Abstract
We report a quantitative theoretical analysis of long-range electron transfer through sensitizer wires bound in the active-site channel of cytochrome P450cam. Each sensitizer wire consists of a substrate group with high binding affinity for the enzyme active site connected to a ruthenium-diimine through a bridging aliphatic or aromatic chain. Experiments have revealed a dramatic dependence of electron transfer rates on the chemical composition of both the bridging group and the substrate. Using combined molecular dynamics simulations and electronic coupling calculations, we show that electron tunneling through perfluorinated aromatic bridges is promoted by enhanced superexchange coupling through virtual reduced states. In contrast, electron flow through aliphatic bridges occurs by hole-mediated superexchange. We have found that a small number of wire conformations with strong donor-acceptor couplings can account for the observed electron tunneling rates for sensitizer wires terminated with either ethylbenzene or adamantane. In these instances, the rate is dependent not only on electronic coupling of the donor and acceptor but also on the nuclear motion of the sensitizer wire, necessitating the calculation of average rates over the course of a molecular dynamics simulation. These calculations along with related recent findings have made it possible to analyze the results of many other sensitizer-wire experiments that in turn point to new directions in our attempts to observe reactive intermediates in the catalytic cycles of P450 and other heme enzymes.
1. Introduction
We have shown that electrons and holes can be delivered rapidly to the active sites of metalloenzymes through substrates (or ligands) linked to redox-active photosensitizers (“sensitizer wires”) [1-12]. Electron transfer (ET) is initiated by optical excitation of the sensitizer at the end of the wire and monitored by spectroscopic methods. Dramatic variations of ET rates through the wires have been observed upon changes in the chemical compositions of both linkers and substrates [1,3,6,10,12].
We would like to know the origin of these large variations in wire ET rates, as detailed understanding will aid the design of improved wires for the study of biological redox reactions.
There have been many investigations of the effects of molecular fluctuations on ET rates in bridge-mediated systems [13-25]. Bridge dynamics can have profound effects on ET kinetics in biological systems, as, for example, in conformationally gated processes [13,15,16,20,21]. Gating occurs when the coupling between two cofactors is increased dramatically as a result of large-scale nuclear motions and the rate of charge transfer is greatly increased. Bridge dynamics also can enhance electronic coupling through equilibrium fluctuations of the torsional angles along the length of the bridge [14,15,17-19,21].
We have calculated the rates of electron transfer through Ru-diimine sensitizer wires to the heme of cytochrome P450. Using a combination of molecular dynamics and electronic coupling calculations, we have found that subtle rearrangements modulate electron flow from the Ru-diimine sensitizer through a wire to the heme. We have analyzed the effects of these rearrangements on the experimentally observed rates as well as other factors that might lead to more rapid electron transfer in these protein:wire conjugates.
2. Structures
Rapid heme reduction is successfully achieved through two similar photoprocesses in the case of five related Ru(II) sensitizer wires. In the first process, electronically excited Ru(II), or Ru(II)*, directly reduces the heme. The driving force of the reaction (-ΔGo) is dependent on the Ru(II) complex employed in the experiment. Here we have examined two complexes: [Ru(2,2′-bipyridine)2L]2+ (bpyRu) and [Ru(4,4′,5,5′-tetramethylbipyridine)2L]2+ (tmbpyRu). The (bpyRu)3+/2+* potential is -0.62 V vs NHE; (tmbpyRu)3+/2+* is -0.75 V vs NHE [6]. In the second process, Ru(II)* is reductively quenched by an external redox partner, followed by Ru(I) to Fe(III) ET to produce an Fe(II) heme. The (bpyRu)2+/+ potential is -1.24 V vs NHE) [1].
