Abstract
It is a commonly held view that oxidatively-induced DNA lesions are repaired by the base excision repair (BER) pathway, whereas DNA lesions induced by UV light and other “bulky” chemical adducts are repaired by the nucleotide excision repair (NER) pathway. While this distinction is generally accurate, the 8,5’ cyclopurine deoxynucleosides represent an important exception, in that they are formed in DNA by the hydroxyl radical, but are specifically repaired by NER, not by BER. They are also strong blocks to nucleases and polymerases, including RNA polymerase II in human cells. In this review, I will discuss the evidence that these lesions are in part responsible for the neurodegeneration that occurs in some XP patients, and what additional evidence would be necessary to prove such a role. I will also consider other DNA lesions that might be involved in XP neurologic disease. Finally, I will also discuss how our recent studies of these lesions have generated novel insights into the process of transcriptional mutagenesis in human cells, as well as the value of studying these lesions not only for a better understanding of NER, but also for other aspects of human health and disease.
1. Introduction
The 8,5’-cyclopurine nucleosides1 were originally discovered by workers interested in the effects of radiation on DNA [1–3]. Some work on these lesions continued through the mid-1980 and 90s [4,5], but interest in them then declined, perhaps due in part to the identification and popularization of 8-oxo-7,8-dihydro-2’-deoxyguanosine (8-oxo-dG) as a marker of oxidative DNA damage.
Interest in the 8,5’-cyclopurine-2’-deoxynucleosides (cyPudNs) was revived in 2000 by reports from two groups [6,7] demonstrating that cyPudNs are substrates for the nucleotide excision repair (NER) pathway, but not for other repair pathways. These observations led both groups to propose that these lesions may be responsible, at least in part, for the neurodegeneration suffered by xeroderma pigmentosum (XP) patients who lack the capacity to carry out NER. The interest in identifying oxidative DNA lesions repaired by NER was stimulated by the fact that the wavelengths of UV light responsible for generating the DNA lesions known to be repaired by NER could not penetrate into the human brain, and therefore could not be responsible for XP neurologic disease [8].
In a recent review, I described the clinical and neuropathological aspects of XP neurologic disease, and the evidence that cyPudNs are involved in this disease [9]. Therefore, in the present work, I will summarize those arguments and findings, but focus primarily on more recent data not covered in the last review, as well as other data related to these unique oxidatively induced DNA lesions.
2. Cyclopurine deoxynucleosides: chemistry, formation, and structural considerations
2.1 Chemistry of CyPudN formation
CyPudNs are DNA lesions that result from the attack of the hydroxyl radical on DNA and specifically on 2’-deoxyadenosine (dA) and 2’-deoxyguanosine (dG) [10]. CyPudNs have been shown to form from the hydroxyl generated by ionizing radiation and from the Fenton reaction [11,12], which is the most relevant source of hydroxyl radicals in vivo. These lesions do not result from singlet oxygen or other types of reactive oxygen species.
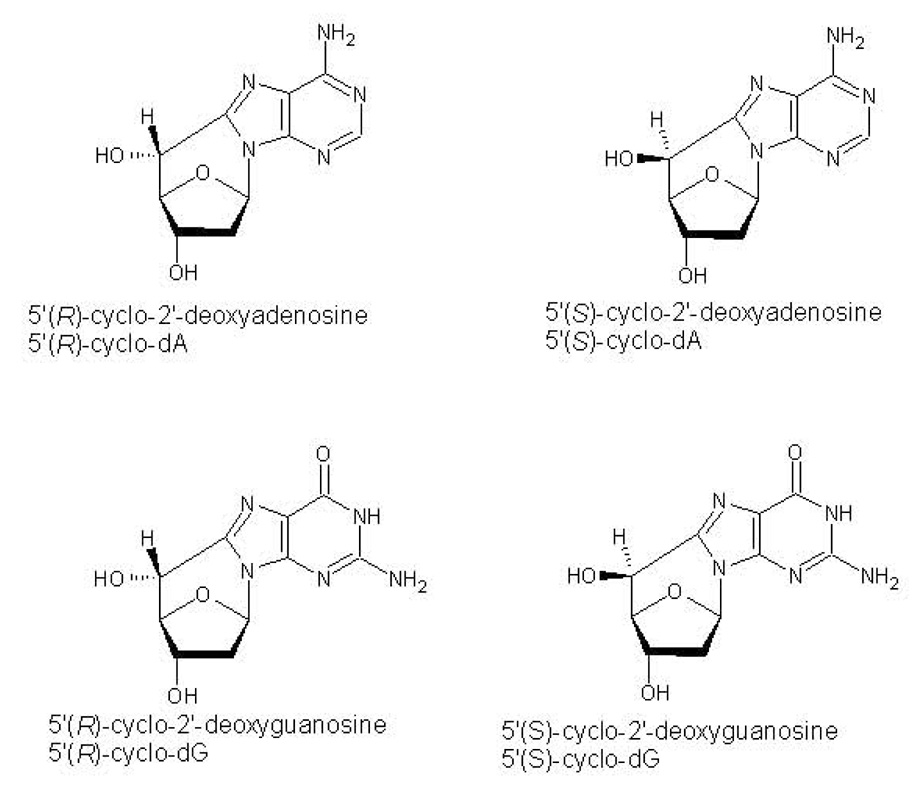
Both 8,5’-cyclo-2’-deoxyadenosine (cyclo-dA) and 8,5’-cyclo-2’-deoxyguanosine (cyclo-dG) can exist in either of two diastereomers, denoted 5’R and 5’S (see figure 1). Interestingly, some studies have indicated that the R and S forms of these lesions have different biological properties [7,13], observations that may ultimately give insight into the mechanistic basis of enzymes that act on DNA.
1.
Chemical structures of the R and S forms of the cyPudNs.
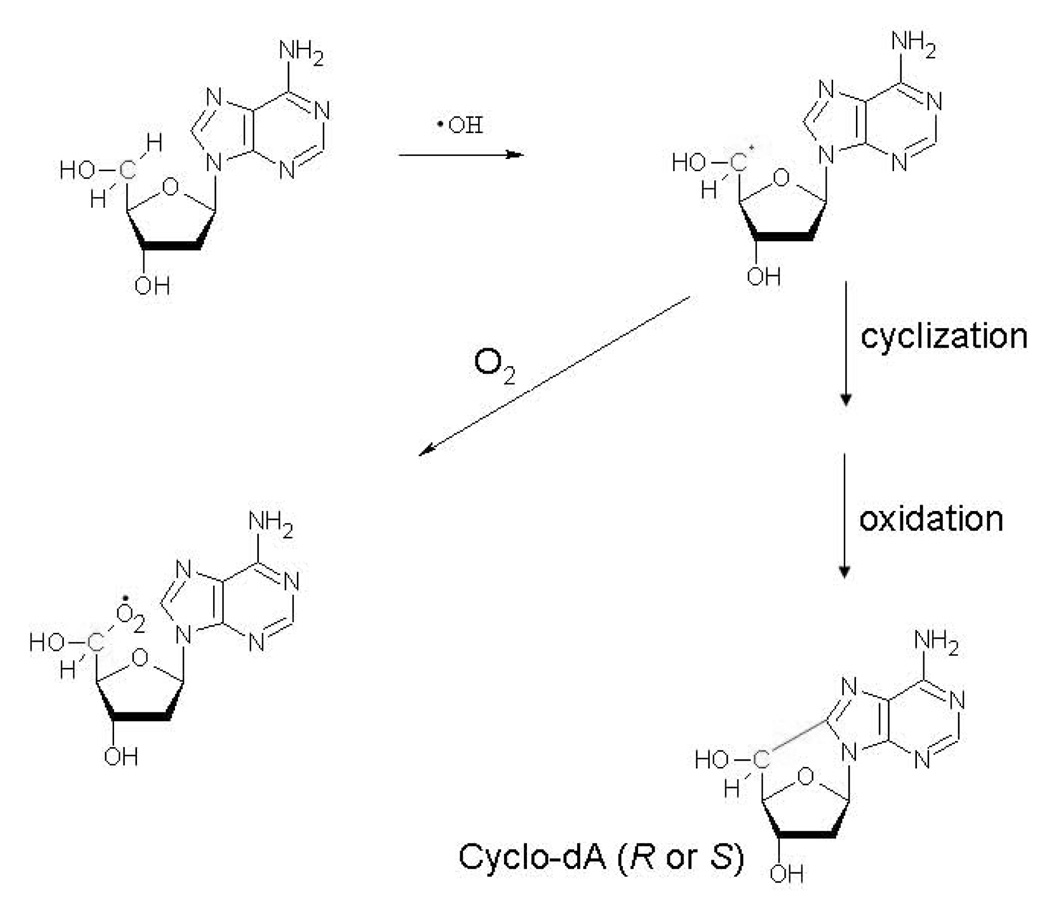
The basic mechanism of formation of cyPudNs involves a 2-step process [14], as illustrated in Fig. 2. In the first step, the hydroxyl radical reacts with the sugar moiety of the nucleoside by abstracting an H atom from C5’, resulting in the formation of the C5’ radical of the sugar moiety. This radical attacks the double bond between N7 and C8 of the purine base, ultimately resulting in the formation of the covalent 8, 5’ bond. The R isomers of both cyclo-dA and cyclo-dG were predominantly formed when single-stranded DNA was irradiated, whereas the S isomers predominated in double-stranded DNA [4].
2.
The mechanism of cyPudN formation, and the role of molecular oxygen in preventing lesion formation.
