatment with 5-Fluororotic acid to select against strains retaining the URA-expressing plasmid.
Protein Expression and Purification
BL21-DE3 bacteria were transformed with GST, His, or His-MBP expression constructs. Transformants were cultured in LB medium with antibiotic selection to A600 ∼1.0 and induced with 0.5 mm isopropyl-1-thio-β-d-galactopyranoside for 3 h at 37 °C (Cps1 NTD- and DPAP B NTD-based GST constructs) or overnight at 22 °C (Rsp5 constructs and Sna3 CTD-GST constructs). Pellets were lysed in 1 ml of buffer (phosphate-buffered saline, pH 7.2 for GST constructs or 50 mm phosphate buffer, pH 7.2 for His and His-MBP constructs) per 50-ml culture volume by probe sonication. Lysates were cleared by centrifugation at 18,000 × g. GST constructs were purified on 5-μl bed volume glutathione-Sepharose (GE Healthcare) per ∼100 μl lysate for 1 h at room temperature, followed by three washes in ubiquitin reaction buffer (see below) plus 0.5% Triton X-100. His- and His-MBP constructs were purified by affinity chromatography according to the manufacturer's protocols using Ni2+ HiTrap HP FPLC (Amersham Biosciences), Ni2+-nitrilotriacetic acid resin (Qiagen), or Nickel spin columns (Qiagen). His- and His-MBP proteins were quantified by Bio-Rad protein assay using bovine serum albumin standards.
Protein-Protein Interaction Studies
Translational fusions between GST and baits were bound to glutathione-Sepharose beads, washed as indicated above, and incubated with ∼5 pmol of His- or His-MBP-Rsp5 constructs for 1 h at 4 °C or 30 min at 22 °C as indicated in figure legends. Protein concentrations of His-Rsp5 and His-MBP-Rsp5 were determined using the Bio-Rad Protein Assay reagent. Reactions were conducted in a total volume of 500 μl in ubiquitin reaction buffer (20 mm HEPES pH 6.8, 50 mm KOAc) with Triton X-100 to 0.5%. Ubiquitination reaction components including yeast E1, UbcH5a, ubiquitin (Boston Biochem), and dithiothreitol, but not MgATP, were added to the binding reaction to mimic the conditions of the ubiquitination reaction. Following the reaction, beads were washed twice with reaction buffer plus detergent and once in buffer lacking detergent. Beads were heated for 3 min at 95 °C in sample buffer to elute bound material. 1/10 of the total reaction was loaded for SDS-PAGE followed by anti-His or anti-MBP Western blot to detect bound material. 1/100 of the total reaction was loaded for Coomassie Brilliant Blue R-250 (Bio-Rad) staining to verify equivalent loading of GST baits.
Autoubiquitination Assay
Reactions containing 125 nm yeast E1, 500 nm UbcH5a, 3 mm MgATP, 2.5 μg of ubiquitin, 400 ng of biotin-ubiquitin (Boston Biochem), and ∼250 nm His-Rsp5 or His-MBP-Rsp5 in a reaction volume of 20 μl were incubated for 30 min at 22 °C. Protein concentrations of His-Rsp5 and His-MBP-Rsp5 were determined using the Bio-Rad Protein Assay reagent. Reactions were stopped with 20 μl of sample buffer containing βME, heated, and loaded on 4–20% gradient gels (Bio-Rad). Ubiquitinated species were detected with avidin-horseradish peroxidase (HRP, Boston Biochem). Reaction buffer contained 20 mm HEPES (pH 6.8), 50 mm KOAc, and 10 μm dithiothreitol.
In Vitro Substrate Ubiquitination
Reaction conditions were identical to GST pulldowns with ubiquitination reaction components including E1 to 5 nm, UbcH5a to 20 nm, 2.5 μg ubiquitin, 400 ng of biotin-ubiquitin, dithiothreitol to 10 μm, MgATP to 120 μm, and E3 to 10 nm. Wash conditions following the ubiquitination reaction included 500 mm KOAc to displace copurifying proteins. Substrate ubiquitination was detected by avidin-HRP.
Live Cell Imaging
Cells grown in minimal media were used for fluorescence microscopy. Micrographs were captured using a fluorescence microscope Olympus IX70 (Center Valley, PA) with fluorescein isothiocyanate or eGFP filters and a digital camera (Coolsnap HQ;Photometrics, Tucson, AZ) and were deconvolved using Delta Vision software (Applied Precision, Issaquah, WA).
Yeast Protein Isolation
Proteins were immunoprecipitated as described in Ref. 9 using anti-Cps1 polyclonal (6) or anti-GFP monoclonal antibody AV JL-8 (BD Bioscience). Ubiquitinated species were detected with anti-ubiquitin monoclonal antibody (Zymed Laboratories Inc., South San Francisco, CA) or monoclonal anti-HA-II (Covance) following SDS-PAGE. An aliquot of the same material was run in parallel and probed by anti-GFP or anti-Cps1 Western blot to verify equivalent loading. Pulse-chase analysis was conducted as described in Ref. 9. Following SDS-PAGE, dried gels were exposed to phosphorimager screens. Screens were processed using the Storm 840 system (GE Healthcare), and signal was quantified using ImageQuant software (GE Healthcare). Kinetics of vacuolar delivery were assessed by comparing the signal from full-length GFP-Cps1 or precursor Cps1 remaining at each time point following the addition of chase.
RESULTS
The Sequence PVEKAPR Constitutes a Sufficient Rsp5 Interaction Motif
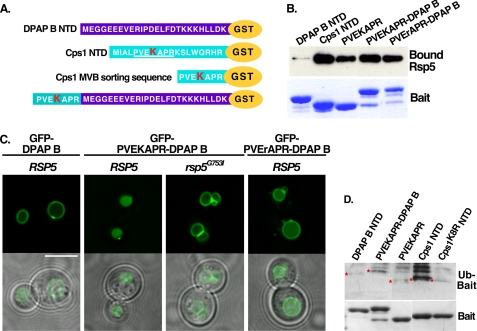
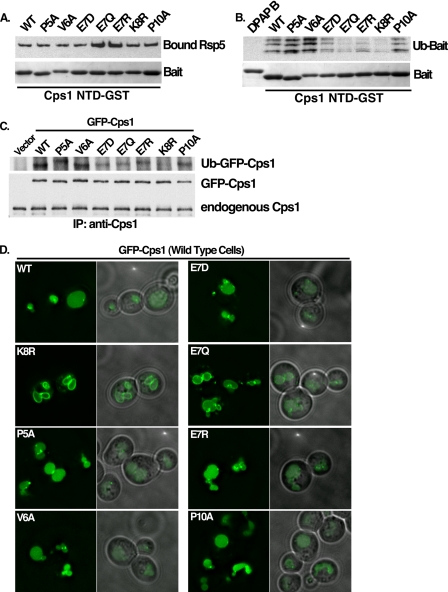
To determine if the PVEKAPR sorting sequence within Cps1 is sufficient to interact with Rsp5, the ability of constructs containing the sequence PVEKAPR to bind Rsp5 was examined in vitro. We have previously demonstrated the ability of Rsp5 to interact directly with the cytoplasmic tail of Cps1 fused to GST (Cps1 NTD-GST) (9). This observation was extended by analyzing the interaction between Rsp5 and a chimera containing the sequence PVEKAPR fused to dipeptidyl aminopeptidase B (PVEKAPR-DPAP B-GST) or PVEKAPR fused directly to GST (PVEKAPR-GST) (Fig. 1A). DPAP B is not an MVB cargo, but can be converted into one through the addition of the Cps1 MVB-targeting sequence (6). As previously observed, the cytoplasmic portion of DPAP B did not interact with Rsp5 in vitro, while both PVEKAPR-DPAP B-GST and PVEKAPR-GST were observed to interact directly with Rsp5 ((9) and Fig. 1B). The lysine within this Cps1 sequence (K8) is required for both ubiquitin modification and MVB targeting (6). Mutation of this lysine to arginine (PVErAPR-DPAP B-GST) did not reduce Rsp5 binding (Fig. 1B), consistent with the idea that defects in Cps1K8R MVB targeting are not due to an inability to recruit the ligase but rather an inability to accept ubiquitin modification. These results indicate that the sequence PVEKAPR promotes a direct interaction with Rsp5 and that the lysine residue within this sequence is not requisite for the interaction.