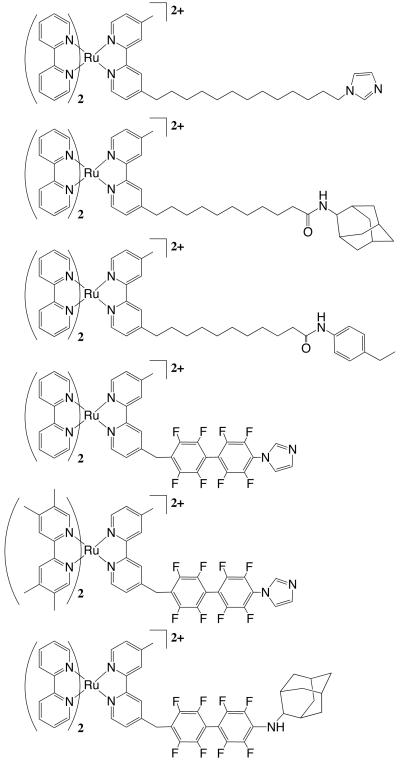
The sensitizer wires that have been investigated are shown in Figure 1. Two types of hydrophobic bridges have been examined: aliphatic hydrocarbon chains (C11 and C13) and fluorinated biphenyls (F8bp). Three types of terminal substrate groups have been investigated: imidazole (Im), adamantane (Ad) and ethylbenzene (EB). One difference in the terminal groups that is significant for electron transfer is the ability of imidazole to coordinate directly to the Fe atom in Fe-porphyrins, while interactions of adamantane or ethylbenzene with the Fe-porphyrin are noncovalent in nature. We have performed calculations on [bpyRu-F8bp-Ad]2+, which showed no heme reduction in flash/quench experiments, because it is similar to [bpyRu-F8bp-Im]2+ and [tmbpyRu-F8bp-Im]2+, whose electronic excited states are capable of directly reducing the Fe(III) heme on submicrosecond timescales.
Figure 1.
Sensitizer wires (top to bottom: bpyRu-C13-Im, bpyRu-C11-Ad, bpyRu-C11-Eb, bpyRu-F8bp-Im, tmbpyRu-F8bp-Im, and bpyRu-F8bp-Ad). Synthesis and characterization of these wires are described in references 1 and 6.
3. Molecular Dynamics Simulations
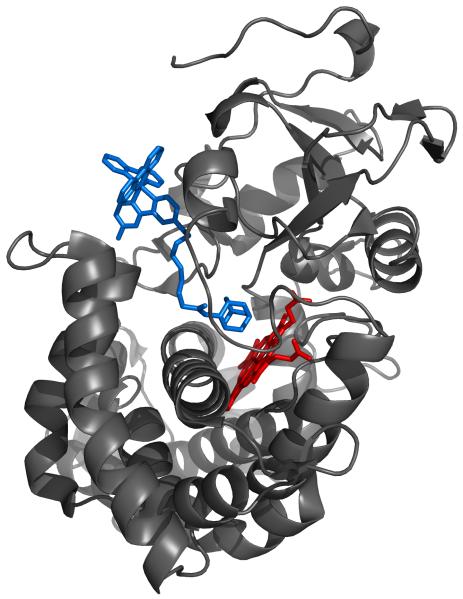
Crystal structures are available for P450 conjugates with bpyRu-F8bp-Ad and bpyRu-C9-Ad (Figure 2) wires [2]. Other wires were constructed using molecular mechanics modeling. Substantial flexibility of the wires in conjugates with P450 is expected (as indicated by the presence of “alternative” structures in the X-ray determinations of P450 with bpyRu-F8bp-Ad and bpyRu-C9-Ad); therefore, molecular dynamics simulations were used to generate a representative set of configurations for the protein:wire conjugates.
Figure 2.
Structure of bpyRu-C9-Ad bound to P450cam (pdb code: 1QMQ) [2].
Simulations of P450:wire conjugates were performed with the AMBER 7 suite of programs [26]. Parameters for protein amino acids and the Fe-porphyrin were taken from the Amber-94 force field [27]. Parameters for the carbon and hydrogen atoms of Ad and hydrocarbon bridges were taken as in aliphatic amino-acid sidechains such as valine and leucine. Atomic charges for atoms of fluorinated biphenyls (F8bp) were set using Hartree-Fock ab initio calculations with a 6-31G* basis set and the Merz-Kollman charge-fitting scheme [28]. The equilibrium geometry of F8bp was taken from an X-ray structure [2]. Force-field parameters for a torsional angle defining the relative orientation of fluorinated phenyl rings were ones that fit the potential energy profile obtained by constrained energy minimization of F8bp (the torsional angle value was a parameter in quantum chemical calculations) using the MP2 method and a 6-31G* basis set. The initial geometry for bpyRu-F8bp-Ad was taken from X-ray data. Initial geometries for other structures were generated by replacing the wire in the X-ray structure of bpyRu-F8bp-Ad or bpyRu-C9-Ad followed by energy minimization to remove unfavorable atom-atom contacts. 500 ps molecular dynamics trajectories were generated for conditions of constant pressure (1 atm) and constant temperature (300 K). The first 100 ps of MD trajectories were discarded as nonequilibrated and snapshots were then collected at regular intervals along remaining portions of the trajectories.
