A long-sought goal in structural biology has been the imaging of membrane proteins in their membrane environments. This goal has been achieved through the use of electron crystallography1 in those special cases where a protein forms highly-ordered arrays in lipid bilayers. It has also been achieved by NMR methods1 in proteins up to 50 kDa in size, although milligram quantities of protein and isotopic labeling is required. For structural analysis of large soluble proteins in microgram quantities an increasingly powerful method that does not require crystallization is single-particle reconstruction from electron microscopy of cryogenically-cooled samples (cryo-EM)2. We now report the first single-particle cryo-EM study of a membrane protein, the human large-conductance calcium- and voltage- activated potassium channel3 (BK), in a lipid environment. The new method is called random spherically-constrained (RSC) single-particle reconstruction. BK channels, members of the six-transmembrane-segment (6TM) ion channel family, were reconstituted at low density into lipid vesicles (liposomes), and their function was verified by a potassium flux assay. Vesicles were also frozen in vitreous ice and imaged in an electron microscope. From images of 8,400 individual protein particles a three-dimensional reconstruction of the BK channel and also its membrane environment was obtained at a resolution of 1.7 to 2.0 nm. Not requiring the formation of crystals, the RSC approach promises to be useful in the structural study of many other membrane proteins as well.
The BK channel3 has many physiological roles; it controls firing patterns in neurons, modulates the tone of blood vessels, and in some animals is an element of the electrical resonator in the ear. Among ion channels it has served as a model system because of its remarkable ion-permeation properties4 and its accessibility for studies of allosteric control of gating3,5. It is formed as a tetramer of α-subunits expressed from the Slo1 gene6, which in the human genome is called KCNMA1. Alternative splicing7 of the Slo1 transcript results in channels having differing conductance and gating properties, while co-expression with various β-subunits results in channels having differing Ca2+ sensitivity and degrees of inactivation6. Like other members of the 6TM ion-channel family, it has voltage-sensor domains (VSDs) that confer the primary sensitivity to membrane potential. The BK channel and other members of the Slo family α-subunits also contain regulator of conductance for K+ (RCK) domains in the large intracellular C-terminal region; these confer8 the sensitivity to Ca2+ and form a “gating ring”. Unlike most 6TM α-subunits, Slo1 contains an additional transmembrane segment S0 and has an extracellular N-terminus (Fig. 1a).
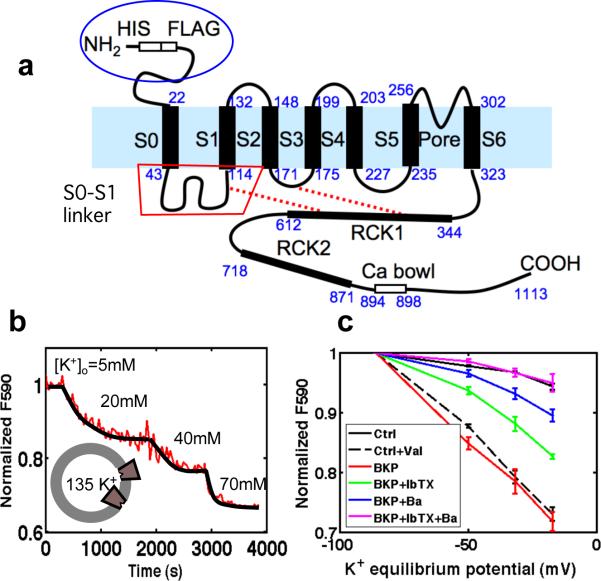
Fig. 1.
BK channel structure and specific potassium permeability. a, Topology and domain structure of the hSlo α-subunit of the BK channel. Residue numbers of native hSlo are shown in blue; the His and FLAG tags add an additional 14 residues to the N-terminus in our construct. b, Fluorescence assay of proteoliposome membrane potential. c, Normalized fluorescence as a function of calculated potassium equilibrium potential. Signals from empty POPC liposomes (Ctrl) or liposomes in the presence of 1 nM Valinomycin, a K+ ionophore (Ctrl+Val) are compared with those from BK proteoliposomes alone or with external addition of the blockers 10 mM Ba2+ or 30 μM iberiotoxin.