FIGURE 1.
The amino acid sequence PVEKAPR is sufficient for Rsp5 interaction. A, scheme representation of the cytoplasmic N-terminal sequences of the indicated proteins, which were expressed as C-terminal GST fusions to mimic the presentation of these proteins at the membrane. The Cps1 acceptor lysine is highlighted in red. B, GST constructs described in A were bound to glutathione-Sepharose beads and incubated with purified His-MBP-Rsp5. Bound material was detected with anti-MBP antibody (Rsp5) or Coomassie Brilliant Blue staining (GST baits). C, live cell imaging of the indicated GFP-tagged cargo was examined in wild type or rsp5Δ yeast co-expressing either DsRed-Rsp5WT or a hypoactive HECT domain mutant (G753I) as the only source of Rsp5. Scale bar represents 5 microns. D, indicated GST fusions were bound to glutathione-Sepharose beads and incubated with the ubiquitination reaction components (E1, E2, MgATP, ubiquitin, biotin-ubiquitin) and His-MBP-tagged Rsp5. Ubiquitin-modified species were visualized by avidin-HRP to evaluate the ability of Rsp5 to modify these peptides in vitro.
While GFP-DPAP B is not ubiquitinated, fusion of PVEKAPR to DPAP B (GFP-PVEKAPR-DPAP B) is sufficient to confer its ubiquitination and re-direction into the MVB pathway (6). To determine if MVB sorting of the GFP-PVEKAPR-DPAP B chimera depends on Rsp5 function, analogous to GFP-Cps1 (12), we compared the trafficking of GFP-PVEKAPR-DPAP B in yeast solely expressing Rsp5WT or Rsp5G753I. The Rsp5G753I mutation resides within the HECT domain and leads to a reduction in catalytic activity (supplemental Fig. S1). Strains harboring certain mutations within the Rsp5 HECT domain display defects in MVB targeting of cargoes while retaining function of the MVB pathway ((9, 12, 31) and data not shown). While GFP-Cps1 and GFP-PVEKAPR-DPAP B were visualized in the lumen of the vacuole in RSP5 cells (as expected in the case of proper Rsp5-dependent MVB sorting), both were mislocalized to the limiting membrane of the vacuole in rsp5G753I cells (Fig. 1C and Refs. 9, 12). Together, these results indicate that the PVEKAPR sequence promotes Rsp5-mediated MVB sorting in the context of Cps1 and the PVEKAPR-DPAP B chimera.
The cytoplasmic domain of DPAP B contains four lysine residues (Fig. 1A), raising the possibility that these could serve as ubiquitin acceptor sites facilitating the MVB targeting of GFP-PVEKAPR-DPAP B. We addressed this possibility by generating a chimera in which the acceptor lysine within the Cps1 sorting motif was mutated to arginine (GFP-PVErAPR-DPAP B). Despite the presence of additional lysine residues within DPAP B, GFP-PVErAPR-DPAP B accumulated at the limiting membrane of the vacuole (Fig. 1C). This is consistent with the lysine acceptor site within the Cps1 sorting motif (K8) remaining the relevant acceptor lysine in the context of the chimera and further suggests that the mode of interaction between Rsp5 and GFP-PVEKAPR-DPAP B is directly analogous to the interaction between Rsp5 and Cps1 in vivo.
Although we have previously demonstrated a direct interaction between Cps1 and Rsp5 in vitro using recombinant proteins (9), the PY-containing cofactor Bsd2 has been shown to promote Cps1 ubiquitination by Rsp5 (17, 30). To determine if the direct interaction we observed between Cps1 and Rsp5 is sufficient to lead to Cps1 ubiquitination in the absence of Bsd2, we first examined the ability of Rsp5 to ubiquitinate Cps1 in vitro. Constructs outlined in Fig. 1A were subjected to in vitro ubiquitination assays and modified species were visualized by Western blotting (Fig. 1D). While DPAP B-GST and Cps1K8R NTD-GST were not ubiquitinated, Cps1 NTD-GST was ubiquitinated by Rsp5 in this assay. Furthermore, constructs containing PVEKAPR (PVEKAPR-GST and PVEKAPR-DPAP B-GST) were also ubiquitinated by Rsp5 (Fig. 1D). We conclude that the sequence PVEKAPR is sufficient to promote recognition by Rsp5 and Rsp5-dependent ubiquitination in vitro. Complementary in vivo analyses are presented below.
HECT C-lobe Recognizes Cps1 Sorting Sequence
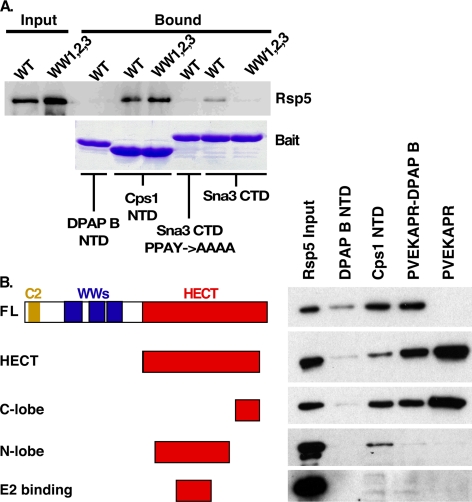
Contacts between Rsp5 WW domains and PY motifs within substrates or substrate adaptors mediate many previously described Rsp5-substrate interactions (9, 17–20, 22–30, 37–41). To determine if Rsp5 WW domains mediate a non-canonical interaction with the Cps1 NTD, we investigated the impact of mutations in the PY-motif interaction sites within the three Rsp5 WW domains (Rsp5WW1,2,3) on the Cps1 interaction. Rsp5WW1,2,3 displayed no defect relative to Rsp5WT in the context of binding to the Cps1 NTD (Fig. 2A). This result is in contrast to observations with the Rsp5 substrate Sna3, in which mutation of the Rsp5 WW domains or the Sna3 PY motif abolished the WW-PY mediated interaction between Sna3 and Rsp5 from yeast lysate (Fig. 2A). Comparable results were generated using bacterially produced Rsp5 and Rsp5WW1,2,3 (supplemental Fig. S1E). These results suggest that the direct interaction between Rsp5 and the Cps1 NTD is not mediated by canonical WW domain interactions.