4. Electronic Coupling Calculations
To compute electronic donor-acceptor interactions we used an energy splitting method where eigenstates of a model electronic Hamiltonian of the system that corresponds to the donor and acceptor electronic states are tuned into quasi-degeneracy by a Hamiltonian perturbation (in this work by applying an electric field in the donor-acceptor direction) [29-31]. The minimal energy splitting of these two eigenstates of the system is equal to twice the donor-acceptor electronic coupling ΔEmin=2HDA. In Hartree-Fock calculations of donor-acceptor systems, HDA can be found from the energy splitting of eigenstates (molecular orbitals) of the one-electron Hamiltonian (Fockian) of the system. For our calculations we compute the energy splitting for both electron and hole transfer. To perform these calculations we either add an extra electron (for the case of electron transfer) or remove an electron (for the case of hole transfer) from the system in accordance with Koopman’s theorem and literature precedent [32-36].
In the ET experiments, the heme at the active site of P450 cycles between Fe(III) and Fe(II) redox states [1-12]. In this redox transition the electronic density changes mainly on the iron atom; therefore, it is likely that both the highest occupied molecular orbital of the Fe(II)-porphyrin and the lowest unoccupied molecular orbital of Fe(III)-porphyrin will be Fe-localized. We assume in our calculations that the occupied Fe-localized orbitals (dxy, dxz, dyz) are equally involved in ET; and, the average ET coupling will be the root mean square (RMS) of ET couplings computed for individual Fe-porphyrin d orbitals.
For the case of computing HDA for electron transfer, the Ru-diimine donor was represented by a single dihydropyrazine ring. The HOMO of dihydropyrazine has the same symmetry and nodal structure as the LUMO of pyridine; thus, the dihydropyrazine HOMO is a reasonable model for the donor one-electron state.
In the second approach for calculating HDA for hole transfer, electrons were removed (to a formal Fe(VI) state). A large compensatory point charge (-5e) was placed on Fe; the resulting energies of Fe-localized unoccupied molecular orbitals are roughly -4 eV, in agreement with the expected electron binding energies of the acceptor with a reduction potential near -0.3 V vs NHE [37] (0.0 V vs NHE approximately corresponds to a -4.5 eV electron binding energy) [38]. The Ru-diimine donor electronic state was modeled in these calculations by the LUMO of the pyridinium cation. The computed value of the pyridinium cation LUMO energy was -3.5 eV in the Hartree-Fock calculations, in agreement with the reduction potential of the donor (~ -0.62 V for the excited state and -1.2 V for the anion donor (vs NHE)) [39].
5. Bridge Conformational Dynamics
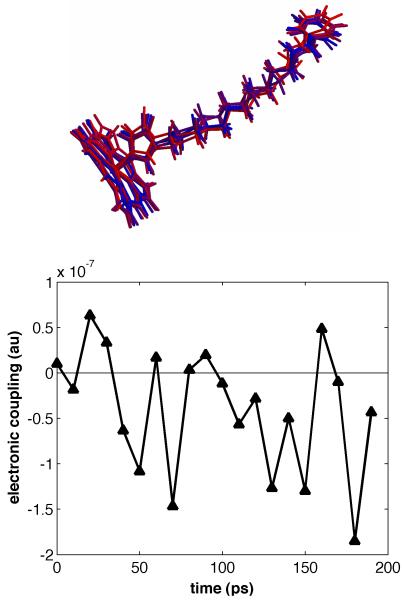
The molecular bridges we have investigated are quite flexible, and multiple conformations are observed in MD calculations. The importance of taking into account molecular bridge fluctuations while computing ET rates has been emphasized several times in the literature [13-23]. Figure 3 (top) shows snapshots of MD trajectories of the P450 complex with a bpyRu-C13-Im wire. We found that the computed electronic coupling varies substantially for the different conformations of the P450:wire conjugate. Figure 3 (bottom) shows the Fe to Ru ET-coupling (HDA) values computed along the MD trajectory. Typically, the computed ET couplings vary by about an order of magnitude from configuration to configuration (corresponding to two orders of magnitude difference in the computed nonadiabatic ET rates). There are geometries with even smaller ET coupling values, some smaller than the maximal coupling by a factor of 30. The observed rates can be accounted for by the contribution of a few configurations that exhibit strong donor-acceptor couplings. It follows that a computed ET rate will almost certainly be much smaller than the experimental value if the coupling is computed in a randomly chosen configuration (or in a geometry derived from X-ray structural analysis). Indeed, appropriate averaging here is complicated, since a few geometries favoring ET are dominant. This issue was found to be especially important for wires with Ad and EB terminal groups that interact with the heme via weak van der Waals interactions, as only certain conformations show good overlap between the orbitals of the terminal group and those of the iron atom.