Recently, Chatgilialoglu and colleagues have carried out a series of further mechanistic studies of cyPudNs formation [15–20]. In addition to discovering better synthetic routes for cyPudNs, these studies have yielded a number of important mechanistic insights into the stereochemistry and energetics of cyPudNs formation, including the determination of a rate constant for the cyclization reaction (1.6 × 105/s) [20]. Because the second step in cyPudNs formation is relatively slow, it can be modified by intracellular components. Molecular oxygen (O2) can react with the C5’ radical at diffusion-controlled rates, resulting in the production of a peroxyl radical rather than a cyPudN (Fig. 2). However, because the O2 concentration in the eukaryotic cell nucleus is relatively low [21,22], the inhibitory effect of O2 on cyclization may be of limited importance in this compartment. On the other hand, the inhibitory effect of O2 on cyclization could be very important in the mitochondria, given the flux of O2 through this organelle. At present, there is no information on whether cyPudNs can form in mitochondrial DNA.
In addition to O2, Chatgilialoglu and colleagues have proposed that intracellular glutathione could theoretically prevent cyPudN formation by reacting with the C5’ radical intermediate [20]. Such a “repair” reaction could represent a biologically relevant protective role against the formation of cyPudNs or any other lesions.
2.2 Structural Alterations in CyPudN
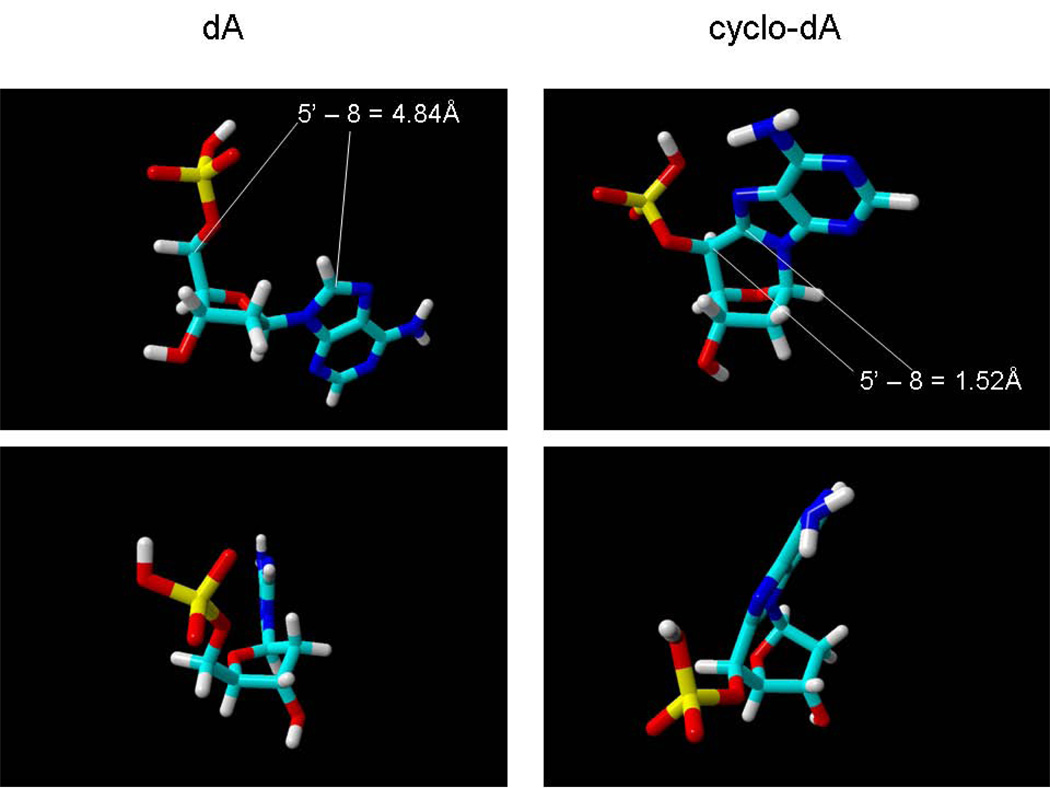
Figure 3 shows energy-minimized models [23] of dA and cyclo-dA, viewed from different orientations, to illustrate the most important structural alterations caused by the additional 8,5’ bond. First, the additional bond locks the base into an anti configuration, and displaces C8 of the purine base by approximately 3Å from its position in dA (Fig.3, top panels). In a dsDNA molecule, this would have the effect of pulling the base towards the sugar-phosphate backbone, away from its hybridization partner. Such an effect would explain the reduction in melting temperature of DNA containing cyPudNs observed experimentally [24]. The second notable feature is unusual sugar pucker, which is imposed upon the molecule by the covalent 8, 5’ bond. Finally, the position of the adenine base relative to the sugar, including the χ angle, is altered, which would affect base stacking interactions, and may therefore play a role in damage recognition [25].
3.
Energy-minimized models of dA (left panels) and 8, 5’-(S) cyclo-dA (right panels) viewed from different perspectives. Modeling was carried out using YASARA-WHATIF (www.Yasara.com)[23]. In the top panels, the distance between the 5’ and 8 carbons is indicated. For comparison, the measured distance between the 5’ and 8 carbons in a crystal structure of cyclo-A is 1.513 Å [26].
Although the structure shown in Fig. 3 is based on energy minimization in silico, it is consistent with many of the structural features of cyPudNs derived from direct measurements of 8,5’-cycloadenosine [26]. As such, this model can serve as a useful framework for understanding the biological effects of cyPudNs, and the development of testable hypotheses regarding effects of these lesions on enzymes.
3. XP Neurologic Disease
3.1 Overview
As discussed above, a major reason for the current interest in cyPudNs is their possible role in XP neurologic disease. In this section, I will briefly outline XP neurological disease, and then review the evidence for a role for cyPudNs in this disease in the next section. The clinical and neuropathological manifestations of XP neurological disease were described in detail in the last review [9], and the reader is referred to that paper and references therein for a more detailed description. In addition, the relationship between XP, NER, and neurological disease is discussed in the contributions of Nouspikel and Niedernhofer, in this Volume.
As pointed out by Friedberg et al [27], the first XP patient with features of neurological disease (“idiocy”, dysarticulation) was described by Neisser [28]. Subsequently, DeSanctis and Cacchione described three patients with the cutaneous features of xeroderma pigmentosum, as well as dwarfism, neurologic disease, and immature sexual development [29]. They also carried out a postmortem neuropathological examination of the brain of one of the patients, and noted the loss of neurons in the cerebellum and cerebral cortex. As a result, the term DeSanctis-Cacchione syndrome is sometimes used to describe patients with XP and severe neurological abnormalities. However, the use of this term can sometimes be confusing (see [9]). Therefore, in this paper I will use the term XP neurologic disease [30], which is preferred because it is a specifically defined clinical entity, and can also encompass later-onset forms of the disease which can develop in patients with normal growth and sexual development [31].
3.2 XP Complementation Groups and Neurologic Disease
XP neurologic disease is observed in patients from complementation groups A, C, and D [30,31]. Late-onset neurologic disease has been reported in XPF patients with hypomorphic mutations [32,33], however, the progression of neurological symptoms in these patients is somewhat different than classic XP neurologic disease. The most severe and earliest onset cases of XP neurologic disease are seen in XPA patients. There is a relatively common “founder” XPA mutation in the Japanese population [34,35] that results in severe XP neurologic disease, with an age of onset between 7–13 years. Other combinations of XPA alleles can result in disease onset even earlier, or much later, depending upon the biochemical consequences of the particular mutations [36–39]. XP neurologic disease is observed in group D patients [36] who have mutations that specifically affect the NER function of the XPD protein [40]. The onset of neurologic disease in these patients is typically in the teens or early 20s. In contrast, XPD mutations that affect other functions of the protein, such as kinase activity of interaction with other TFIIH components, result in either XP-CS or trichiothiodystrophy (TTD), two qualitatively different disorders [40] (see also below). Genotype-phenotype relationships in complementation group D are very complex [41], which may be due in part to the ability of different mutant “null” alleles to complement each other [42].
It is often not appreciated that XPC patients can also develop neurologic disease, although it is of late onset and clinically asymptomatic. However, disease progression can be followed by careful clinical testing, particularly of the patients hearing ability [43]. It is worth emphasizing here that the well-known distinction between TC-NER (which required the CSA and CSB proteins) and GG-NER (which requites the XPC protein) applies to dividing cells. In contrast, terminally differentiated cells, including neurons, have a different pattern of NER within the genome, termed domain-associated repair (DAR). In DAR, both the transcribed and non-transcribed strands of active genes are repaired, whereas NER in non-transcribed DNA sequences is essentially undetectable [44]. Because DAR requires the XPC protein [45], the neurologic disease observed in XPC patients may be related to the role of XPC in DAR. Based on the proposal of Nouspikel et al, [45] (see also Nouspikel, this Volume) a plausible explanation for neurodegeneration in XPC is that, because the non-transcribed strand of DNA serves as a template for repair, failure to repair the non-transcribed strand will eventually impact repair of the transcribed strand.
3.3 XP Neurologic Disease is a Primary Neurodegeneration, and is Qualitatively Different Than CS or TTD Neurologic Disease
XP neurologic disease is a primary neurodegeneration, in that there is no evidence of other factors such as amyloid deposits or spongiform encephalopathy. The disease is characterized by the loss of large neurons, not only in many regions of the brain and spinal cord, but also in the peripheral nervous system (for review see [9]). In fact, the earliest manifestations of XP neurologic disease are sensorineural hearing loss and areflexia, likely due to degeneration of neurons in the dorsal root ganglia which are part of the peripheral nervous system. Moreover, in the neuropathological study of the XPC patient described above, direct evidence of neuropathology was only observed in the peripheral nervous system [46].
It is important to stress that XP neurologic disease is qualitatively different than the neurologic disease that affects patients with Cockayne syndrome (CS) and XP patients in complementation groups B, D, and G that have the combined features of XP and CS [31]. In XP neurologic disease, it is the neurons that are primarily affected. In contrast, in CS, the most prominent degenerative change that is observed is an unique form of demyelination (leukodystrophy) called tigroid demyelination [31]. This observation indicates that it is the myelin-forming glial cells (oligodendrocytes) that are primarily affected in the brains of CS patients. Many CS patients also develop calcium deposits in the brain, particularly in the basal ganglia and cerebellum. Neither tigroid demyelination nor calcification is observed in XP neurological disease. TTD patients also have myelination defects [41] and may have brain calcification; thus, TTD neurologic disease is more similar to CS neurologic disease than to XP neurologic disease.