FIGURE 2.
Rsp5 HECT C-lobe mediates interaction with Cps1 MVB sorting sequence. A, lysates from yeast expressing either TAP-tagged wild type or the indicated WW-domain mutant Rsp5 were incubated with the indicated GST constructs to determine if Cps1 is a WW-domain interacting substrate. Bound TAP-Rsp5 was detected with anti-actin polyclonal antibody following gel electrophoresis. B, protein-protein interaction utilizing the indicated His-MBP-Rsp5 truncations and the indicated bait-GST constructs. Bound material was visualized by anti-MBP Western blotting.
A set of Rsp5 truncations was generated to investigate regions required for interacting with PVEKAPR (Fig. 2B). In vitro interaction studies were performed under conditions mimicking the in vitro ubiquitination assay using these Rsp5 truncations and a variety of baits: DPAP B NTD-GST, Cps1 NTD-GST, PVEKAPR-DPAP B-GST, and PVEKAPR-GST. Under these conditions interactions between full-length Rsp5 and Cps1 NTD-GST or PVEKAPR-DPAP B-GST were observed as before, while Rsp5 interaction with PVEKAPR-GST was greatly reduced (Fig. 2B). In contrast, the HECT domain construct robustly interacted with PVEKAPR-GST, as well as the other two constructs containing the PVEKAPR sequence (Fig. 2B). Additional truncations of the HECT construct were generated to refine our understanding of interacting regions. While N-terminal HECT subdomains (e.g. N-lobe and E2 binding) failed to display the interaction profile of HECT, the C-lobe construct retained the HECT binding profile (Fig. 2B). While other regions of Rsp5 may contribute to Cps1 recognition in the context of full-length protein, these results implicate the C-terminal lobe of the HECT domain as responsible for directly interacting with PVEKAPR.
N-lobe of Rsp5 HECT Domain Contributes to Cps1 Interaction
While the above results indicate that the C-lobe of Rsp5 specifically interacts with the sequence PVEKAPR, PVEKAPR-GST was defective for interaction with full-length Rsp5 at temperatures conducive to enzyme function (22 °C) (Fig. 2B) and was not ubiquitinated at the level of Cps1 NTD-GST (Fig. 1D). Furthermore, the N-terminal portion (N-lobe) of the HECT domain contributed to Cps1 NTD recognition (Fig. 2B). To explore the hypothesis that a region within the HECT N-lobe contributes to Cps1 binding, and thus a functional association between Cps1 and Rsp5, we sought to identify residues within this domain required for Cps1 interaction that are not required for Rsp5 catalytic activity. We reasoned that highly divergent residues within the N-lobe, either by sequence or structure, might impact substrate binding while not affecting HECT activity.
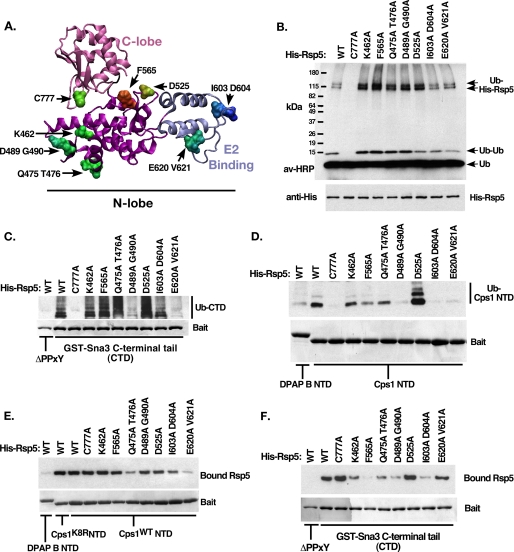
Comparison of HECT structures (42–44) indicated a high level of structural alignment overall between homologs (vector alignment search tool, VAST, (45)), with a limited number of regions representing significant deviation from the consensus structure within both the N-lobe and the C-lobe (not shown). Diversity at the sequence level was determined by sequence alignment of HECT homologs (supplemental Table S1, ConSeq, Ref. 46), and residues likely to be solvent exposed were identified by plotting conservation scores onto a homology-based structural model of the Rsp5 HECT domain (SwissProt (47), not shown). Seven mutations within the Rsp5 N-lobe, as well as the active site cysteine, were selected for further analysis, as identified in Fig. 3A (C777A, K462A, F565A, Q475A T476A, D489A G490A, D525A, I603A D604A, E620A V621A).
FIGURE 3.
Characterization of Rsp5 N-lobe mutants. A, homology structural model of the Rsp5 HECT domain indicating the location of the C-lobe (pink) and E2 binding domain (blue). Residues selected for site-directed mutation are indicated. B, in vitro autoubiquitination reactions were performed using the indicated form of Rsp5. Ubiquitinated species were detected using avidin-HRP. Bands corresponding to free ubiquitin, di-ubiquitin, and 2 forms of ubiquitinated His-Rsp5 are indicated. C, C-terminal cytoplasmic tail of the PY-motif containing Rsp5 substrate Sna3 was immobilized and included in the ubiquitination reaction as in B. Following the 30-min reaction time, beads were washed with a high-salt buffer to remove bound Rsp5 and other copurifying proteins. Ubiquitinated bound material was visualized as in B, while baits were visualized by Coomassie Brilliant Blue staining. The PY-mutant form of Sna3 tail was used as a negative control. Bands corresponding to ubiquitinated GST-Sna3 C-terminal domain (CTD) are indicated. D, ubiquitination reactions were performed as in C, except the reaction substrates were the cytoplasmic tails of DPAP B NTD (non-Rsp5 substrate) or Cps1 NTD (Rsp5 HECT-interacting substrate) fused to GST. E, Rsp5 binding reactions were performed to correlate the ability of His-Rsp5 mutants to bind Cps1 NTD, reaction conditions were kept identical to D, except that MgATP was omitted to prevent the modification of His-Rsp5. Low salt wash buffer was used to preserve noncovalent interactions. Detection of bound Rsp5 was by anti-His Western blot. F, Rsp5 binding reactions were performed as in E, except with the indicated GST constructs.
N-terminal His-tagged forms of full-length wild type or mutant Rsp5 were generated and purified. To assess the impact of the HECT mutations on Rsp5 catalytic activity, in vitro autocatalytic ubiquitination was performed and ubiquitinated species were visualized by Western blotting. With the exception of the active site cysteine mutant, all forms of Rsp5 retained autocatalytic activity (Fig. 3B). As Sna3 represents a substrate that is recognized by Rsp5 in a manner distinct from Cps1, the ability of these mutants to ubiquitinate a WW-dependent substrate (Sna3) was investigated. The PY motif found within the C-terminal cytoplasmic tail of Sna3 (Sna3 CTD) was required for modification by Rsp5 in this assay, as were Rsp5 WW domains, analogous to results seen in vivo (Ref. 9, Fig. 3C, supplemental Fig. S1C). Five of the seven N-lobe mutants tested modified Sna3 at levels equal to or greater than wild type Rsp5 (Fig. 3C), further confirming that these N-lobe mutations do not negatively impact Rsp5 catalytic activity. Lastly, the ability of these forms of Rsp5 to ubiquitinate Cps1 NTD-GST in vitro was examined. Of the five mutant Rsp5 species that retained both autocatalytic activity and the ability to ubiquitinate Sna3 only I603A D604A was defective for Cps1 ubiquitination (Fig. 3D). This ubiquitination defect correlated with a detectable reduction in the steady state association between the I603A D604A mutant form of Rsp5 and Cps1 (Fig. 3E). Two other Rsp5 mutant forms, E620A V621A and D489A G490A, were similarly defective for Cps1 ubiquitination, however these mutants also had reduced levels of ubiquitinated Sna3, indicating a more general defect in substrate modification. Therefore the I603A D604A mutant form of Rsp5 showed a defect in Cps1 ubiquitination not associated with a reduction in catalytic activity, and this defect correlated with a decrease in Cps1 binding. Together, these results suggest that residues Ile-603 and Asp-604 are specifically required for Cps1 recognition.