Figure 3.
Top: bpyRu-C13-Im conformations taken from snapshots along the MD simulation. This figure illustrates the large variety of structures available to a sensitizer wire. Bottom: Average donor-acceptor (pyrazine-Fe) electronic couplings (HDA) for different P450:bpyRu-C13-Im MD snapshots. Calculated HDA values cover a range from 0.2 × 10-7 to around 2 × 10-7 atomic units.
Beratan and coworkers have published an extensive theoretical analysis of electron tunneling through Ru-modified cytochrome b562 [22]. They analyze different Ru-modified proteins, and find two general categories of electronic coupling behavior. In one, ET occurs from the heme edge through the protein to the Ru(III) acceptor; here, there are multiple possible pathways through the protein into Ru(III) orbitals. This is a structure-insensitive regime - while it was found that specific conformations are likely to have greater couplings than other conformations, dynamical averaging along multiple pathways largely negated the dependence of ET rates on specific nuclear coordinates. However, the second case, where electrons tunnel from an axial heme position to Ru(III), does show a structure-dependent limit. Here, conformational fluctuations influence the ET rate, as there is only one entry pathway. This case is similar to the P450:wire system under discussion here.
There are other important cases where ET rates are known to be influenced by molecular dynamics, for example, Troisi and coworkers found that the rate of charge transfer in a C clamp molecule depends on the fluctuations of intervening solvent molecules [19]. Dynamics also have been found to tune the ET rates in certain molecular transport junctions [40,41].
6. Bridge Orbital Energies
Our Hartree-Fock calculated electronic couplings in Ru-wire-Fe-porphyrin systems used two different charge states of the system and different models for the Ru-diimine donor. One model describes superexchange interactions through occupied electronic states of the bridge (hole transfer), while the second model reproduces superexchange interactions through unoccupied one-electron states of the bridge (electron-mediated superexchange). Table 1 gives the ET couplings computed for dxy, dxz, and dyz orbitals of the Fe-porphyrin and an orbital representing the donor (dihydropyrazine for electron transfer and pyridinium ion for hole transfer).
Table 1.