3.3. Evidence That XP Neurologic Disease Results From Defective NER
The evidence that XP neurologic disease results from defective NER is compelling, as it is based on several overlapping and mutually reinforcing lines of evidence. First, the severity of XP neurologic disease correlates with the inability of cells to survive exposure to UV radiation [47], a measure of NER in essential genes. Second, XP neurologic disease is observed in patients in complementation groups A, C, and D. The only known function of the XPA protein is in NER, and although XPD and XPC proteins have additional functions [48,49], the only function that the three proteins have in common is NER. Third, in XPD patients, mutations which inactivate the NER function of XPD result in XP neurologic disease, whereas XPD mutations that result in a loss of XPD protein, and thus loss of the additional functions of XPD (transcription and nuclear receptor kinase activity) result in CS or trichiothiodystrophy (TTD) [40]. Finally, XP variant patients, who manifest the cutaneous features of XP due to mutation in the XPV gene have normal NER, do not develop neurologic disease.
As noted above, the neurologic diseases observed in CS and TTD are qualitatively different than XP neurologic disease. Consistent with these differences, there is increasing evidence that the mechanistic basis of these neurologic diseases are fundamentally different as well. Specifically, Compe et al [50] recently showed that the myelination defects observed in a mouse model of TTD [51] are best explained not by a DNA repair deficiency, but rather by the loss of a co-activator function of TFIIH in thyroid hormone regulated gene expression in the brain. Furthermore, at least some of the clinical manifestations of XPG-CS patients are likely to be due to defects in nuclear hormone dependent transcription, rather than to DNA repair defects [52,53].
4. CyPudNs and XP neurologic disease
4.1. Background
The scientific rationale for the possibility that cyPudNs are involved in XP neurologic disease derives from three primary sources. Robbins and colleagues [8] initially proposed that since UV light cannot access neurons in the human brain, XP neurologic disease is most likely due to the failure to repair DNA damage resulting from an endogenous damage, such as that caused by free radicals [30]. Dizdaroglu and colleagues [4,5,14] showed that both cyclo-dA and cyclo-dG could be formed in DNA, and proposed that these lesions were likely to be substrates for NER but not BER. Subsequently, Lindahl and colleagues [54] showed that exposing plasmid DNA to hydroxyl radicals from either ionizing radiation or the Fenton reaction resulted in the formation of some type of DNA damage that could be specifically repaired by the NER pathway in cell extracts. The DNA molecules used as substrates in that work had been treated with specific DNA glycosylases to remove 8-oxo-dG and thymine glycol, and were covalently closed plasmids, ruling out the possibility that the damage repaired by NER in the experiment were either single-strand or double strand breaks. In the same paper, XP cells were shown to repair single-strand breaks normally [54]. These and other experiments provided the first direct evidence that the hydroxyl radical could generate some class of DNA lesion(s) that are specifically repaired by the NER pathway, and therefore might be involved in XP neurologic disease. Based on these experiments, the authors discussed possible oxidatively induced DNA lesions that might be specific substrates for NER, including cyPudNs.
Against this background, in 2000, different research groups [6,7] showed that cyPudNs are in fact substrates for NER, but are not detectably repaired by glycosylase-initiated base-excision repair, or any other repair pathway detectable in human cell extracts. Both groups therefore proposed that XP neurologic disease may result from the inability to repair cyPudNs [6,7].
4.2. The Case for CyPudNs as Neurodegenerative DNA Lesions in XP
As discussed in detail in the earlier publication [9], and expanded upon below, the case for cyPudNs as causative DNA lesions in XP neurologic disease can be summarized as follows:
1. CyPudNs are substrates for NER that is defective in XP patients who develop XP neurologic disease [6,7].
2. CyPudNs are not substrates for any other DNA repair pathway in mammalian cells [6,7].
3. CyPudNs are strong blocks to gene expression in XP cells [6,55], a biological effect that is compatible with causing neurodegeneration. They also stimulate the production of mutant RNA transcripts in human cells [56], as discussed in more detail below.
4. CyPudNs are chemically stable, endogenous DNA adducts [57–59]. As pointed out by Lindahl [60], DNA lesions that are inherently chemically unstable could not accumulate to a significant extent even in the absence of repair.
5. Recent data supporting a role for cyPudNs in XP neurologic disease
Within the past year, two additional studies have been published which provide further support to the role of cyPudNs in XP neurological disease, and these are reviewed below.
5.1. The Glycosidic Bond in Cyclo-dA is Stabilized Against Spontaneous Hydrolysis
As described above, efforts to detect repair of cyPudNs by either glycosylase-initiated BER or direct repair have been unsuccessful (see [61] for a recent attempt). Thus, the totality of the evidence indicates that NER is the only enzymatic pathway that can remove cyPudNs from DNA. Given the existence of NER in all forms of life from microorganisms to humans, it is difficult to envision what the evolutionarily selective pressure would be for the enzymatic “back-up” pathway for cyPudNs repair. However, the possibility that some organism may have an enzymatic activity cannot be formally ruled out.
In the absence of NER or a specific DNA repair pathway, one conceivable mechanism that could operate to remove cyPudNs from DNA would be a combined degradation/repair mechanism, in which a non-enzymatic chemical alteration of the cyPudNs would render it susceptible to “gratuitous” repair by an alternate DNA repair pathway which is primarily involved in the repair of some other type of DNA lesion. One possibility for such a chemical alteration would be hydrolysis of the 1’–9 N-glycosidic bonds that link the purine base to the sugar moiety in DNA. This bond is known to be relatively unstable under physiological conditions [60]. Indeed it is the instability of this chemical bond that is responsible for the estimated loss of approximately 20,000 purine residues per cell per day in a mammalian cell [62]. In view of the evidence that many other chemical base modifications can destabilize the glycosidic bonds in DNA [63,64], we considered the possibility that the glycosidic bond might also be destabilized in cyPudNs. If so, and if the resulting hydrolyzed form of cyPudN could be subsequently repaired (e.g. by an AP endonuclease), such a coupled degradation-repair mechanism could be of biological significance.
To address the first step in this hypothetical mechanism, we [65] developed a method to detect cyclo-dA containing a hydrolyzed glycosidic bond, and used this method to compare the stability of the glycosidic bond in cyclo-dA against acid hydrolysis, relative to the situation in dA. The use of acid hydrolysis in these experiments was selected because spontaneous glycosidic bond hydrolysis is acid-catalyzed [64]. However, while the combined degradation/repair model described above requires that the glycosidic bond in cyclo-dA to be destabilized, we found that the glycosidic bond in 8’5’ bond in cyclo-dA is in fact more stable than in dA against acid hydrolysis, by a factor of approximately 40-fold [65]. Therefore, this observation essentially rules out the possibility that spontaneous hydrolysis of the glycosidic bond of cyPudNs in DNA occurs in vivo to any biologically significant extent. By extension, therefore, the question of whether AP endonuclease or any other enzyme could process an altered cyclo-dA residue resulting from spontaneous glycosidic bond hydrolysis becomes irrelevant, and this combined degradation/gratuitous repair mechanism can therefore be effectively excluded from further consideration as a realistic mechanism to remove cyPudNs from DNA in the absence of NER.
5.2. Cyclo-dA Strongly Blocks Transcription, and Also Stimulates Transcriptional Mutagenesis
In our initial studies of the effects of DNA lesions on gene expression in XPA cells, we demonstrated that although a single cyclo-dA or a single thymine dimer were strong blocks to transcription, the blockage was not absolute [6]. Specifically, in these original studies we detected an approximately 75% reduction in luciferase activity as a result of a single cyclo-dA or single thymine dimer on the transcribed strand of an luciferase reporter gene [6]. In that work, it was difficult to determine whether the residual luciferase activity we detected was due to bypass of the lesion during transcription, or some other process.
In view of the demonstration that the XPV gene product could bypass thymine dimers and other “replication-blocking” DNA lesions [66,67], as well as a discovery of transcriptional mutagenesis by Doetsch and colleagues [68], we developed a method to search for transcriptional mutagenesis resulting from bypass of cyclo-dA and other lesions in human cells [56]. Using this method, we discovered two novel types of mutant RNA transcripts resulting from the bypass of the cyclo-dA lesion (see Fig. 4).
4.
RNA transcripts resulting from bypass of cyclo-dA in human cells [56]. The double-stranded DNA template that was transfected into cells is shown at the bottom. The location of the cyclo-dA lesion is indicated by the red italic A.
In the first type, which we refer to as a 5' A mutation, the polymerase correctly incorporates UMP opposite cyclo-dA, but then misincorporates an AMP opposite the next template dA residue 5’ to cyclo-dA. The second type contains deletions of 7, 13 or 22 nucleotides beginning after polymerase incorporation of UMP opposite the lesion. Because these transcripts contained deletions of multiple nucleotides, we refer to them multiple nucleotide deletions (MNDs) [56].
Importantly, we also detected multiple nucleotide deletions in RNA from XPA cells transfected with a thymine dimer containing construct. In this case, the transcripts contained a 12 nucleotide deletion following incorporation of A opposite the 3’ T of the TT-dimer. Thus, MNDs resulting from the TT-dimer result from a very similar mechanism as observed with cyclo-dA. However, the exact sequences of the transcripts derived from the two lesions are in fact different. This is as expected given that the transcripts resulted from two different DNA lesions located in slightly different sequence contexts, and rules out the possibility that the MNDs we detected from the TT-dimer construct were the result of contamination. Furthermore, the fact that we detected MND transcripts from both the cyclo-dA- and TT dimer-containing constructs rules out the possibility that such transcripts resulted from a rare population of mutant plasmids used to prepare the constructs or from some peculiarity with the specific lesion-containing oligonucleotides. Rather, these results indicate that the formation of multiple nucleotide deletions are a general outcome, albeit a rare one, of the interaction between RNA polymerase II and a helix-distorting DNA lesion in a human cell.