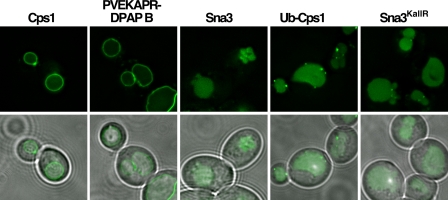
The impact of the I603A D604A mutation on Rsp5 function in vivo was examined by analyzing GFP-tagged MVB cargoes in cells expressing only Rsp5I603A D604A. Both GFP-Cps1 and GFP-PVEKAPR-DPAP B localized to the limiting membrane of the vacuole (Fig. 4), indicative of failure to enter intralumenal vesicles during MVB sorting. By contrast Ub-GFP-Cps1, Sna3-GFP and Sna3KallR-GFP were all observed within the vacuolar lumen (Fig. 4). These cargoes do not require ubiquitin modification by Rsp5 to enter the MVB pathway ((8, 9, 18, 19)), although Sna3KallR-GFP requires the ability to associate with an active form of Rsp5 for this ubiquitin-independent sorting event to occur (9). Therefore, this result is consistent with GFP-Cps1 and GFP-PVEKAPR-DPAP B being specifically defective for ubiquitin-dependent MVB sorting in the mutant Rsp5 background. Although the steady state association between Rsp5I603A D604A and Sna3 CTD was markedly reduced in vitro compared with wild type Rsp5 (Fig. 3F), Sna3KallR was sorted into the MVB pathway in the background expressing only this mutant form of Rsp5 (Fig. 4). This suggests that Rsp5I603A D604A retains appreciable catalytic activity as well as sufficient ability to associate with Sna3 in vivo. Taken together, these results indicate that in addition to PVEKAPR-C-lobe contacts, residues within the N-lobe of the Rsp5 HECT domain make contributions relevant for Cps1 recognition and subsequent ubiquitination in vitro and in vivo.
FIGURE 4.
Rsp5I603A D604A displays cargo-specific MVB sorting defects. Steady-state localization of GFP-tagged MVB cargo proteins were examined by fluorescence microscopy for their ability to sort into the MVB pathway in yeast expressing only the I603A D604A form of Rsp5.
MVB Sorting Sequence Residues Required for a Functional Interaction with Rsp5
To determine the contributions of residues within the PVEKAPR MVB sorting sequence to the Rsp5 interaction, the ability of recombinant Rsp5 to bind and ubiquitinate forms of the cytoplasmic domain of Cps1 containing point mutations within the Cps1 MVB sorting sequence was assayed (Fig. 5, A and B). Binding studies revealed that E7Q and E7R mutant forms of Cps1 displayed an apparent increase in Rsp5 association (Fig. 5A). In contrast, these mutant forms were defective for in vitro Rsp5-mediated ubiquitination (Fig. 5B). In vivo ubiquitination mirrored in vitro ubiquitination results, in which E7 mutants displayed reduced levels of ubiquitinated Cps1 (Figs. 5C). These results imply a role for the charged residue E7 in transferring ubiquitin between Rsp5 and Cps1, despite the observation that binding between Cps1 and Rsp5 is not decreased.
FIGURE 5.
Residues within Cps1 MVB sorting sequence required for a functional interaction with Rsp5. A, Cps1 NTD-GST mutant proteins were tested for their ability to recruit His-Rsp5 in vitro under conditions identical to the ubiquitin reaction (−MgATP) in B. Bound Rsp5 was detected by anti-His western and equivalent loading of GST baits was by Coomassie Brilliant Blue staining. B, Rsp5 was assessed for its ability to ubiquitinate mutant forms of Cps1 in vitro. After a 30-min ubiquitination reaction in the presence of immobilized substrate, beads were washed with buffer containing 500 mm KOAc to remove noncovalently bound material. Bound, ubiquitinated material was detected with avidin-HRP. Equivalent loading of GST baits was confirmed by Coomassie Brilliant Blue staining. C, wild type or mutant forms of full-length GFP-Cps1 were co-expressed with HA-ubiquitin in a pep12Δ yeast background to allow detection of in vivo ubiquitin-modified Cps1. Immunoprecipitation was performed with anti-Cps1 polyclonal antibody and ubiquitinated species were detected with anti-HA Western blotting. Anti-GFP Western blot of the same material serves as a loading control. D, same GFP-tagged Cps1 mutants as in C above were examined by fluorescence microscopy for their ability to sort into the MVB pathway in a wild type yeast background as determined by GFP fluorescence in the vacuolar lumen.
The impact of these mutations on MVB targeting of Cps1 was assessed within the context of full-length GFP-Cps1 by visualization in wild type cells. Fig. 5D reveals that none of the point mutants tested displayed a dramatic MVB missorting phenotype by comparison to the lysine acceptor mutant form (K8R), although the E7Q mutation displayed a weak phenotype, revealing a modest accumulation of GFP signal at the limiting membrane of the vacuole. Together, these results can be interpreted as having two implications: that a partial defect in ubiquitination of Cps1 may not be manifested as a dramatic MVB sorting defect, and/or additional factors may impact Cps1 targeting into the MVB pathway. Of these two, the later appears more likely given the finding that Bsd2 facilitates Cps1 ubiquitination and MVB sorting in a distinct genetic background (17, 30).