Donor-acceptor couplings (HDA) for electron and hole transfer were computed as described in Section 4. Electron transfer rates are computed from Eq. (1) using the maximum calculated HDA value (|HDA|2 = maximum[HDA(electron transfer)2, HDA(hole transfer)2] divided by the square root of six to account for all of the bipyridine ligands) and assuming λ=0.9 eV. Driving forces and experimental rates are taken from references 1 and 6
| Wire | MD RMS HDA (electron transfer) (eV) |
MD RMS HDA (hole transfer) (eV) |
Driving Force, -ΔG (eV) |
ET rate Computed (s-1) |
ET rate Experimental (s-1) |
|---|---|---|---|---|---|
| [bpyRu-C13-Im]2+ | 1.5×10-6 | 5.1×10-7 | 0.9 | 1.6×104 | 2×104 [1] |
| [bpyRu-C11-Ad]2+ | 1.7×10-6 | 8.6×10-7 | 0.9 | 2.1×104 | 2×104 [1] |
| [bpyRu-C11-EB]2+ | 6.7×10-7 | 3.5×10-7 | 0.9 | 3.3×103 | 2×104 [1] |
| [bpyRu-F8bp-Im]2+ | 8.3×10-5 | 2.4×10-4 | 0.32 | 1.1×107 | 4×106 [6] |
| [tmbpyRu-F8bp-Im]2+ | 8.3×10-5 | 2.4×10-4 | 0.45 | 4.7×107 | 3×107 [6] |
| [bpyRu-F8bp-Ad]2+ | 2.6×10-6 | 1.1×10-6 | 0.32 | 1.3×103 | NA |
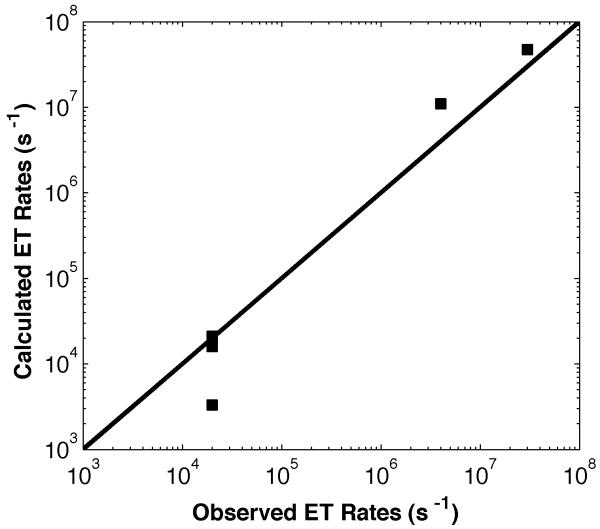
Electron transfer rates computed using the high-temperature nonadiabatic form of the Marcus expression (1) [42-45] and donor-acceptor couplings obtained from the quantum chemical calculations are set out in Table 1 along with experimental values.
| (1) |
Rates averaged over thermally accessible geometries agree reasonably well (within a factor of 4) with the observed rates in P450:wire conjugates (Table 1, Figure 4). For [bpyRu-F8bp-Im]2+ and [tmbpyRu-F8bp-Im]2+, superexchange interactions are dominated by coupling through unoccupied electronic states of the aromatic bridge (probably because of the inductive effect of F-substitution, which lowers bridge energy levels). For the saturated hydrocarbon bridge C13-Im, the main contributor to donor-acceptor coupling is hole superexchange. The hole and electron contributions to the superexchange coupling differ by about a factor of two.
Figure 4.
It is interesting to note that calculations predict slower electron transfer through the [bpyRu-F8bp-Ad]2+ wire. The Ru(II) excited state likely decays back to the ground state before electron transfer can occur. There are two main differences between this wire and F8bp analogues. The first is that it has an adamantyl terminal group that does not coordinate directly to the heme. The absence of terminal group-heme coordination likely reduces the overall donor-acceptor coupling. The second difference is that coupling for [bpyRu-F8bp-Ad]2+ is primarily due to hole superexchange, as is the case for all of the sensitizer wires with aliphatic bridge groups. We calculate that [bpyRu-F8bp-Ad]2+ ET is slower than [bpyRu-C11-Ad]2+ ET, -ΔG0 is less than λ for [bpyRu-F8bp-Ad]2+, while -ΔG0 is equal to λ for [bpyRu-C11-Ad]2+.
We turn now to comparisons of our work on P450cam with related sensitizer-wire experiments involving inducible nitric oxide synthase (iNOS) [7-12]. Photoinduced Fe(III) heme reduction in the iNOS systems is much faster than in P450cam even though the donor-acceptor distances are similar (Table 2). For conjugates of iNOS with [Re-Im-F8bp-Im]+ and [Re-Im-C3-F8bp-Im]+, we proposed that the redox process is initiated by rapid reduction of Re(I)* by a nearby tryptophan residue, analogous to the first step in an experimentally validated azurin [Cu(I)-Trp-Re(I)*] hopping model system [8,46]. We also found that an iNOS Fe(III) heme can be reduced rapidly by a surface-bound [bpyRu-F9bp]2+ sensitizer wire (k = 2 × 107 s-1) [12]. which could represent another case where multiple pathways couple the sensitizer to the edge of the heme [22].
Table 2.