Both 5’A and MND transcripts resulting from cyclo-dA were observed in cells from an XPA patient, an XPD patient, and a CSB patient [56]. Interestingly, however, we detected a small but statistically significant decrease in the ratio of MND to 5’A transcripts in cells from the CSB patient compared to the XPA and XPD patients, suggesting that the CSB protein may modulate the probability of MND transcript formation.
5.2.1. Transcriptional Mutagenesis: Mechanistic Considerations
From a mechanistic standpoint, the question of how MND transcripts are generated is particularly interesting. Inspection of the sequences of the MNDs resulting from cyclo-dA showed that these transcripts could be explained by the polymerase first incorporating UMP opposite the lesion, followed by a dislocation of the template strand resulting in the newly incorporated UMP being opposed to a downstream template dA residue, which could then act as a primer for continued transcription. The sequence of the MND transcript resulting from the TT-dimer could be explained by an analogous mechanism. For a working model of such a mechanism and more details, see [56].
Another notable feature of the effects of cyclo-dA on transcription and human cells that we detect is that in all three types of transcripts produced, i.e. wild-type, 5' A mutations, and MNDs, the polymerase incorporates the correct nucleotide, i.e. UMP opposite cyclo-dA. Thus, at the level of transcription, cyclo-dA does not directly stimulate misincorporation. The lack of direct misincorporation opposite cyclo-dA can be understood in terms of the structure of the lesion. As noted above, the main effect of the additional bond is to displace the adenosine base back towards the sugar phosphate backbone, away from the incoming rNTP. However, the additional C8-C5’ bond does not directly adduct any of the atoms in the adenine base that are involved in Watson-Crick base pairing.
Another implication of our results is that they indicate the existence of a mechanistic branchpoint occurring after the polymerase incorporates UMP opposite cyclo-dA. In kinetic terms, this implies that incorporation of UMP opposite cyclo-dA is “fast” relative to subsequent steps leading to lesion bypass. Ongoing studies in our laboratory, using a minimal transcription system, are generally consistent with this scheme (Cheng, Jia, and Brooks, manuscript in preparation). Ultimately, much insight into these mechanistic questions will be gained by direct observation of the polymerase transcribing up to and past the cyclo-dA lesion, which can be achieved using X-ray crystallography.
5.3 Transcriptional Mutagenesis versus Transcription Blocking: Implications for XP Neurologic Disease
Considering the relative significance of transcription blockage versus transcriptional mutagenesis by cyPudNs in relation to XPA neurologic disease, the transcription blocking effect would appear to be much more important, for several reasons. First, the transcription blocking effect is quantitatively more important. Our original and subsequent studies have consistently shown that a single cyclo-dA lesion reduces luciferase activity by approximately 75% in XP cells and in vitro studies using purified RNA polymerases or nuclear extracts are consistent with this magnitude of transcription blockage (unpublished observations). Secondly, a transcription-blocking DNA lesion will have the same effect on gene expression wherever it is located in transcribed sequence (5’ or 3’ untranslated sequence, intron or exon). In contrast, TM will only result in an abnormal protein if it causes a change in the amino acid sequence of the encoded protein, which is not possible if it is located within an untranslated sequence, or the wobble position of a codon. In this regard, it is worth noting that in human genes, introns can be very large, i.e. 10 kB or greater.
It is theoretically possible that transcriptional mutagenesis from bypass of cyPudNs could cause the continued production of some very stable type of toxic mutant protein, such as amyloid or prion protein, which could accumulate, leading to neuronal death. However, neuropathological studies have not detected amyloid accumulation in the brains of XP patients, nor is there any evidence of spongiform encephalopathy which is characteristic of prion diseases [69]. For all these reasons, therefore, although the production of mutant or non-functional proteins due to TM could certainly contribute to neurodegeneration in cells in which they are produced, the neurodegeneration observed in XP patients is most likely primarily due to the transcription-blocking effects of cyPudNs as well as other endogenous transcription-blocking DNA lesions that are repaired by NER as well.
While stalling of RNAPII at DNA lesions can trigger P53-dependent apoptosis [70–72], it remains to be determined whether neurodegeneration in XP is due to this mechanism, as opposed to a progressive inactivation of essential gene products ultimately resulting in neuronal death. This topic, as well as different models of the neuronal death process in XP, are discussed in more detail in the previous review [9].
6. Other DNA Lesions and XP Neurologic Disease
6.1. Lesions Repaired by BER
Given the evidence that XP neurologic disease results from defective NER, the most likely mechanisms is the accumulation of DNA lesions that are specifically repaired by NER, but not by other pathways such as BER. Therefore, experiments designed to identify such lesions [54,73] are of particular relevance and interest in this context. It follows that DNA lesions which are primarily repaired by pathways other than NER, such as BER, are unlikely to play a role in XP neurologic disease. Since NER has an almost infinite substrate range, it is not surprising that 8-oxo-dG, thymine glycol, and urea are substrates for NER in vitro, [74] even undamaged DNA can be processed to some extent by NER in vitro [75]. However, in vivo studies have shown that cells from XP NER deficient XP patients repair 8-oxo-dG normally [40,76] (although cells from the rare XPD patients that display the clinical features of CS exhibit an unusual strand-break response to response to DNA containing 8-oxo-dG ; see [40,77]).
Recently, direct evidence against the hypothesis that neurodegeneration resulting from NER deficiency is due to the loss of the back-up repair of 8-oxo-dG comes from studies in mice [78]. When Csb−/− mice (which do not develop neurodegeneration) were crossed with Xpc−/− mice, an early onset neurodegeneration was observed, particularly in the cerebellum. In contrast, when the Csb−/− mice were crossed with Ogg1−/− mice, no neurodegeneration was observed. It seems difficult to argue that the neurodegeneration observed in the Csb−/− Xpc−/− mice was due to the loss of NER acting as a “back-up” repair pathway for 8-oxo-dG removal, when complete inactivation of the main repair pathway for 8-oxo-dG had no such effect.
6.2. Propano-deoxyguanosine adducts
Aside from the cyPudNs, more realistic candidate neurodegenerative DNA lesions in XP are the exocyclic propano-deoxyguanosine (PdG) adducts derived from lipid aldehydes such as crotonaldehyde, acrolein, malondialdehyde and 4-hydroxynonenal [79,80]. The malondialdehyde PdG adduct can arise in DNA from base propenals [81]. Like cyPudNs, and in contrast to 8-oxo-dG, these lesions are specifically repaired by NER [82], and they are substantial blocks to transcription by RNA polymerase II, at least in vitro [83]. However, PdG adducts can undergo chemical rearrangements into alternate forms [84] that are substrates for DNA repair reactions other than NER. Also, these lesions are adducted to guanine via nitrogen-carbon bonds, which are less stable than the carbon-carbon bonds in cyPudNs. For both reasons, these lesions are likely to be less stable in DNA over time in the absence of NER than the cyPudNs. Still, increased levels of transcription blockage by unrepaired PdG lesions may also contribute to XP neurologic disease.
6.3. Intrastrand DNA Cross-Links Resulting From Oxidative Stress
Several DNA intrastrand cross-links resulting from the attack of free radicals on DNA have been described [85–89]. These lesions appear structurally analogous to the canonical types of DNA lesions repaired by NER i.e. the thymine dimers and 6–4 product. One of these crosslinks has been shown to be a substrate for the bacterial NER system in vitro [90]. Based on their structures, it is highly likely that such lesions would be strong blocks to transcription in human cells. While at present there is no published evidence that they are endogenously formed, the fact that some of these lesions can be induced in DNA under Fenton reaction conditions [91,92] indicates that they can occur in vivo. An intriguing possibility is that one or more of these may correspond to endogenous DNA modifications (I-compounds) detected by the 32P-postlabelling assay [93]. Based on chemical composition, some I-compounds resulting from oxidative stress were previously proposed to represent intrastrand DNA crosslinks involving purines [94–96]. We were able to identify a subset of these I-compounds as dinucleotides containing cyclo-dA [57], but the chemical structure of others remain unidentified.
6.4. Other Endogenous DNA Lesions?
Finally, it is entirely possible that other endogenous DNA lesions can be formed in neurons that require NER for their removal, and have yet to be discovered. The products of oxidative damage to DNA have been thoroughly investigated [97], and it seems likely that the majority of oxidatively induced DNA lesions that can be formed have been identified. However, many other cellular components can react with DNA to form stable adducts, including lipids (see above), neurotransmitters and their derivatives [98,99], steroid hormones [100], and leukotrienes [101]. Many of these are cell-type specific, and therefore, DNA adducts resulting from them would only affect a subset of cells. Until recently, most of these adducts have been identified by reacting specific chemicals of interest with nucleosides or DNA. However, a more global, unbiased approach to endogenous DNA adduct detection would be much more efficient and useful.
A very interesting and important step in this regard has been taken by Kanaly et al [102], who have applied HPLC-MS-MS to the DNA “adductome” of human tissue samples. In addition to detecting several known DNA adducts, including the PdG adducts, the authors have identified numerous additional adducts, some of which are tissue-specific. Further studies of this type, especially applied to tissue samples from repair-deficient individuals, should identify other endogenous DNA adducts that are substrates for NER, and thus of potential relevance for XP neurological disease.