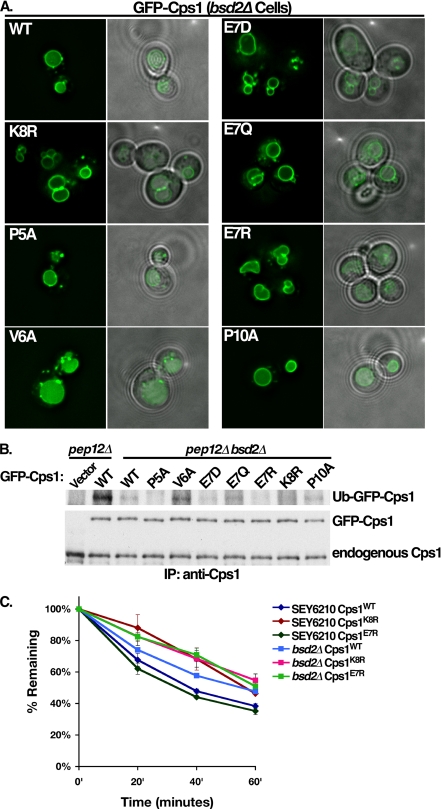
Bsd2 has been reported to act as an adaptor between Rsp5 and Cps1. It is therefore reasonable that in vitro ubiquitination defects observed did not clearly manifest as in vivo MVB sorting defects due to Bsd2 contributions to the Rsp5-Cps1 functional interaction. To investigate this possibility, GFP-Cps1 mutants were analyzed in an isogenic bsd2Δ strain. While GFP-Cps1 localized to the vacuolar lumen in wild type cells (Fig. 5D), partial mislocalization to the limiting membrane of the vacuole was observed in bsd2Δ cells (Fig. 6A). In contrast to the localization observed in wild type cells, GFP-Cps1 PVEKAPR mutants, with the exception of V6A, displayed missorting phenotypes in the bsd2Δ strain (Fig. 6A). This trafficking defect correlated with a decrease in ubiquitination in vivo, as wild type GFP-Cps1 displayed a marked reduction in ubiquitination in bsd2Δ cells as compared with BSD2 cells (Fig. 6B). The only mutant that displayed appreciable ubiquitination was the V6A form (Fig. 6B), consistent with its ability to sort into the MVB pathway in the bsd2Δ background (Fig. 6A) and enhanced ubiquitination of this mutant form in vitro (Fig. 5B).
FIGURE 6.
Bsd2 enhances Cps1 ubiquitination and MVB sorting. A, indicated full-length GFP-Cps1 (wild type and mutant) constructs were expressed in an isogenic background in which the BSD2 gene had been deleted. B, same GFP-Cps1 constructs were immunoprecipitated and visualized as in 5C from yeast lacking both PEP12 and BSD2 to assess the ability of Rsp5 to ubiquitinate these forms of Cps1 in the absence of Bsd2. C, maturation kinetics of wild type, E7R, or K8R forms of GFP-Cps1 were compared in the wild type background compared with the isogenic strain lacking BSD2 (bsd2Δ). Yeast cells were labeled with [35S]cysteine and methionine. Following a chase with unlabeled amino acids, cell lysates were immunoprecipitated with polyclonal anti-Cps1 antibody. The GFP moiety is rapidly cleaved following delivery to the vacuolar lumen, therefore kinetics of delivery to the lumen of the vacuole were assessed by monitoring the disappearance of the full-length form of GFP-Cps1 at the indicated time points by phosphorimaging.
Kinetic analysis of GFP-Cps1 maturation (an indicator of MVB-mediated vacuolar delivery and processing) was also utilized. This revealed a delay in the maturation rate of wild type Cps1 in bsd2Δ cells compared with wild type cells (Fig. 6C), consistent with both the partial sorting defect observed (Fig. 6A) and the defect in ubiquitination (Fig. 6B). Additionally, GFP-Cps1E7R processing was indistinguishable from GFP-Cps1 in wild type cells, while in bsd2Δ cells it was delayed comparably to the lysine acceptor mutant (GFP-Cps1K8R) (Fig. 6C). Together, these results support a model wherein Bsd2 facilitates ubiquitination and MVB sorting of GFP-Cps1. However, Bsd2 is not required for Cps1 to interact with Rsp5 in a manner that leads to Cps1 ubiquitination and subsequent MVB sorting.
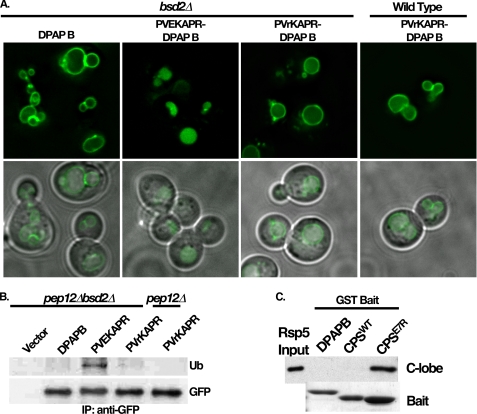
To specifically address PVEKAPR contributions to Rsp5 interaction in vivo, we examined the ability of the GFP-PVEKAPR-DPAP B chimera to be delivered into the MVB pathway in the absence of Bsd2. GFP-PVEKAPR-DPAP B was observed within the lumen of the vacuole in the bsd2Δ background (Fig. 7A), suggesting that Bsd2 is not required for PVEKAPR-dependent MVB sorting. As the Cps1E7R NTD-GST construct displayed defective ubiquitination in vitro (Fig. 5B), this mutation was introduced into the GFP-PVEKAPR-DPAP B chimera (GFP-PVrKAPR-DPAP B), and the impact on MVB targeting was investigated. GFP-PVrKAPR-DPAP B was observed within the limiting membrane of the vacuole in both wild type and bsd2Δ cells (Fig. 7A). Furthermore, while GFP-PVEKAPR-DPAP B was ubiquitinated in the bsd2Δ background, GFP-PVrKAPR-DPAP B displayed ubiquitination defects in both BSD2 and bsd2Δ backgrounds (Fig. 7B). Therefore, both in vitro and in vivo data support the assertion that the glutamate residue within the sequence PVEKAPR plays a role in ubiquitination by Rsp5. This was further addressed by investigating the ability of the C-lobe construct to bind Cps1 NTD-GST constructs with and without this glutamate. Fig. 7C shows that the C-lobe construct displays dramatically enhanced binding to Cps1E7R NTD relative to the wild type version of this bait. Full-length Rsp5 also displayed enhanced binding to this construct (Fig. 5A), suggesting that the E7R mutation hinders ubiquitination of Cps1 by Rsp5 in vivo and in vitro by stabilizing or otherwise promoting a non-productive interaction between the mutant form of Cps1 and the Rsp5 C-lobe.
FIGURE 7.
Cps1 NTD mutant E7R specifically impacts Rsp5 C-lobe binding. A, GFP-PVEKAPR-DPAP B and GFP-PVrKAPR-DPAP B chimeras were expressed in the bsd2Δ background and visualized by fluorescence microscopy. B, ubiquitination status of DPAP B or the indicated DPAP B chimeras was assessed as described in Fig. 5C, with the exception that immunoprecipitation and detection was using anti-GFP monoclonal antibody. C, ability of Cps1E7R NTD-GST to recruit His-MBP-C-lobe was compared with Cps1WT NTD-GST and DPAP B NTD. Detection of bound Rsp5 was by anti-MBP Western blot, bait was visualized by Coomassie Brilliant Blue staining following SDS-PAGE.
Bsd2 PY Motifs Are Required to Enhance Rsp5 WW-domain-mediated Cps1 Ubiquitination and Sorting
Bsd2 contains one canonical and one non-canonical PY motif (30). Previous studies have demonstrated that Bsd2 interacts with Rsp5 in a manner that depends on both PY motifs and Rsp5 WW domains to facilitate Cps1 ubiquitination and MVB sorting (30). In addition, our results are consistent with previous reports that Bsd2 modulates Cps1 ubiquitination and MVB sorting (Fig. 6). While it has been hypothesized that Bsd2 and Cps1 interact through polar residues within the membrane (17, 30), we were unable to generate transmembrane mutant forms of GFP-Cps1E7R that phenocopied the MVB sorting defect observed in bsd2Δ cells, although a partial defect was apparent (supplemental Fig. S2, A–C). Regardless of the mechanism by which Bsd2 interacts with Cps1, we examined whether Bsd2 PY motifs and Rsp5 WW domains are strictly required for Cps1 modification and MVB sorting in vivo.