Results of sensitizer-wire/protein experiments with cytochrome P450cam and iNOS. Superscripts denote references. Subscripts: (a) distances inferred from [Re-Im-F9bp]+; (b) the heme was reduced using a flash/quench method; (c) the heme was reduced using a flash/quench method but a rate was not determined; (d) the heme was reduced directly from Ru(II)* or Re(I)*
| Wire | RRu-Fe (Å) | Rate of Heme Reduction (s-1) | |
|---|---|---|---|
| cytochrome P450cam | [bpyRu-C11-Im]2+ | NA | NA |
| [bpyRu-C13-Im]2+ | 21.2[2] | 2×104 [1]b | |
| [bpyRu-C9-Ad]2+ | 21.4[2] | NA | |
| [bpyRu-C11-Ad]2+ | 21.0[2] | 2×104 [1]b | |
| [bpyRu-C7-EB]2+ | 19.5[2] | 1×103 [3]b | |
| [bpyRu-C9-EB]2+ | 19.4[2] | NA [3]c | |
| [bpyRu-C10-EB]2+ | 19.9[2] | NA [3]c | |
| [bpyRu-C11-EB]2+ | 20.1[2] | 2×104 [1]b | |
| [bpyRu-C12-EB]2+ | 20.5[2] | NA [3]c | |
| [bpyRu-C13-EB]2+ | 20.6[2] | NA [3]c | |
| [bpyRu-F8bp-Im]2+ | 22.1[2] | 4.4×106 [6]d | |
| [tmbpyRu-F8bp-Im]2+ | 18.1[6] | 2.8×107 [6]d | |
| [Ru-F8bp-Ad]2+ | 22.1[6] | NA | |
| [tmbpyRu-F9bp]2+ | 17.0[6] | NA | |
| iNOS | [Re-Im-F8bp-Im]+ | 18.0[7]a | 7×109 [8]d |
| [Re-Im-C3-F8bp-Im]+ | 18.0[7]a | 3×109 [8]d | |
| [Re-Im-C8-ArgNO2]+ | 25.5[10] | > 1×106 [10]d | |
| [Re-Im-F9bp]+ | 18.0[7] | NA | |
| [tmbpyRu-F9bp]2+ | 20.0[7] | 2×107 [12]b |
7. Conclusions and the Path Forward
Our calculations of electron transfer in complexes of P450 with molecular wires account for dramatic differences in the rates through bridges of different chemical compositions, thereby demonstrating the predictive power of ab initio Hartree-Fock theory. We show that electron flow is modulated by a minority of accessible complex geometries with stronger donor-acceptor electronic couplings; therefore, inclusion of multiple complex geometries generated in molecular dynamics simulations will be required for reliable theoretical estimates of electron transfer rates in systems with structural flexibility. Comparison of donor-acceptor coupling values computed using energy splittings of occupied and unoccupied molecular orbitals indicates that superexchange interactions through aromatic fluorinated-phenyl-imidazole bridges involve virtual reduced states (electron-type superexchange), while hole superexchange accounts for the coupling of systems with aliphatic hydrocarbon bridges.
In work that is taking us in a new direction, we are employing polyfluorinated-phenyl side-binding sensitizer wires to phototrigger ET from a Ru(II)-diimine to an Fe(III) heme [12]. These side-binding wires are very attractive for studying heme enzyme catalysis because they do not block entry of substrates to the active center. We are currently following this lead by constructing modified enzymes with sensitizers attached covalently to specific surface positions, as this will allow strategic placement of aromatic residues to control electron flow by facilitating multistep tunneling processes between donor and acceptor sites [8,46]).