7. CyPudNs and XP Neurologic Disease: Summary and Remaining Issues
In this review, I have sought to outline some basic aspects of how cyPudNs are formed, their biological properties and repair, and the evidence that they are involved in the pathogenesis of XP neurologic disease. In making the case for the cyPudNs as neurodegenerative DNA lesions in XP, I am not claiming that they are the only lesions responsible; rather, my argument is that based on a reasonable set of criteria they represent the best candidates of any currently known DNA lesions for a role in XP neurologic disease. As more data becomes available about some of the other DNA lesions, such as the oxidatively induced DNA crosslinks, the case that these lesions may also play a role in XP neurologic disease may be strengthened. In addition, as indicated above, it is likely that additional DNA lesions will be identified in the human brain that are substrates for NER, and therefore involved in XP neurologic disease as well. A comprehensive catalog of the human brain “adductome” is therefore important; not only for XP neurologic disease, but for understanding the other neurologic disease resulting from DNA repair defects (see other contributions in this Volume).
An essential criterion that must be met before a role for cyPudNs in XP neurologic disease can be accepted is that the lesions should be shown to accumulate in brain tissue from XP patients or mice relative to controls. While this seems straightforward to test, in fact this question is substantially complicated by two factors: the heterogeneity of NER within the genome, and the cellular heterogeneity of the brain. The GG-NER pathway does not function effectively in neurons or other terminally differentiated cells, even those with normal NER genes [44]. Since assays of DNA damage in total DNA samples reflect GG-NER, it follows that one would not expect to see a difference between normal and XP samples when measuring DNA lesion levels in a portion of the genome where repair does not function even in normal individuals. With regard to cellular heterogeneity, in XP neurologic disease, it is the neurons that are most severely affected. However, neurons make up only about 10% of the cells in the brain, the rest being glial cells. So, measuring DNA adducts in total brain DNA means that one is measuring DNA adducts primarily in non-transcribed sequences in glial cell DNA. This problem is exacerbated even further in postmortem brain tissue from patients with XP neurologic disease, in which, by definition, much neuronal death has already occurred. When neurons die, glial cells, which retain the capacity to divide, proliferate.
Taking these points into considerations, to rigorously test the role of cyPudNs or other DNA lesions in XP neurological disease, it will be necessary to develop is a method to quantitate lesion levels on the transcribed strand of active genes in neurons, analogous to the classic gene and strand-specific NER assays At present, there is no method available for doing this, but further improvements in DNA adduct measurement techniques, coupled to methods for cellular fractionation and enrichment for specific sequences, should allow these limitations to be overcome in the future. An antibody that can specifically detect cyPudNs would be particularly useful in this regard, and efforts to develop such a reagent are ongoing.
8. Regulation of Repair of CyPudNs Following Ionizing Radiation (IR)
8.1 Is NER Inducible by IR?
Finally, I would like to highlight two recent papers [103,104] which raise interesting questions about the regulation of the NER pathway, and also highlight some unique advantages to assaying cyPudNs to investigate this pathway. In these studies, human keratinocytes were exposed to oxidative stress from ionizing radiation or a chemical (KBr) to increase levels of cyPudNs. Then, the decline in levels of cyPudNs back to basal levels was taken as a measure of repair of these lesions. The authors found that keratinocytes from both an XPC patient and a CSA patient were deficient in repair of cyPudNs using this method. While the defective in repair in XPC cells is consistent with the known role of XPC in global genome NER, defective repair in CSA is somewhat surprising, since it is thought to be involved in TC-NER. However, there are data suggesting a role for the CS proteins in GGR [105].
The other interesting aspect of these studies is that the basal levels of cyPudNs were not different in XPC vs. WT cells. Basal levels of DNA lesions in repair-proficient cells are generally assumed to reflect a steady-state between lesion formation and repair. However, the lack of a difference in basal cyPudN levels between the WT and XPC cells is inconsistent with this idea. One interpretation of these results is that these cells utilize DAR (i.e. NER only functions in transcribed DNA) in the absence of exogenous DNA damage, and GG-NER is induced by IR. Although DAR was originally discovered in terminally differentiated cells, Nouspikel et al [45] proposed that it may also apply to some proliferating cells as well. Also, the concept that GG-NER is inducible by DNA damage is not new; studies by Ford and Hanawalt showed that GG-NER of thymine dimers and chemical adducts in human cells is dependent upon the P53 gene activation [106].
8.2 Is GG-NER Operative in Cells That Are Not Exposed to Exogenous DNA Damage?
The classic methods that have been used to study NER (cell survival assays, unscheduled DNA synthesis, and recovery of RNA synthesis) involve exposing cells to exogenous DNA damage. There is no doubt that such assays have generated many important and fundamental insights into DNA repair and skin cancer in humans. However, with such assays alone, it is not possible to determine the status of NER in the absence of exogenous DNA damage. Because the cyPudNs are endogenous lesions that are specific substrates for NER, assaying these lesions provides a method of assessing GG-NER in cells without exposing them to any exogenous DNA damage. Therefore, the results of such assays can provide a novel perspective to the complex and controversial issue of GG-NER in quiescent and terminally differentiated cells [107,108].
9. General Summary and Future Perspectives
In summary, cyPudNs are a unique and interesting class of endogenous DNA lesions, in that they are generated in DNA by oxidative stress, but are specifically repaired by the NER pathway. As such, the biological properties of these lesions are much more like those of other NER substrates such as thymine dimers than to other commonly studied oxidatively induced DNA lesions such as thymine glycol or 8-oxo-dG. In addition to being the best candidates amongst currently known DNA lesions for playing a causative role in XP neurologic disease, studies on cyPudNs have also yielded unexpected insights into topics of more general interest, i.e. transcriptional mutagenesis [56]. A full understanding of how lesions provoke the novel types of TM events we have identified may ultimately provide insight into basic transcriptional mechanisms. In addition, while beyond the scope of this review, published data indicates that these lesions may be of significance to other medically relevant aspects of biology such as breast cancer [109,110], chemotherapy [111], and biomarkers of environmental pollution [112]. It is likely that studies of this class of DNA lesions will increase in the future, and in turn yield additional insights of relevance to human health.
Acknowledgements
I thank Dr. Miral Dizdaroglu for advice on the figures, and many helpful discussions, and Lori Cooper and Cheryl Marietta for helpful comments and careful proofreading of the manuscript
Footnotes
The term cyclopurine nucleosides can refer to either the ribonucleoside or deoxyribonucleoside. The primary focus of this paper is on the DNA adducts, the 8,5’-cyclopurine-2’-deoxynucleosides, abbreviated cyPudNs.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Keck K. Bildung von cyclonudleotiden bei bestrahlung wassiriger losungen von purinenucleotiden. Z Naturforsch. 1968;23b:1034–1043. [PubMed] [Google Scholar]
- 2.Mariaggi N, Cadet J, Teoule R. Cyclisation radicalaire de la desoxy-2'-adenosine en solution aqueuse, sous l'effet du rayonnement gamma. Tetrahedron. 1976;32:2385–2387. [Google Scholar]
- 3.Raleigh JA, Kremers W, Whitehouse R. Radiation chemistry of nucleotides: 8,5'-cyclonucleotide formation and phosphate release initiated by hydroxyl radical attack on adenosine monophosphates. Radiat Res. 1976;65:414–422. [PubMed] [Google Scholar]
- 4.Dirksen ML, Blakely WF, Holwitt E, Dizdaroglu M. Effect of DNA conformation on the hydroxyl radical-induced formation of 8,5'-cyclopurine 2'-deoxyribonucleoside residues in DNA. Int J Radiat Biol. 1988;54:195–204. doi: 10.1080/09553008814551631. [DOI] [PubMed] [Google Scholar]