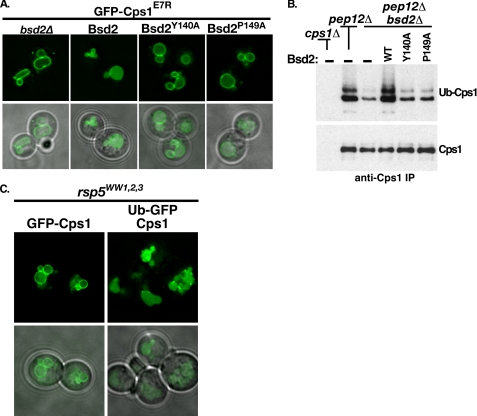
The E7R mutant form of GFP-Cps1 was observed to be sensitive to the presence of Bsd2; trafficking like wild type Cps1 in the wild type yeast background (Figs. 5D and 6C) and missorting in the bsd2Δ background (Fig. 6, A and C). To examine the contribution of Bsd2 PY motifs to Cps1 MVB sorting, we expressed GFP-Cps1E7R in bsd2Δ yeast co-expressing either empty vector or BSD2 promoter-driven wild type, Y140A, or P149A Bsd2 constructs. As seen before, GFP-Cps1E7R localized to the limiting membrane of the vacuole in the bsd2Δ background (Fig. 8A). Bsd2WT complemented this MVB sorting defect (Fig. 8A). In contrast, neither PY-motif mutant form of Bsd2 complemented the GFP-Cps1E7R MVB sorting defect (Fig. 8A). These observations paralleled in vivo ubiquitination status of endogenous Cps1; Bsd2WT was able to restore Cps1 ubiquitination while both Bsd2Y140A and Bsd2P149A displayed Cps1 ubiquitination levels similar to vector alone (Fig. 8B). Therefore, both Bsd2 PY motifs are required to facilitate Cps1 ubiquitination and MVB sorting of GFP-Cps1. Interestingly, endogenous Cps1 ubiquitination was clearly detectable in the bsd2Δ background transformed with vector alone (Fig. 8B). This result is consistent with results from the in vitro Cps1 ubiquitination assay, which shows that Cps1 modification occurs in the absence of Bsd2 (Figs. 1D, 3E, 5B). Together these data support the idea that Rsp5 can ubiquitinate Cps1 directly in vivo, but this modification is enhanced by Bsd2 PY motif interactions with Rsp5.
FIGURE 8.
Bsd2 PY motifs are required to enhance WW-domain mediated Cps1 ubiquitination and sorting. A, Bsd2-sensitive cargo GFP-Cps1E7R was visualized by fluorescence microscopy in the bsd2Δ background co-expressing wild type or two different PY mutant forms of Bsd2. B, pep12Δ or pep12Δ bsd2Δ strains were transformed with vector alone or plasmids expressing the indicated forms of Bsd2 under control of the BSD2 promoter. Endogenous Cps1 was immunoprecipitated using anti-Cps1 polyclonal antibody and de-glycosylated to improve detection of ubiquitinated species. Detection of ubiquitin-Cps1 was by anti-ubiquitin monoclonal antibody. The same material was detected by anti-Cps1 polyclonal antibody as a loading control. C, an rsp5Δ strain expressing Rsp5 in which all three WW domains had been mutated was transformed with the indicated MVB cargo and visualized by fluorescence microscopy.
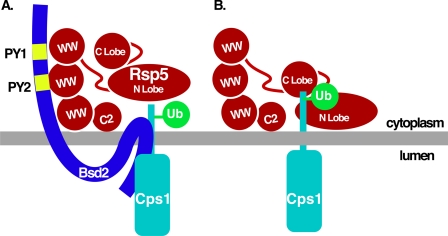
WW domain mutant forms of Rsp5 are defective for MVB targeting of cargoes such as Cps1 (9, 12, 15, 18), presumably due to the inability to utilize PY-containing adaptors such as Bsd2. To rule out the possibility that Rsp5 WW domain mutations lead to a general MVB sorting defect, we examined the ability of a ubiquitin-GFP-Cps1 translational fusion to be sorted into the MVB pathway in an Rsp5WW1,2,3 mutant background (Fig. 8C). In contrast to GFP-Cps1, which was retained at the limiting membrane, ubiquitin-GFP-Cps1 was delivered to the vacuolar lumen in the Rsp5 WW domain mutant background (Fig. 8C). This result suggests that the MVB sorting defects observed in Rsp5 WW mutant backgrounds is specifically the result of cargo ubiquitination defects (direct or adaptor-mediated), rather than a general defect the in the function of the MVB pathway. Together, these data support a model that Bsd2 PY motif-Rsp5 WW domain interactions enhance Cps1 modification (Fig. 9A), though Cps1 and Rsp5 can interact directly through HECT domain-Cps1 sorting sequence contacts (Fig. 9B).
FIGURE 9.
Models for adaptor-assisted and direct Rsp5-Cps1 interaction. A, model for the Bsd2-facilitated interaction between Rsp5 and Cps1. Bsd2 PY motifs interact with Rsp5 WW domains to facilitate Cps1 ubiquitination. B, Rsp5 interacts directly with Cps1 leading to Cps1 ubiquitination. The Cps1 sorting sequence PVEKAPR interacts with the Rsp5 C-lobe in a manner that requires the Glu7 residue within the sorting sequence, while additional Cps1 residues interact with the Rsp5 N-lobe.
DISCUSSION
Here we have described a direct interaction between the Nedd4-family ligase Rsp5 and one of its substrates, Cps1. This interaction involves contacts between Cps1 and both lobes of the Rsp5 HECT domain. This is in contrast to previously described interactions between HECT ligases and their substrates that require regions of the ligase N-terminal to the catalytic domain such as WW domains, capable of interacting directly with substrate PY motifs or PY-containing adaptor proteins (9, 18–20, 22–26). In particular, these results highlight the ability of Rsp5 to interact with this substrate directly or via an adaptor-enhanced mode, providing multiple levels of modulating ligase-substrate interactions and outcome.
The amino acid sequence PVEKAPR within Cps1 was necessary and sufficient to recruit the C-lobe of the Rsp5 HECT domain. This observation assigns this binding surface within Cps1, which contains the ubiquitin acceptor lysine, to the catalytic cysteine-containing C-lobe of Rsp5 during productive interaction. It is interesting to note that the amino acid sequence PVEKAPR, or variants thereof, can be found in a number of yeast proteins, suggesting this may represent a conserved mode of Rsp5 interaction. However, the Cps1 NTD also interacted with the N-lobe of the Rsp5 HECT domain. The PVEKAPR peptide was not sufficient to promote this interaction, suggesting that in addition to the PVEKAPR-HECT C-lobe interaction, contacts involving an extended region of the Cps1 cytoplasmic domain and the Rsp5 N-lobe contribute to optimal ubiquitination. Site-directed mutagenesis aimed at identifying the location of binding between Cps1 and the N-lobe revealed a single Rsp5 N-lobe mutant, I603A D604A, that specifically reduced Cps1 binding and ubiquitination while retaining general catalytic activity. This mutation resides within the same subdomain presumed to be involved in E2 recruitment (E2BD), at the distal end of the N-lobe, predicted to be proximal to the E2 catalytic cysteine (42). The simplest interpretation of these results is that Cps1 binds simultaneously to the C-lobe and the E2 binding region of the N-lobe and that both interactions are important for Cps1 modification.