Membrane proteins were extracted from HEK293 cells stably expressing FLAG-tagged human Slo1 (hSlo). They were purified by an anti-FLAG affinity column, and reconstituted into 1-Palmitoyl-2-Oleoyl-sn-Glycero-3-Phosphocholine (POPC) liposomes with detergent removal via gel filtration. The resulting proteoliposomes were separated from empty liposomes and free hSlo protein through a discontinuous gradient centrifugation process (Supplementary Fig. 1a, b). The 125 kDa hSlo protein is seen by SDS-PAGE (Supplementary Fig. 1c). The reconstitution was adjusted to yield an average protein content of 1–2 BK channels per 30 nm proteoliposome.
The function of reconstituted BK channels was assayed using the cationic fluorescent dye JC-1 to monitor K+-induced changes in membrane potential9,10. Proteoliposomes loaded with 135 mM KCl were diluted into 5 mM KCl and the red fluorescence of JC-1 aggregates was measured. A subsequent decrease in fluorescence as external KCl concentration was increased indicates K+-selective permeability (Fig. 1b).
Iberiotoxin (IbTx)11 is a highly selective blocker of BK channels, binding to the extracellular face of the pore. Added to the external solution, IbTx partially reduced the fluorescence signal from the proteoliposomes, as did Ba2+, which blocks the pore from the intracellular side12. The combination of both blockers reduced the K+ flux to control levels (Fig. 1c, Supplementary Fig. 2). We conclude that BK channels were inserted in both orientations in the vesicle membranes, with the majority of channels oriented inside-out.
Single-particle reconstruction of unstained cryo-EM specimens typically requires the acquisition of 104 to 105 particle images. Acquiring this many images of protein particles in liposomes is challenging because at most only a few tens of liposomes are present in a typical micrograph, which spans <1 μm2 of specimen area. To obtain a uniform, high density of BK proteoliposomes to optimize data collection, we used a 2D streptavidin crystal as an affinity surface in the cryo-EM specimens. Proteoliposomes, doped with a few copies of biotinylated lipid and osmotically swollen to ensure a spherical shape, were allowed to attach to the crystal (Fig. 2a) before blotting and rapid-freezing of the specimen. Low-dose electron-microscope (EM) images (Supplementary Fig. 3a) show periodic information from the 2D crystal that can be used as an image-quality reference13. For further processing we computationally removed the crystal information from images (Fig. 2b).
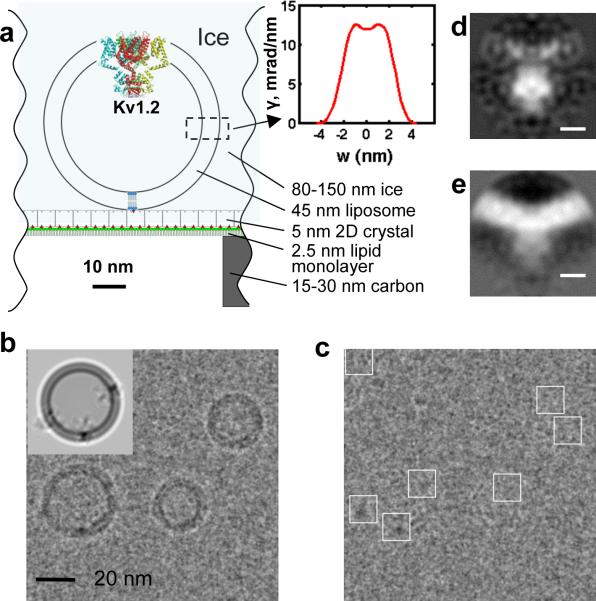
Fig. 2.
Cryo-EM specimen and image processing. a, Scale drawing of the tethered proteoliposome system. The inset shows the electron-scattering profile of the POPC bilayer. b, EM image with the periodic crystal information removed. The inset shows a simulation in which the membrane profile and three copies of the BK structure are oriented to reproduce the proteoliposome image underneath. c, The same micrograph after subtraction of modeled membranes. BK channel particles were selected manually (white boxes). d,e, Central sections of the 3D reconstruction of BK channels after subtraction of the membrane density and after a patch of membrane was computationally restored, respectively; scale bar is 5 nm.