- 5.Dizdaroglu M, Dirksen ML, Jiang HX, Robbins JH. Ionizing-radiation-induced damage in the DNA of cultured human cells. Identification of 8,5-cyclo-2-deoxyguanosine. Biochem J. 1987;241:929–932. doi: 10.1042/bj2410929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brooks PJ, Wise DS, Berry DA, Kosmoski JV, Smerdon MJ, Somers RL, Mackie H, Spoonde AY, Ackerman EJ, Coleman K, Tarone RE, Robbins JH. The oxidative DNA lesion 8,5' -(S)-cyclo-2'-deoxyadenosine is repaired by the nucleotide excision repair pathway and blocks gene expression in mammalian cells. J Biol Chem. 2000;275:22355–22362. doi: 10.1074/jbc.M002259200. [DOI] [PubMed] [Google Scholar]
- 7.Kuraoka I, Bender C, Romieu A, Cadet J, Wood RD, Lindahl T. Removal of oxygen free-radical-induced 5',8-purine cyclodeoxynucleosides from DNA by the nucleotide excision-repair pathway in human cells. Proc Natl Acad Sci USA. 2000;97:3832–3837. doi: 10.1073/pnas.070471597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andrews A, Barrett S, Robbins J. Xeroderma Pigmentosun neurological abnormalities correlate with the colony forming ability after ultraviolet irradiation. Proc Natl Acad Sci USA. 1978;75:1984–1988. doi: 10.1073/pnas.75.4.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brooks PJ. The case for 8,5'-cyclopurine-2'-deoxynucleosides as endogenous DNA lesions that cause neurodegeneration in xeroderma pigmentosum. Neuroscience. 2007;145:1407–1417. doi: 10.1016/j.neuroscience.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.von Sonntag C. The Chemical Basis of Radiation Biology. London: Taylor and Francis; 1987. [Google Scholar]
- 11.Henle ES, et al. Oxidative damage to DNA constituents by iron-mediated fenton reactions. The deoxyguanosine family. J Biol Chem. 1996;271:21177–21186. [PubMed] [Google Scholar]
- 12.Murata-Kamiya N, Kamiya H, Muraoka M, Kaji H, Kasai H. Comparison of oxidation products from DNA components by gamma-irradiation and Fenton-type reactions. J Radiat Res (Tokyo) 1997;38:121–131. doi: 10.1269/jrr.38.121. [DOI] [PubMed] [Google Scholar]
- 13.Kuraoka I, Robins P, Masutani C, Hanaoka F, Gasparutto D, Cadet J, Wood RD, Lindahl T. Oxygen free radical damage to DNA. Translesion synthesis by human DNA polymerase eta and resistance to exonuclease action at cyclopurine deoxynucleoside residues. J Biol Chem. 2001;276:49283–49288. doi: 10.1074/jbc.M107779200. [DOI] [PubMed] [Google Scholar]
- 14.Dizdaroglu M. Free-radical-induced formation of an 8,5'-cyclo-2'-deoxyguanosine moiety in deoxyribonucleic acid. Biochem J. 1986;238:247–254. doi: 10.1042/bj2380247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chatgilialoglu C, Caminal C, Altieri A, Vougioukalakis GC, Mulazzani QG, Gimisis T, Guerra M. Tautomerism in the guanyl radical. J Am Chem Soc. 2006;128:13796–13805. doi: 10.1021/ja062636h. [DOI] [PubMed] [Google Scholar]
- 16.Russo M, Jimenez LB, Mulazzani QG, D'Angelantonio M, Guerra M, Miranda MA, Chatgilialoglu C. Chemical radiation studies of 8-bromo-2'-deoxyinosine and 8-bromoinosine in aqueous solutions. Chemistry. 2006;12:7684–7693. doi: 10.1002/chem.200600040. [DOI] [PubMed] [Google Scholar]
- 17.Navacchia ML, Chatgilialoglu C, Montevecchi PC. C5'-adenosinyl radical cyclization. A stereochemical investigation. J Org Chem. 2006;71:4445–4452. doi: 10.1021/jo060197z. [DOI] [PubMed] [Google Scholar]
- 18.Jimenez LB, Encinas S, Miranda MA, Navacchia ML, Chatgilialoglu C. The photochemistry of 8-bromo-2'-deoxyadenosine. A direct entry to cyclopurine lesions. Photochem Photobiol Sci. 2004;2:1042–1046. doi: 10.1039/b410939b. [DOI] [PubMed] [Google Scholar]
- 19.Chatgilialoglu C, Duca M, Ferreri C, Guerra M, Ioele M, Mulazzani QG, Strittmatter H, Giese B. Selective generation and reactivity of 5'-adenosinyl and 2'-adenosinyl radicals. Chemistry. 2004;10:1249–1255. doi: 10.1002/chem.200305488. [DOI] [PubMed] [Google Scholar]
- 20.Chatgilialoglu C, Guerra M, Mulazzani QG. Model studies of DNA C5' radicals. Selective generation and reactivity of 2'-deoxyadenosin-5'-yl radical. J Am Chem Soc. 2003;125:3839–3848. doi: 10.1021/ja029374d. [DOI] [PubMed] [Google Scholar]
- 21.Zander R. Cellular oxygen concentration. Adv Exp Med Biol. 1976;75:463–467. doi: 10.1007/978-1-4684-3273-2_54. [DOI] [PubMed] [Google Scholar]
- 22.Joenje H. Genetic toxicology of oxygen. Mutat Res. 1989;219:193–208. doi: 10.1016/0921-8734(89)90001-5. [DOI] [PubMed] [Google Scholar]
- 23.Vriend G. WHAT IF: a molecular modeling and drug design program. J Mol Graph. 1990;8:52–56. doi: 10.1016/0263-7855(90)80070-v. 29. [DOI] [PubMed] [Google Scholar]
- 24.Romieu A, Gasparutto D, Molko D, Cadet J. Site-Specific Introduction of (5'S)-5',8-Cyclo-2'-deoxyadenosine into Oligodeoxyribonucleotides. J. Org. Chem. 1998;63:5245–5249. [Google Scholar]
- 25.Yang W. Poor base stacking at DNA lesions may initiate recognition by many repair proteins. DNA Repair (Amst) 2006;5:654–666. doi: 10.1016/j.dnarep.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 26.Haromy TP, Raleigh J, Sundaralingam M. Enzyme-bound conformations of nucleotide substrates. X-ray structure and absolute configuration of 8,5'-cycloadenosine monohydrate. Biochemistry. 1980;19:1718–1722. doi: 10.1021/bi00549a031. [DOI] [PubMed] [Google Scholar]
- 27.Friedberg E, Walker G, Siede W, Wood R, Schultz R, Ellenberger T. DNA Repair and Mutagenesis. Washington DC: ASM press; 2005. [Google Scholar]
- 28.Neisser A. Ueber das 'Xeroderma pigmentosum' (Kaposi): Lioderma essentialis cum melanosi et telangiectasia. Jahrschr. Dermatol. Syphil. 1883:47–62. [Google Scholar]
- 29.De Sanctis C, Cacchione A. L'Idiozia xerodermica. Riv. Sper. Freniat. 1932;56:269–292. [Google Scholar]
- 30.Robbins JH. A childhood neurodegeneration due to defective DNA repair: a novel concept of disease based on studies xeroderma pigmentosum. J Child Neurol. 1989;4:143–146. doi: 10.1177/088307388900400215. [DOI] [PubMed] [Google Scholar]
- 31.Rapin I, Lindenbaum Y, Dickson DW, Kraemer KH, Robbins JH. Cockayne syndrome and xeroderma pigmentosum. Neurology. 2000;55:1442–1449. doi: 10.1212/wnl.55.10.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moriwaki S, Nishigori C, Imamura S, Yagi T, Takahashi C, Fujimoto N, Takebe H. A case of xeroderma pigmentosum complementation group F with neurological abnormalities. Br J Dermatol. 1993;128:91–94. doi: 10.1111/j.1365-2133.1993.tb00154.x. [DOI] [PubMed] [Google Scholar]
- 33.Sijbers AM, van Voorst Vader PC, Snoek JW, Raams A, Jaspers NG, Kleijer WJ. Homozygous R788W point mutation in the XPF gene of a patient with xeroderma pigmentosum and late-onset neurologic disease. J Invest Dermatol. 1998;110:832–836. doi: 10.1046/j.1523-1747.1998.00171.x. [DOI] [PubMed] [Google Scholar]
- 34.Hirai Y, Kodama Y, Moriwaki S, Noda A, Cullings HM, Macphee DG, Kodama K, Mabuchi K, Kraemer KH, Land CE, Nakamura N. Heterozygous individuals bearing a founder mutation in the XPA DNA repair gene comprise nearly 1% of the Japanese population. Mutat Res. 2006;601:171–178. doi: 10.1016/j.mrfmmm.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 35.Satokata I, Tanaka K, Miura N, Miyamoto I, Satoh Y, Kondo S, Okada Y. Characterization of a splicing mutation in group A xeroderma pigmentosum. Proc Natl Acad Sci U S A. 1990;87:9908–9912. doi: 10.1073/pnas.87.24.9908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robbins JH, Brumback RA, Mendiones M, Barrett SF, Carl JR, Cho S, Denckla MB, Ganges MB, Gerber LH, Guthrie RA, et al. Neurological disease in xeroderma pigmentosum. Documentation of a late onset type of the juvenile onset form. Brain. 1991;114:1335–1361. doi: 10.1093/brain/114.3.1335. [DOI] [PubMed] [Google Scholar]
- 37.Maeda T, Sato K, Minami H, Taguchi H, Yoshikawa K. Chronological difference in walking impairment among Japanese group A xeroderma pigmentosum (XP-A) patients with various combinations of mutation sites. Clin Genet. 1995;48:225–231. doi: 10.1111/j.1399-0004.1995.tb04094.x. [DOI] [PubMed] [Google Scholar]
- 38.Sidwell RU, Sandison A, Wing J, Fawcett HD, Seet JE, Fisher C, Nardo T, Stefanini M, Lehmann AR, Cream JJ. A novel mutation in the XPA gene associated with unusually mild clinical features in a patient who developed a spindle cell melanoma. Br J Dermatol. 2006;155:81–88. doi: 10.1111/j.1365-2133.2006.07272.x. [DOI] [PubMed] [Google Scholar]
- 39.States JC, Myrand SP. Splice site mutations in a xeroderma pigmentosum group A patient with delayed onset of neurological disease. Mutat Res. 1996;363:171–177. doi: 10.1016/0921-8777(96)00004-3. [DOI] [PubMed] [Google Scholar]
- 40.Lehmann AR. The xeroderma pigmentosum group D (XPD) gene: one gene two functions, three diseases. Genes Dev. 2001;15:15–23. doi: 10.1101/gad.859501. [DOI] [PubMed] [Google Scholar]