8. Acknowledgements
We thank David Beratan for helpful discussions. This work was supported by NIH (GM078792 to MRH and DK019038 to HBG and GM068461 to JRW) and NSF (CHE-0802907 to HBG and JRW). MR thanks the MURI program of the AFOSR and the Chemistry Division of the NSF for support. This paper is dedicated to Ralph Pearson-friend, mentor, colleague, scientist and honest broker.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
9. References
- [1].Wilker JJ, Dmochowski IJ, Dawson JH, Winkler JR, Gray HB. Angew. Chem. Int. Ed. 1999;38:90–92. [Google Scholar]
- [2].Dmochowski IJ, Crane BR, Wilker JJ, Winkler JR, Gray HB. Proc. Natl. Acad. Sci. USA. 1999;96:12987–90. doi: 10.1073/pnas.96.23.12987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Dmochowski IJ. Probing Cytochrome P450 with Sensitizer-linked Substrates. California Institute of Technology; 2000. [Google Scholar]
- [4].Dunn AR, Dmochowski IJ, Bilwes AM, Gray HB, Crane BR. Proc. Natl. Acad. Sci. USA. 2001;98:12420–25. doi: 10.1073/pnas.221297998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Dunn AR, Hays AM, Goodin DB, Stout CD, Chiu R, Winkler JR, Gray HB. J. Am. Chem. Soc. 2002;124:10254–55. doi: 10.1021/ja0271678. [DOI] [PubMed] [Google Scholar]
- [6].Dunn AR, Dmochowski IJ, Winkler JR, Gray HB. J. Am. Chem. Soc. 2003;125:12450–56. doi: 10.1021/ja0294111. [DOI] [PubMed] [Google Scholar]
- [7].Dunn AR, Belliston-Bittner W, Winkler JR, Getzoff ED, Stuehr DJ, Gray HB. J. Am. Chem. Soc. 2005;127:5169–73. doi: 10.1021/ja046971m. [DOI] [PubMed] [Google Scholar]
- [8].Belliston-Bittner W, Dunn AR, Nguyen YHL, Stuehr DJ, Winkler JR, Gray HB. J. Am. Chem. Soc. 2005;127:15907–15. doi: 10.1021/ja0543088. [DOI] [PubMed] [Google Scholar]
- [9].Contakes SM, Nguyen YHL, Gray HB, Glazer EC, Hays AM, Goodin DB. Photofunctional Transition Metals Complexes. Springer-Verlag Berlin; Berlin: 2007. pp. 177–203. [Google Scholar]
- [10].Nguyen YHL, Winkler JR, Gray HB. J. Phys. Chem. B. 2007;111:6628–33. doi: 10.1021/jp071405+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Whited CA, Belliston-Bittner W, Dunn AR, Winkler JR, Gray HB. J. Porphyrins Phthalocyanines. 2008;12:971–78. doi: 10.1142/S1088424608000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Whited CA, Belliston-Bittner W, Dunn AR, Winkler JR, Gray HB. J. Inorg. Biochem. 2009;103:906–11. doi: 10.1016/j.jinorgbio.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Graige MS, Feher G, Okamura MY. Proc. Natl. Acad. Sci. USA. 1998;95:11679–84. doi: 10.1073/pnas.95.20.11679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Balabin IA, Onuchic JN. Science. 2000;290:114–17. doi: 10.1126/science.290.5489.114. [DOI] [PubMed] [Google Scholar]
- [15].Berlin YA, Burin AL, Siebbeles LDA, Ratner MA. J. Phys. Chem. A. 2001;105:5666–78. [Google Scholar]
- [16].Davis WB, Ratner MA, Wasielewski MR. J. Am. Chem. Soc. 2001;123:7877–86. doi: 10.1021/ja010330z. [DOI] [PubMed] [Google Scholar]
- [17].Stuchebrukhov AA. Adv. Chem. Phys. 2001;118:1–44. [Google Scholar]
- [18].Troisi A, Nitzan A, Ratner MA. J. Chem. Phys. 2003;119:5782–88. [Google Scholar]
- [19].Troisi A, Ratner MA, Zimmt MB. J. Am. Chem. Soc. 2004;126:2215–24. doi: 10.1021/ja038905a. [DOI] [PubMed] [Google Scholar]
- [20].Weiss EA, Tauber MJ, Kelley RF, Ahrens MJ, Ratner MA, Wasielewski MR. J. Am. Chem. Soc. 2005;127:11842–50. doi: 10.1021/ja052901j. [DOI] [PubMed] [Google Scholar]