Ideally a 3D reconstruction would contain an entire proteoliposome, complete with BK channel and spherical membrane. Unfortunately the variability of liposome size precludes the merging of their images; instead we fitted and subtracted a model14 of the membrane contribution to each image and reconstructed the protein particles alone (Fig. 2c). In the random spherically-constrained (RSC) method used here, the determination of the angles of orientation of each particle is greatly aided15 by the spherical vesicle geometry. The apparent position of the particle in a projection image, relative to the vesicle center, specifies two of the three Euler angles (Supplementary Fig. 4a) within a fourfold ambiguity. Our 3D reconstructions relied on the Fourier-space reconstruction strategy of Grigorieff16 but with geometric constraints applied to the angular search. The 3D reconstructions imposed C4 symmetry and utilized subsets of the 8400 particle images from 644 micrographs. The resulting EM density map (Fig. 2d) shows a low density in the transmembrane region, as expected from the subtraction of the modeled membranes. A parallel reconstruction, performed using the same angle assignments but with a patch of membrane density restored to each particle image, illustrates the curved membrane (Fig. 2e). The resolution of a reconstruction from the entire dataset was estimated to be 1.7–2.0 nm by the Fourier shell correlation (Supplementary Fig. 4b). Differences in vesicle size, and therefore membrane curvature, appear to have little effect on the channel structure (see Supplementary Fig. 5).
Two-thirds of the Slo1 protein mass is in the large cytoplasmic C-terminal domain; we therefore assign the large particle mass that was usually found external to a proteoliposome to be the C-terminal domain of an inside-out BK channel. With the membrane potential close to zero and free calcium in the nanomolar range, we expect that the derived structure reflects a closed channel. Only 3% of voltage sensors are activated5 under these conditions.
The transmembrane region of BK, containing the pore and voltage-sensor domains (Fig. 3) is similar in extent to that seen in recent X-ray crystal structures of 6TM potassium channels: the voltage-gated channel Kv1.2 structure shows an open (or possibly depolarized-inactivated) state17 while MlotiK1, a prokaryotic ligand-gated channel, is seen in its closed state18. Because the membrane-subtraction process modifies the densities of the protein at the membrane-aqueous interface, it is best to use the membrane-restored EM map (Fig. 3d) for a comparison of the extracellular face of BK with the other 6TM channel structures. BK shows protrusions corresponding to the turret region of the pore domain (red square) and the S2 helix of the voltage-sensor (green circle). There is good correspondence of these features to both the Kv1.2 and MlotiK1 X-ray structures, and at this resolution, there is expected to be little difference in the extracellular face between open and closed states. BK however shows a much larger protrusion at the periphery of the VSD (blue octagon). The additional helix S0 and the extracellular ~40 N-terminal residues are expected to give a feature of this size.
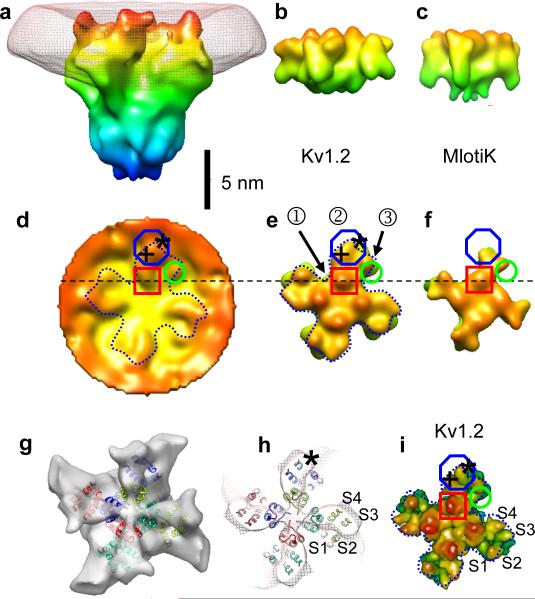
Fig. 3.