- 41.Kraemer KH, Patronas NJ, Schiffmann R, Brooks BP, Tamura D, DiGiovanna JJ. Xeroderma pigmentosum, trichothiodystrophy and Cockayne syndrome: a complex genotype-phenotype relationship. Neuroscience. 2007;145:1388–1396. doi: 10.1016/j.neuroscience.2006.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andressoo JO, Jans J, de Wit J, Coin F, Hoogstraten D, van de Ven M, Toussaint W, Huijmans J, Thio HB, van Leeuwen WJ, de Boer J, Egly JM, Hoeijmakers JH, van der Horst GT, Mitchell JR. Rescue of progeria in trichothiodystrophy by homozygous lethal Xpd alleles. PLoS Biol. 2006;4:e322. doi: 10.1371/journal.pbio.0040322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robbins JH, Brumback RA, Moshell AN. Clinically asymptomatic xeroderma pigmentosum neurological disease in an adult: evidence for a neurodegeneration in later life caused by defective DNA repair. Eur Neurol. 1993;33:188–190. doi: 10.1159/000116932. [DOI] [PubMed] [Google Scholar]
- 44.Nouspikel T. DNA repair in differentiated cells: some new answers to old questions. Neuroscience. 2007;145:1213–1221. doi: 10.1016/j.neuroscience.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 45.Nouspikel TP, Hyka-Nouspikel N, Hanawalt PC. Transcription domain-associated repair in human cells. Mol Cell Biol. 2006;26:8722–8730. doi: 10.1128/MCB.01263-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robbins JH, Kraemer KH, Merchant SN, Brumback RA. Adult-onset xeroderma pigmentosum neurological disease--observations in an autopsy case. Clin Neuropathol. 2002;21:18–23. [PubMed] [Google Scholar]
- 47.Andrews A, Barrett S, Robbins J. Xeroderma Pigmentosum neurological abnormalities correlate with the colony forming ability after ultraviolet irradiation. Proc Natl Acad Sci USA. 1978;75:1984–1988. doi: 10.1073/pnas.75.4.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Egly JM. The 14th Datta Lecture. TFIIH: from transcription to clinic. FEBS Lett. 2001;498:124–128. doi: 10.1016/s0014-5793(01)02458-9. [DOI] [PubMed] [Google Scholar]
- 49.Araki M, Masutani C, Takemura M, Uchida A, Sugasawa K, Kondoh J, Ohkuma Y, Hanaoka F. Centrosome protein centrin 2/caltractin 1 is part of the xeroderma pigmentosum group C complex that initiates global genome nucleotide excision repair. J Biol Chem. 2001;276:18665–18672. doi: 10.1074/jbc.M100855200. [DOI] [PubMed] [Google Scholar]
- 50.Compe E, Malerba M, Soler L, Marescaux J, Borrelli E, Egly JM. Neurological defects in trichothiodystrophy reveal a coactivator function of TFIIH. Nat Neurosci. 2007;10:1414–1422. doi: 10.1038/nn1990. [DOI] [PubMed] [Google Scholar]
- 51.de Boer J, de Wit J, van Steeg H, Berg RJ, Morreau H, Visser P, Lehmann AR, Duran M, Hoeijmakers JH, Weeda G. A mouse model for the basal transcription/DNA repair syndrome trichothiodystrophy. Mol Cell. 1998;1:981–990. doi: 10.1016/s1097-2765(00)80098-2. [DOI] [PubMed] [Google Scholar]
- 52.Ito S, Kuraoka I, Chymkowitch P, Compe E, Takedachi A, Ishigami C, Coin F, Egly JM, Tanaka K. XPG stabilizes TFIIH, allowing transactivation of nuclear receptors: implications for Cockayne syndrome in XP-G/CS patients. Mol Cell. 2007;26:231–243. doi: 10.1016/j.molcel.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 53.Friedberg EC, Wood RD. New insights into the combined Cockayne/xeroderma pigmentosum complex: human XPG protein can function in transcription factor stability. Mol Cell. 2007;26:162–164. doi: 10.1016/j.molcel.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 54.Satoh M, Jones C, Wood R, Lindahl T. DNA excision-repair defect of xeroderma pigmentosum prevents removal of a class of oxygen free radical induced base lesions. Proc. Nat'l Acad Sci. 1993;90:6335–6339. doi: 10.1073/pnas.90.13.6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marietta C, Gulam H, Brooks PJ. A single 8,5'-cyclo-2'-deoxyadenosine lesion in a TATA box prevents binding of the TATA binding protein and strongly reduces transcription in vivo. DNA Repair. 2002;1:967–975. doi: 10.1016/s1568-7864(02)00148-9. [DOI] [PubMed] [Google Scholar]
- 56.Marietta C, Brooks PJ. Transcriptional bypass of bulky DNA lesions causes new mutant RNA transcripts in human cells. EMBO Rep. 2007;8:388–393. doi: 10.1038/sj.embor.7400932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Randerath K, Zhou GD, Somers RL, Robbins JH, Brooks PJ. A 32P-postlabeling assay for the oxidative DNA lesion 8,5'-cyclo-2'-deoxyadenosine in mammalian tissues: evidence that four type II I-compounds are dinucleotides containing the lesion in the 3' nucleotide. J Biol Chem. 2001;276:36051–36057. doi: 10.1074/jbc.M105472200. [DOI] [PubMed] [Google Scholar]
- 58.Dizdaroglu M, Jaruga P, Rodriguez H. Identification and quantification of 8,5'-cyclo-2'-deoxy-adenosine in DNA by liquid chromatography/ mass spectrometry. Free Radic Biol Med. 2001;30:774–784. doi: 10.1016/s0891-5849(01)00464-6. [DOI] [PubMed] [Google Scholar]
- 59.Jaruga P, Theruvathu J, Dizdaroglu M, Brooks PJ. Complete release of (5'S)-8,5'-cyclo-2'-deoxyadenosine from dinucleotides, oligodeoxynucleotides and DNA, and direct comparison of its levels in cellular DNA with other oxidatively induced DNA lesions. Nucleic Acids Res. 2004;32:e87. doi: 10.1093/nar/gnh087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–714. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 61.Nyaga SG, Jaruga P, Lohani A, Dizdaroglu M, Evans MK. Accumulation of oxidatively induced DNA damage in human breast cancer cell lines following treatment with hydrogen peroxide. Cell Cycle. 2007;6:1472–1478. [PubMed] [Google Scholar]
- 62.Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev Genet. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- 63.Cavalieri EL, Rogan EG. A unifying mechanism in the initiation of cancer and other diseases by catechol quinones. Ann N Y Acad Sci. 2004;1028:247–257. doi: 10.1196/annals.1322.029. [DOI] [PubMed] [Google Scholar]
- 64.Gates KS, Nooner T, Dutta S. Biologically relevant chemical reactions of N7-alkylguanine residues in DNA. Chem Res Toxicol. 2004;17:839–856. doi: 10.1021/tx049965c. [DOI] [PubMed] [Google Scholar]
- 65.Theruvathu JA, Jaruga P, Dizdaroglu M, Brooks PJ. The oxidatively induced DNA lesions 8,5'-cyclo-2'-deoxyadenosine and 8-hydroxy-2'-deoxyadenosine are strongly resistant to acid-induced hydrolysis of the glycosidic bond. Mech Ageing Dev. 2007;128:494–502. doi: 10.1016/j.mad.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Masutani C, Araki M, Yamada A, Kusumoto R, Nogimori T, Maekawa T, Iwai S, Hanaoka F. Xeroderma pigmentosum variant (XP-V) correcting protein from HeLa cells has a thymine dimer bypass DNA polymerase activity. Embo J. 1999;18:3491–3501. doi: 10.1093/emboj/18.12.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 68.Viswanathan A, You HJ, Doetsch PW. Phenotypic change caused by transcriptional bypass of uracil in nondividing cells. Science. 1999;284:159–162. doi: 10.1126/science.284.5411.159. [DOI] [PubMed] [Google Scholar]
- 69.Aguzzi A, Heikenwalder M. Pathogenesis of prion diseases: current status and future outlook. Nat Rev Microbiol. 2006;4:765–775. doi: 10.1038/nrmicro1492. [DOI] [PubMed] [Google Scholar]
- 70.Yamaizumi M, Sugano T. U.v.-induced nuclear accumulation of p53 is evoked through DNA damage of actively transcribed genes independent of the cell cycle. Oncogene. 1994;9:2775–2784. [PubMed] [Google Scholar]
- 71.Ljungman M, Zhang F. Blockage of RNA polymerase as a possible trigger for u.v. light-induced apoptosis. Oncogene. 1996;13:823–831. [PubMed] [Google Scholar]
- 72.Derheimer FA, O'Hagan HM, Krueger HM, Hanasoge S, Paulsen MT, Ljungman M. RPA and ATR link transcriptional stress to p53. Proc Natl Acad Sci U S A. 2007;104:12778–12783. doi: 10.1073/pnas.0705317104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Roldan-Arjona T, Sedgwick B. DNA base damage induced by ionizing radiation recognized by Escherichia coli UvrABC nuclease but not Nth or Fpg proteins. Mol Carcinog. 1996;16:188–196. doi: 10.1002/(SICI)1098-2744(199608)16:4<188::AID-MC2>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 74.Reardon JT, Bessho T, Kung HC, Bolton PH, Sancar A. In vitro repair of oxidative DNA damage by human nucleotide excision repair system: possible explanation for neurodegeneration in xeroderma pigmentosum patients. Proc Natl Acad Sci U S A. 1997;94:9463–9468. doi: 10.1073/pnas.94.17.9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Branum ME, Reardon JT, Sancar A. DNA repair excision nuclease attacks undamaged DNA. A potential source of spontaneous mutations. J Biol Chem. 2001;276:25421–25426. doi: 10.1074/jbc.M101032200. [DOI] [PubMed] [Google Scholar]
- 76.Runger TM, Epe B, Moller K. Repair of ultraviolet B and singlet oxygen-induced DNA damage in xeroderma pigmentosum cells. J Invest Dermatol. 1995;104:68–73. doi: 10.1111/1523-1747.ep12613504. [DOI] [PubMed] [Google Scholar]
- 77.Theron T, Fousteri MI, Volker M, Harries LW, Botta E, Stefanini M, Fujimoto M, Andressoo JO, Mitchell J, Jaspers NG, McDaniel LD, Mullenders LH, Lehmann AR. Transcription-associated breaks in xeroderma pigmentosum group D cells from patients with combined features of xeroderma pigmentosum and Cockayne syndrome. Mol Cell Biol. 2005;25:8368–8378. doi: 10.1128/MCB.25.18.8368-8378.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Laposa RR, Huang EJ, Cleaver JE. Increased apoptosis, p53 up-regulation, and cerebellar neuronal degeneration in repair-deficient Cockayne syndrome mice. Proc Natl Acad Sci U S A. 2007;104:1389–1394. doi: 10.1073/pnas.0610619104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Burcham PC. Genotoxic lipid peroxidation products: their DNA damaging properties and role in formation of endogenous DNA adducts. Mutagenesis. 1998;13:287–305. doi: 10.1093/mutage/13.3.287. [DOI] [PubMed] [Google Scholar]