- [21].Valis L, Wang Q, Raytchev M, Buchvarov I, Wagenknecht HA, Fiebig T. Proc. Natl. Acad. Sci. USA. 2006;103:10192–95. doi: 10.1073/pnas.0600957103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Prytkova TR, Kurnikov IV, Beratan DN. Science. 2007;315:622–25. doi: 10.1126/science.1134862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Beratan DN, Skourtis SS, Balabin IA, Balaeff A, Keinan S, Venkatramani R, Xiao D. Acc. Chem. Res. 2009 doi: 10.1021/ar900123t. #. #. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Balabin IA, Beratan DN, Skourtis SS. Phys. Rev. Lett. 2008;101:158102. doi: 10.1103/PhysRevLett.101.158102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Skourtis SS, Lin J, Beratan DN. In: Modern Methods for Theoretical Physical Chemistry of Biopolymers. Starikov EB, Lewis JP, Tanaka S, editors. Elsevier; Boston: 2006. [Google Scholar]
- [26].Case DA, Pearlman DA, Cladwell JW, Cheatham TE, III, Wang J, Ross WS, Simerling C, Darden T, Merz KM, Stanton RV, Cheng A, J VJ, Crowley M, Tsui V, Gohlke H, Radmer R, Duan Y, Pitera J, Massova I, Seibel GL, Singh UC, Weiner P, Kohlman PA. AMBER7 - Package of molecular simulation programs. Department of Pharmaceutical Chemistry, University of California at San Francisco; 2002. [Google Scholar]
- [27].Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. J. Am. Chem. Soc. 1995;117:5179–97. [Google Scholar]
- [28].Besler BH, Merz KM, Kollman PA. J. Comput. Chem. 1990;11:431–39. [Google Scholar]
- [29].Larsson S. J. Chem. Soc. Farad. T. 2. 1983;79:1375–88. [Google Scholar]
- [30].Newton MD. Coord. Chem. Rev. 2003;238:167–85. [Google Scholar]
- [31].Prytkova TR, Kurnikov IV, Beratan DN. J. Phys. Chem. B. 2005;109:1618–25. doi: 10.1021/jp0457491. [DOI] [PubMed] [Google Scholar]
- [32].Paddon-Row MN, Jordan KD. J. Am. Chem. Soc. 1993;115:2952–60. [Google Scholar]
- [33].Berlin YA, Hutchison GR, Rempala P, Ratner MA, Michl J. J. Phys. Chem. A. 2003;107:3970–80. [Google Scholar]
- [34].Newton MD. Theor. Chem. Acc. 2003;110:307–21. [Google Scholar]
- [35].Newton MD. Chem. Rev. 1991;91:767–92. [Google Scholar]
- [36].Liang CX, Newton MD. J. Phys. Chem. 1992;96:2855–66. [Google Scholar]
- [37].Gunsalus IC, Meeks JR, Lipscomb JD. Ann. N.Y. Acad. Sci. 1973;212:107–21. doi: 10.1111/j.1749-6632.1973.tb47590.x. [DOI] [PubMed] [Google Scholar]
- [38].Pearson RG. J. Am. Chem. Soc. 1986;108:6109–14. [Google Scholar]
- [39].Sligar SG, Gunsalus IC. Proc. Natl. Acad. Sci. USA. 1976;73:1078–82. doi: 10.1073/pnas.73.4.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Mayor M, Weber HB. Angew. Chem. Int. Ed. 2004;43:2882–84. doi: 10.1002/anie.200301733. [DOI] [PubMed] [Google Scholar]
- [41].Basch H, Cohen R, Ratner MA. Nano Lett. 2005;5:1668–75. doi: 10.1021/nl050702s. [DOI] [PubMed] [Google Scholar]
- [42].Newton MD, Sutin N. Annu. Rev. Phys. Chem. 1984;35:437–80. [Google Scholar]
- [43].Marcus RA. Rev. Mod. Phys. 1993;65:599–610. [Google Scholar]
- [44].Barbara PF, Meyer TJ, Ratner MA. J. Phys. Chem. 1996;100:13148–68. [Google Scholar]
- [45].Gray HB, Winkler JR. Proc. Natl. Acad. Sci. USA. 2005;102:3453–549. doi: 10.1073/pnas.0408029102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Shih C, Museth AK, Abrahamsson M, Blanco-Rodriguez AM, Di Bilio AJ, Sudhamsu J, Crane BR, Ronayne KL, Towrie M, Vlcek A, Jr., Richards JH, Winkler JR, Gray HB. Science. 2008;320:1760–62. doi: 10.1126/science.1158241. [DOI] [PubMed] [Google Scholar]