Structure of the transmembrane region. a, Surface rendering of the membrane-subtracted, inside-out BK channel map, obtained from 3400 images of particles in large vesicles. Superimposed is the membrane density (mesh). Maps were filtered to a resolution of 2.0 nm; isosurfaces are colored according to the z-coordinate. b, c, Surface renderings of Kv1.2 and MlotiK X-ray structures, filtered to 1.7 nm resolution for comparison. d, Extracellular aspect of the membrane-restored reconstruction of BK. e–f, Corresponding extracellular views of Kv1.2 and MlotiK. g, Extracellular aspect of the membrane-subtracted BK map (solid) with transmembrane helices of the docked Kv1.2 structure superimposed. h, Section (2 nm thick) of the membrane-subtracted BK map (mesh) near the membrane center with the corresponding Kv1.2 helices superimposed. i, Surface rendering of the Kv1.2 transmembrane region, but with a resolution of 0.3 nm.
In a plane at the center of the membrane, features in the EM protein density map are little affected by membrane subtraction; there the envelope of each VSD is seen to correspond well to the four-helix bundle of the Kv1.2 structure (Fig. 3h). Compared to Kv1.2, the EM map contains additional density at the VSD periphery (see also Fig. 4e) which can account for the additional S0 helix. This location, which would place it in contact with S2 and S3, is consistent with a recent crosslinking study19. On the other hand, the EM map does not match the configuration of VSD helices in the KvAP crystal structure20, which is thought to reflect a non-native conformation.
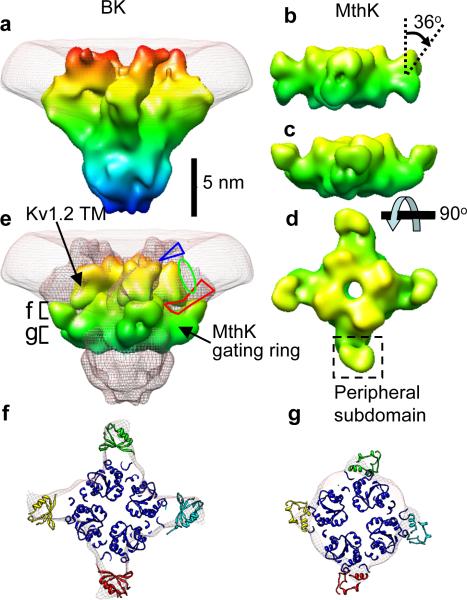
Figure 4.
Structure of the gating ring. a, Side view of BK and membrane, rotated 45° about the vertical axis from the view in Fig. 3a. b, Surface rendering of the “closed” MthK gating ring filtered to 1.7 nm resolution. c–d, The MthK gating ring modified by a 36° rotation of the peripheral domains. e, View of the BK map (mesh) with Kv1.2 transmembrane region and MthK gating ring docked. All models are colored according to the z-coordinate. The blue triangle and red polygon indicate possible locations for the N-terminal region and the S0–S1 linker, respectively. The green oval is the proposed location of the S0 helix. f, g, “Top” views of the BK map and docked MthK gating ring, in the sections marked in part e.
The gating ring, a calcium sensor region formed by RCK domains, is apparent in the EM map of BK. In the prokaryotic Ca2+-activated K+ channel MthK, the gating ring consists of eight identical RCK domains. In BK, each α-subunit is thought to contain two RCK domains, yielding a total of eight RCKs in the tetrameric channel complex8,21,22. The inner domain of MthK gating ring, the structure formed by helices A to F and associated beta strands, is well conserved in nearly all RCK sequences8. The RCK peripheral domain, which produces the four protruding regions in the MthK gating ring, is formed by helices G to J and their associated beta strands and is variable among different species8. As expected, the well-conserved inner domain of the closed MthK gating ring23 is readily docked into the EM map of BK (Fig. 4 e and f). The periphery of the MthK gating ring does not match the strong continuous density near the transmembrane region of BK. This density can be better matched by tilting the peripheral domains of MthK gating ring by 36° (Fig. 4c and d). Even so, the tilted MthK peripheral domains contain excess mass compared to the EM map (Fig. 4g), consistent with the idea that in BK the RCK2 domain is truncated and is about 100 amino acid residues shorter than RCK1.