- 80.Blair IA. Lipid hydroperoxide-mediated DNA damage. Exp Gerontol. 2001;36:1473–1481. doi: 10.1016/s0531-5565(01)00133-4. [DOI] [PubMed] [Google Scholar]
- 81.Dedon PC, Plastaras JP, Rouzer CA, Marnett LJ. Indirect mutagenesis by oxidative DNA damage: formation of the pyrimidopurinone adduct of deoxyguanosine by base propenal. Proc Natl Acad Sci U S A. 1998;95:11113–11116. doi: 10.1073/pnas.95.19.11113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Johnson KA, et al. Repair of propanodeoxyguanosine by nucleotide excision repair in vivo and in vitro. J Biol Chem. 1997;272:11434–11438. doi: 10.1074/jbc.272.17.11434. [DOI] [PubMed] [Google Scholar]
- 83.Cline SD, Riggins JN, Tornaletti S, Marnett LJ, Hanawalt PC. Malondialdehyde adducts in DNA arrest transcription by T7 RNA polymerase and mammalian RNA polymerase II. Proc Natl Acad Sci U S A. 2004;101:7275–7280. doi: 10.1073/pnas.0402252101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mao H, Schnetz-Boutaud NC, Weisenseel JP, Marnett LJ, Stone MP. Duplex DNA catalyzes the chemical rearrangement of a malondialdehyde deoxyguanosine adduct. Proc Natl Acad Sci U S A. 1999;96:6615–6620. doi: 10.1073/pnas.96.12.6615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang Q, Wang Y. Generation of 5-(2'-deoxycytidyl)methyl radical and the formation of intrastrand cross-link lesions in oligodeoxyribonucleotides. Nucleic Acids Res. 2005;33:1593–1603. doi: 10.1093/nar/gki301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gu C, Wang Y. Thermodynamic and in vitro replication studies of an intrastrand G[8-5]C cross-link lesion. Biochemistry. 2005;44:8883–8889. doi: 10.1021/bi050036+. [DOI] [PubMed] [Google Scholar]
- 87.Xerri B, Morell C, Grand A, Cadet J, Cimino P, Barone V. Radiation-induced formation of DNA intrastrand crosslinks between thymine and adenine bases: A theoretical approach. Org Biomol Chem. 2006;4:3986–3992. doi: 10.1039/b609134b. [DOI] [PubMed] [Google Scholar]
- 88.Bellon S, Gasparutto D, Saint-Pierre C, Cadet J. Guanine-thymine intrastrand cross-linked lesion containing oligonucleotides: from chemical synthesis to in vitro enzymatic replication. Org Biomol Chem. 2006;4:3831–3837. doi: 10.1039/b609460k. [DOI] [PubMed] [Google Scholar]
- 89.Box HC, Dawidzik JB, Budzinski EE. Free radical-induced double lesions in DNA. Free Radic Biol Med. 2001;31:856–868. doi: 10.1016/s0891-5849(01)00653-0. [DOI] [PubMed] [Google Scholar]
- 90.Gu C, Zhang Q, Yang Z, Wang Y, Zou Y, Wang Y. Recognition and incision of oxidative intrastrand cross-link lesions by UvrABC nuclease. Biochemistry. 2006;45:10739–10746. doi: 10.1021/bi060423z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hong H, Cao H, Wang Y, Wang Y. Identification and quantification of a guanine-thymine intrastrand cross-link lesion induced by Cu(II)/H2O2/ascorbate. Chem Res Toxicol. 2006;19:614–621. doi: 10.1021/tx060025x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cao H, Wang Y. Quantification of oxidative single-base and intrastrand cross-link lesions in unmethylated and CpG-methylated DNA induced by Fenton-type reagents. Nucleic Acids Res. 2007;35:4833–4844. doi: 10.1093/nar/gkm497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Randerath K, Randerath E, Zhou GD, Li D. Bulky endogenous DNA modifications (I-compounds) -possible structural origins and functional implications. Mutat Res. 1999;424:183–194. doi: 10.1016/s0027-5107(99)00018-4. [DOI] [PubMed] [Google Scholar]
- 94.Carmichael PL, She MN, Phillips DH. Detection and characterization by 32P-postlabelling of DNA adducts induced by a Fenton-type oxygen radical-generating system. Carcinogenesis. 1992;13:1127–1135. doi: 10.1093/carcin/13.7.1127. [DOI] [PubMed] [Google Scholar]
- 95.Randerath K, Randerath E, Smith CV, Chang J. Structural origins of bulky oxidative DNA adducts (type II I-compounds) as deduced by oxidation of oligonucleotides of known sequence. Chem Res Toxicol. 1996;9:247–254. doi: 10.1021/tx950085v. [DOI] [PubMed] [Google Scholar]
- 96.Lloyd DR, Phillips DH, Carmichael PL. Generation of putative intrastrand cross-links and strand breaks in DNA by transition metal ion-mediated oxygen radical attack. Chem Res Toxicol. 1997;10:393–400. doi: 10.1021/tx960158q. [DOI] [PubMed] [Google Scholar]
- 97.Evans MD, Dizdaroglu M, Cooke MS. Oxidative DNA damage and disease: induction, repair and significance. Mutat Res. 2004;567:1–61. doi: 10.1016/j.mrrev.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 98.Levay G, Bodell WJ. Detection of dopamine--DNA adducts: potential role in Parkinson's disease. Carcinogenesis. 1993;14:1241–1245. doi: 10.1093/carcin/14.6.1241. [DOI] [PubMed] [Google Scholar]
- 99.Reddy MV, Storer RD, Laws GM, Armstrong MJ, Barnum JE, Gara JP, McKnight CG, Skopek TR, Sina JF, DeLuca JG, Galloway SM. Genotoxicity of naturally occurring indole compounds: correlation between covalent DNA binding and other genotoxicity tests. Environ Mol Mutagen. 2002;40:1–17. doi: 10.1002/em.10088. [DOI] [PubMed] [Google Scholar]
- 100.Akanni A, Abul-Hajj YJ. Estrogen-nucleic acid adducts: reaction of 3,4-estrone-o-quinone radical anion with deoxyribonucleosides. Chem Res Toxicol. 1997;10:760–766. doi: 10.1021/tx970026c. [DOI] [PubMed] [Google Scholar]
- 101.Hankin JA, Murphy RC. Mass spectrometric quantitation of deoxyguanosine and leukotriene A4-deoxyguanosine adducts of DNA. Anal Biochem. 2004;333:156–164. doi: 10.1016/j.ab.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 102.Kanaly RA, Matsui S, Hanaoka T, Matsuda T. Application of the adductome approach to assess intertissue DNA damage variations in human lung and esophagus. Mutat Res. 2007 doi: 10.1016/j.mrfmmm.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 103.D'Errico M, Parlanti E, Teson M, Degan P, Lemma T, Calcagnile A, Iavarone I, Jaruga P, Ropolo M, Pedrini AM, Orioli D, Frosina G, Zambruno G, Dizdaroglu M, Stefanini M, Dogliotti E. The role of CSA in the response to oxidative DNA damage in human cells. Oncogene. 2007;26:4336–4343. doi: 10.1038/sj.onc.1210232. [DOI] [PubMed] [Google Scholar]
- 104.D'Errico M, Parlanti E, Teson M, de Jesus BM, Degan P, Calcagnile A, Jaruga P, Bjoras M, Crescenzi M, Pedrini AM, Egly JM, Zambruno G, Stefanini M, Dizdaroglu M, Dogliotti E. New functions of XPC in the protection of human skin cells from oxidative damage. Embo J. 2006;25:4305–4315. doi: 10.1038/sj.emboj.7601277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fousteri M, van Hoffen A, Vargova H, Mullenders LH. Repair of DNA lesions in chromosomal DNA impact of chromatin structure and Cockayne syndrome proteins. DNA Repair (Amst) 2005;4:919–925. doi: 10.1016/j.dnarep.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 106.Ford JM. Regulation of DNA damage recognition and nucleotide excision repair: another role for p53. Mutat Res. 2005;577:195–202. doi: 10.1016/j.mrfmmm.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 107.Bielas JH, Heddle JA. Quiescent murine cells lack global genomic repair but are proficient in transcription-coupled repair. DNA Repair (Amst) 2004;3:711–717. doi: 10.1016/j.dnarep.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 108.van der Wees C, Jansen J, Vrieling H, van der Laarse A, Van Zeeland A, Mullenders L. Nucleotide excision repair in differentiated cells. Mutat Res. 2007;614:16–23. doi: 10.1016/j.mrfmmm.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 109.Anderson KM, Jaruga P, Ramsey CR, Gilman NK, Green VM, Rostad SW, Emerman JT, Dizdaroglu M, Malins DC. Structural alterations in breast stromal and epithelial DNA: the influence of 8,5'-cyclo-2'-deoxyadenosine. Cell Cycle. 2006;5:1240–1244. doi: 10.4161/cc.5.11.2816. [DOI] [PubMed] [Google Scholar]
- 110.Rodriguez H, Jaruga P, Leber D, Nyaga SG, Evans MK, Dizdaroglu M. Lymphoblasts of women with BRCA1 mutations are deficient in cellular repair of 8,5'-Cyclopurine-2'-deoxynucleosides and 8-hydroxy-2'-deoxyguanosine. Biochemistry. 2007;46:2488–2496. doi: 10.1021/bi062022p. [DOI] [PubMed] [Google Scholar]
- 111.Birincioglu M, Jaruga P, Chowdhury G, Rodriguez H, Dizdaroglu M, Gates KS. DNA base damage by the antitumor agent 3-amino-1,2,4-benzotriazine 1,4-dioxide (tirapazamine) J Am Chem Soc. 2003;125:11607–11615. doi: 10.1021/ja0352146. [DOI] [PubMed] [Google Scholar]
- 112.Malins DC, Anderson KM, Stegeman JJ, Jaruga P, Green VM, Gilman NK, Dizdaroglu M. Biomarkers signal contaminant effects on the organs of English sole (Parophrys vetulus) from Puget Sound. Environ Health Perspect. 2006;114:823–829. doi: 10.1289/ehp.8544. [DOI] [PMC free article] [PubMed] [Google Scholar]