Although there is clearly an interaction between Cps1 and the Rsp5 HECT domain, it remains to be seen whether direct interaction between a given HECT domain and its cognate substrates is a general requirement for the transfer of ubiquitin. The mechanism for bringing a WW-domain bound substrate in close proximity to the HECT catalytic cysteine has not yet been determined. We observed reduced binding between several Rsp5 HECT domain mutants and the WW-domain interacting substrate Sna3, including one key mutation that led to a dramatic reduction in Sna3 modification (D489A G490A). Nedd4 family ligases display a high level of sequence and structural homology within their WW-domain substrate binding modules. To achieve specificity in substrate selection, one possibility is that determinants within the HECT domain also play a part in PY substrate recognition, either directly via HECT-substrate contacts or indirectly via HECT-WW contacts. Identifying the basis for these changes should expand our understanding of the mechanism employed by HECT ligases to modify many substrates.
Evidence supports a model in which adaptors can contribute to the modulation of HECT ligase function in vivo (27–30, 48). Similarly, while we observed a direct interaction between the PVEKAPR sequence and Rsp5 that was sensitive to mutations therein, the presence of Bsd2 was capable of suppressing mutations within this sequence in vivo (e.g. Cps1E7R). The E7R mutation decreased the ability of Rsp5 to ubiquitinate Cps1 in vivo and in vitro, yet correlated with increased binding between Cps1 and the Rsp5 C-lobe and impaired Cps1 MVB sorting (Fig. 5). Several models could explain these apparently inconsistent observations. Prolonged ligase interactions with substrates may impair the efficiency of ligase function or promote in vivo interactions with deubiquitinating enzymes that are also associated with MVB sorting machinery (49–51). Alternatively, Cps1 may actively contribute to ubiquitin transfer and ligase release. While the mechanism is unclear, Bsd2 may function to increase the local concentration of Rsp5 relative to Cps1, thereby increasing the likelihood of Cps1 ubiquitination and MVB sorting. However, the presence of Bsd2 was unable to compensate for a suboptimal interaction between Cps1 and Rsp5I603A D604A. The Rsp5I603A D604A mutant retained general catalytic activity in vitro and promoted MVB sorting of Sna3KallR, which requires the ability to associate with a catalytically active form of Rsp5 for MVB sorting (9). It therefore appears that the presence of Bsd2 contributes more to the Rsp5-Cps1 interaction than simply increasing the local concentration of these two proteins. For instance, Bsd2 may productively stabilize the interaction between Rsp5 and the PVEKAPR sequence within Cps1. An alternative hypothesis to explain the ability of Bsd2 to suppress Cps1E7R MVB sorting defects is that interaction with Bsd2 facilitates Rsp5 catalytic activity, either directly or indirectly. There is precedent for cofactors activating Nedd4-family ligases, as recent data indicates that Bsd2 homologs activate Nedd4 family members by releasing them from autoinhibitory conformations (52). This hypothesis is also consistent with the finding that disrupting the ability of Nedd4-2 WW-domains to engage in inhibitory interactions with a PY motif located within the Nedd4-2 HECT C-lobe promotes Nedd4-2 autoubiquitination and subsequent degradation (53). Although the precise mechanism by which Bsd2 promotes Cps1 ubiquitination has not been resolved, these observations further support the model that in vivo substrate modification by HECT ligases can be modulated through interaction with cofactors.
Cps1 is a model Rsp5 substrate with the potential to reveal much about HECT domain-substrate interactions and may even be a model for the mechanism of ubiquitin transfer between HECT ligases and their substrates. The HECT catalytic cysteine is located within the C-lobe of the HECT domain, which connects to the larger N- lobe by a flexible linker (42, 43). Based on these findings, the current model for ubiquitin transfer between HECT and E2 catalytic sites proposes that the HECT C-lobe swings across the N-lobe via allowable rotations and extensions of the flexible linker to transfer ubiquitin from the E2 active site cysteine to the HECT active site cysteine (43). The mechanism by which ubiquitin is subsequently transferred to the substrate is unclear. We have described a direct interaction between Cps1 and the Rsp5 HECT domain. Future studies to resolve the mechanism of ubiquitin transfer between HECT ligases and their cognate substrates will be aided by model systems such as this.
Supplementary Material
Acknowledgments
We thank Drs. Brian Davies, Bruce Horazdovsky, and Ed Leof for thoughtful discussions, Dr. Darren Carney and Bob Sikkink for assistance with protein purification, and Dr. Georges Mer for isothermal calorimetry and useful discussions.
This work was supported, in whole or in part, by Grant R01 GM73024 from the National Institutes of Health. This work was also supported by the American Heart Association Grant AHA0430369Z (to D. J. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Table S1.
- MVB
- multivesicular body
- HECT
- homologous to E6-AP C terminus
- NTD
- N-terminal domain
- CTD
- C-terminal domain
- E2BD
- E2 binding domain
- HRP
- horseradish peroxidase
- GST
- glutathione S-transferase
- GFP
- green fluorescent protein
- DPAP B
- dipeptidylaminopeptidase B
- MBP
- maltose binding protein
- TAP
- tandem affinity purification.