The calcium bowl, a high-affinity calcium-binding site that lies after RCK2, might also reside in the gating ring. Alternatively, it could be located below the gating ring in the large mass of density which we assign to the remainder of the protein sequence. This density is of the correct size to encompass the 240 C-terminal residues (Fig. 4e) including the calcium bowl.
The close apposition between the transmembrane region and the gating ring is consistent with functional studies22 demonstrating the formation of a Mg2+ binding site between residues in the transmembrane region and in the gating ring. These residues are D99 in the S0–S1 linker and N172 in S2 of the transmembrane domain, along with residues E374 and E399 in RCK1 of the BK gating ring (Fig. 1a). In the MthK channel the gating ring is connected to the S6 helix in the pore region by a disordered 17-residue linker, leaving a cleft with a maximum width ~1 nm in the open state8; the resulting lateral openings allow ions to access the ion conduction pore. A similar linker is expected to be formed in BK channels, but a cleft of this size would not be visible in our map, even though it must be large enough to accommodate the inactivation domain of the β2 subunit24.
A new single-particle reconstruction technique, RSC cryo-EM, thus provides the first structural model of the Ca2+- and voltage- activated K+ channel. The disposition of the voltage-sensor domains (VSDs) of 6TM channels has been a matter of controversy, but we find that the membrane-embedded channel's VSDs match well with two recent 6TM channel X-ray crystal structures. The calcium-sensing “gating ring” is also visible in the density map, as is the density corresponding to regions unique to this channel protein. The RSC technique has the added advantage that the channel protein is imaged in a membrane environment where channel activity can also be assayed. It should be possible to obtain reconstructions at higher resolution through the use of larger numbers of particle images than we used here.
Methods Summary
Purification and reconstitution of BK protein
Full-length human Slo (hSlo) protein (gi:507922) carrying an N-terminal FLAG tag was obtained from an HEK293 cell line25. Protein was solubilized in dodecylmaltoside and purified using a FLAG antibody affinity column, where the detergent was exchanged with decylmaltoside (DM) before elution with FLAG peptide. BK protein was concentrated and added to DM-solubilized POPC lipid (DM:POPC=3:1) giving a final protein-to-lipid molar ratio of 1:5,000. Also present in the lipid mixture was biotinylated dipalmitoyl-phosphatidyethanolamine (biotin–DPPE), at a low concentration (1/3600 of POPC) calculated to yield about 3 copies per 30 nm liposome. Gel filtration was used to remove detergent. After the liposomes were concentrated, they were floated on a discontinuous Nycodenz gradient. Protein-free liposomes were found in the 3% Nycodenz band, while proteoliposomes appeared at the 5%–15% boundary. Lipid concentrations were determined by measuring phosphate using a colorimeter; protein was determined with the BCA method, and the relative fraction of BK in each layer was determined by densitometry of a Western blot.
Tethering of BK proteoliposomes and cryo-EM imaging
Two-dimensional (2D) streptavidin crystals were grown at room temperature using the procedure described by Wang et al.13 After the 2D streptavidin crystal was transferred to the perforated carbon film, the crystal was incubated with BK proteoliposome suspensions for 10–40 min to allow binding. The sample was blotted at room temperature and immediately fast-frozen in liquid ethane. Samples were imaged at −180 °C in a Tecnai F30 microscope at 300 keV with a 30 μm objective aperture and zero-loss energy filtering. The electron dose at the specimen was 1000–3000 e/nm2. Images were taken at 50,000 magnification and 2–5 μm defocus, and recorded on a GIF 2K×2K UltraScan 1000 FT camera with an effective pixel size of 0.253 nm.
Methods
Membrane potential measurements with JC-1 to assay ion channel activity
5P,5P,6,6P-Tetrachloro-1,1,3,3P-tetraethylbenzimadazolylcarbocyanine iodide (JC-1, Invitrogen) was used to assay ion channel activity using a method similar to that of Chanda and Mathew9. BK proteoliposomes loaded with 135 mM KCl were incubated with channel blockers if desired, and diluted into 5mM KCl solution containing 1.6 μM of JC-1, giving a total lipid concentration of ~6 μM. The fluorescence signal of the J-aggregates (λex=480nm, λem=590nm) was monitored as the external K+ concentration was increased by the addition of 2M KCl solution.