REFERENCES
- 1.Welchman R. L., Gordon C., Mayer R. J. (2005) Nat. Rev. Mol. Cell Biol. 6, 599–609 [DOI] [PubMed] [Google Scholar]
- 2.Huang T. T., D'Andrea A. D. (2006) Nat. Rev. Mol. Cell Biol. 7, 323–334 [DOI] [PubMed] [Google Scholar]
- 3.Hoeller D., Hecker C. M., Dikic I. (2006) Nat. Rev. Cancer 6, 776–788 [DOI] [PubMed] [Google Scholar]
- 4.Hershko A., Ciechanover A. (1998) Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 5.Pickart C. M. (2001) Annu. Rev. Biochem. 70, 503–533 [DOI] [PubMed] [Google Scholar]
- 6.Katzmann D. J., Babst M., Emr S. D. (2001) Cell 106, 145–155 [DOI] [PubMed] [Google Scholar]
- 7.Urbanowski J. L., Piper R. C. (2001) Traffic 2, 622–630 [DOI] [PubMed] [Google Scholar]
- 8.Reggiori F., Pelham H. R. (2001) EMBO J. 20, 5176–5186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oestreich A. J., Aboian M., Lee J., Azmi I., Payne J., Issaka R., Davies B. A., Katzmann D. J. (2007) Mol. Biol. Cell 18, 707–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hein C., Springael J. Y., Volland C., Haguenauer-Tsapis R., André B. (1995) Mol. Microbiol. 18, 77–87 [DOI] [PubMed] [Google Scholar]
- 11.Hicke L., Riezman H. (1996) Cell 84, 277–287 [DOI] [PubMed] [Google Scholar]
- 12.Katzmann D. J., Sarkar S., Chu T., Audhya A., Emr S. D. (2004) Mol. Biol. Cell 15, 468–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morvan J., Froissard M., Haguenauer-Tsapis R., Urban-Grimal D. (2004) Traffic 5, 383–392 [DOI] [PubMed] [Google Scholar]
- 14.Soetens O., De Craene J. O., Andre B. (2001) J. Biol. Chem. 276, 43949–43957 [DOI] [PubMed] [Google Scholar]
- 15.Dunn R., Klos D. A., Adler A. S., Hicke L. (2004) J. Cell Biol. 165, 135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blondel M. O., Morvan J., Dupré S., Urban-Grimal D., Haguenauer-Tsapis R., Volland C. (2004) Mol. Biol. Cell 15, 883–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hettema E. H., Valdez-Taubas J., Pelham H. R. (2004) EMBO J. 23, 1279–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watson H., Bonifacino J. S. (2007) Mol. Biol. Cell 18, 1781–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McNatt M. W., McKittrick I., West M., Odorizzi G. (2007) Mol. Biol. Cell 18, 697–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stawiecka-Mirota M., Pokrzywa W., Morvan J., Zoladek T., Haguenauer-Tsapis R., Urban-Grimal D., Morsomme P. (2007) Traffic 8, 1280–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ingham R. J., Gish G., Pawson T. (2004) Oncogene 23, 1972–1984 [DOI] [PubMed] [Google Scholar]
- 22.Staub O., Dho S., Henry P., Correa J., Ishikawa T., McGlade J., Rotin D. (1996) EMBO J. 15, 2371–2380 [PMC free article] [PubMed] [Google Scholar]
- 23.Harty R. N., Brown M. E., Wang G., Huibregtse J., Hayes F. P. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 13871–13876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shearwin-Whyatt L. M., Brown D. L., Wylie F. G., Stow J. L., Kumar S. (2004) J. Cell Sci. 117, 3679–3689 [DOI] [PubMed] [Google Scholar]
- 25.Rougier J. S., van Bemmelen M. X., Bruce M. C., Jespersen T., Gavillet B., Apothéloz F., Cordonier S., Staub O., Rotin D., Abriel H. (2005) Am. J. Physiol. Cell Physiol. 288, C692–C701 [DOI] [PubMed] [Google Scholar]
- 26.Chang A., Cheang S., Espanel X., Sudol M. (2000) J. Biol. Chem. 275, 20562–20571 [DOI] [PubMed] [Google Scholar]
- 27.Léon S., Erpapazoglou Z., Haguenauer-Tsapis R. (2008) Mol. Biol. Cell 19, 2379–2388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nikko E., Sullivan J. A., Pelham H. R. (2008) EMBO Rep. 9, 1216–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin C. H., MacGurn J. A., Chu T., Stefan C. J., Emr S. D. (2008) Cell 135, 714–725 [DOI] [PubMed] [Google Scholar]
- 30.Sullivan J. A., Lewis M. J., Nikko E., Pelham H. R. (2007) Mol. Biol. Cell 18, 2429–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strochlic T. I., Schmiedekamp B. C., Lee J., Katzmann D. J., Burd C. G. (2008) Mol. Biol. Cell 19, 4694–4706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Odorizzi G., Babst M., Emr S. D. (1998) Cell 95, 847–858 [DOI] [PubMed] [Google Scholar]
- 33.Sikorski R. S., Hieter P. (1989) Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson J. S., Klionsky D. J., Banta L. M., Emr S. D. (1988) Mol. Cell. Biol. 8, 4936–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wendland B., Emr S. D., Riezman H. (1998) Curr. Opin. Cell Biol. 10, 513–522 [DOI] [PubMed] [Google Scholar]
- 36.Burd C. G., Peterson M., Cowles C. R., Emr S. D. (1997) Mol. Biol. Cell 8, 1089–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kasanov J., Pirozzi G., Uveges A. J., Kay B. K. (2001) Chem. Biol. 8, 231–241 [DOI] [PubMed] [Google Scholar]
- 38.Harty R. N., Brown M. E., McGettigan J. P., Wang G., Jayakar H. R., Huibregtse J. M., Whitt M. A., Schnell M. J. (2001) J. Virol. 75, 10623–10629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kato Y., Ito M., Kawai K., Nagata K., Tanokura M. (2002) J. Biol. Chem. 277, 10173–10177 [DOI] [PubMed] [Google Scholar]
- 40.Shcherbik N., Kee Y., Lyon N., Huibregtse J. M., Haines D. S. (2004) J. Biol. Chem. 279, 53892–53898 [DOI] [PubMed] [Google Scholar]
- 41.Stamenova S. D., Dunn R., Adler A. S., Hicke L. (2004) J. Biol. Chem. 279, 16017–16025 [DOI] [PubMed] [Google Scholar]
- 42.Huang L., Kinnucan E., Wang G., Beaudenon S., Howley P. M., Huibregtse J. M., Pavletich N. P. (1999) Science 286, 1321–1326 [DOI] [PubMed] [Google Scholar]
- 43.Verdecia M. A., Joazeiro C. A., Wells N. J., Ferrer J. L., Bowman M. E., Hunter T., Noel J. P. (2003) Mol. Cell 11, 249–259 [DOI] [PubMed] [Google Scholar]
- 44.Ogunjimi A. A., Briant D. J., Pece-Barbara N., Le Roy C., Di Guglielmo G. M., Kavsak P., Rasmussen R. K., Seet B. T., Sicheri F., Wrana J. L. (2005) Mol. Cell 19, 297–308 [DOI] [PubMed] [Google Scholar]
- 45.Gibrat J. F., Madej T., Bryant S. H. (1996) Curr. Opin. Struct. Biol. 6, 377–385 [DOI] [PubMed] [Google Scholar]
- 46.Berezin C., Glaser F., Rosenberg J., Paz I., Pupko T., Fariselli P., Casadio R., Ben-Tal N. (2004) Bioinformatics 20, 1322–1324 [DOI] [PubMed] [Google Scholar]
- 47.Schwede T., Kopp J., Guex N., Peitsch M. C. (2003) Nucleic Acids Res. 31, 3381–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Putz U., Howitt J., Lackovic J., Foot N., Kumar S., Silke J., Tan S. S. (2008) J. Biol. Chem. 283, 32621–32527 [DOI] [PubMed] [Google Scholar]
- 49.Ren J., Kee Y., Huibregtse J. M., Piper R. C. (2007) Mol. Biol. Cell 18, 324–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Row P. E., Prior I. A., McCullough J., Clague M. J., Urbé S. (2006) J. Biol. Chem. 281, 12618–12624 [DOI] [PubMed] [Google Scholar]
- 51.Mizuno E., Kobayashi K., Yamamoto A., Kitamura N., Komada M. (2006) Traffic 7, 1017–1031 [DOI] [PubMed] [Google Scholar]
- 52.Mund T., Pelham H. R. (2009) EMBO Rep. 10, 501–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bruce M. C., Kanelis V., Fouladkou F., Debonneville A., Staub O., Rotin D. (2008) Biochem. J. 415, 155–163 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.