The large permeability of BK channels (~108 ions/s) and the small liposome size means that the membrane potential of a liposome will be established very quickly once a channel opens, as the movement of only ~100 ions is required. We performed the flux assays in nanomolar free Ca2+ and at liposome membrane potentials near zero, to reduce the BK open probability5 to about 10−5. Even so, only one functional channel provides a maximal fluorescence signal from a given liposome. The time scale of charging is so much faster than the time scale of redistribution of JC-1 across the membrane (tens of seconds) that the block of BK channels must be very complete if a reduction in the fluorescence response to K+ gradients is to be observed. Thus we applied channel blockers at about 1000 × Kd in order to reduce the net permeability to control levels.
Image processing
The periodic 2D streptavidin crystal information was removed computationally as described13, and the liposome membrane contribution was removed using a model based on the average POPC membrane profile from 250 micrographs. The model was obtained using the Hankel transform as described14. Images of liposomes smaller than 20 nm were not used. BK particle images were manually picked using EMAN boxer26 and the contrast-transfer function (CTF) parameters were estimated using a home-made Matlab program13 from the power spectrum of each micrograph. The picked BK particle images, with crystal and liposome information removed, were used to determine the 3D structure employing constraints based on the spherical geometry of the proteoliposomes. First, two of the three Euler angles (θ and ψ Supplementary Fig. 4a) were estimated based on the position in the image of the particle with respect to the liposome center. To account for uncertainty in the estimate of the particle center, the two angles were allowed to vary in a range corresponding to a maximum in-plane displacement of the particle center of 1.8 nm. Then reference images (projection images of the 3D BK map from the earlier iteration) that fell within these ranges were computed, using an angular step size of 3°. The references were sorted according to the similarity (cross correlation coefficient) with the particle image, and tested from the best to the worst match until the predicted position based on the angles of that reference was consistent with the observed position in the cryo-EM image. Then the angles of that reference were assigned as the angles of the BK particle. All possible orientations of the BK channels were included in this dataset (Supplementary Fig. 6).
After three cycles of initial search, refinement of the structure was carried out in the same way except that the absolute value of the correlation coefficient, rather than the correlation coefficient itself, was maximized. This serves to reduce the reference bias in the final structure27. Finally, the 3D map was constructed with equal weighting of each particle image, using the least-squares Fourier-space algorithm that is used in the Frealign program16. Because the handedness of the reconstruction from projections was not known, we compared the intra-membrane density of Kv1.2 and the BK map to make the assignment.
The first reconstruction was a 3D electron-scattering map of BK channels with membrane removed (Fig. 2d). Then each particle image was modified by re-inserting the image of a patch of membrane in the following way. Based on the assigned Euler angles and the model of the corresponding lipid vesicle, a spherical sector of membrane centered on the particle was defined. The projection of this membrane patch, modified by the contrast transfer function, was then added back to the particle image and a second 3D map was constructed (Fig. 2e). Because the angle assignments and scaling were maintained between the two reconstructions, the difference quantitatively shows the membrane density. Given estimates28 of the internal potential of protein, lipid membrane and water, the contrast between membrane and protein regions is expected to be only about 50% of that between protein and water.
The proteoliposomes used for structure determination ranged from 20 to 60 nm in diameter (Supplementary Fig. 5h). The question therefore arises, whether the membrane curvature in vesicles of different sizes would affect the observed BK structure. To address this question, two BK structures were reconstructed from small (20–24.5 nm) and large (24.5–60 nm) BK proteoliposomes respectively (Supplementary Fig. 5). For examination of the “extracellular” channel surface the threshold of the EM map was first set such that the “bare” membrane thickness was 5.0 nm. In the vicinity of BK channels, the extracellular views of BK from small and large vesicles were seen to be very similar (Supplementary Fig. 5 a,b). When the threshold was increased, the features in regions 1–3 (Supplementary Fig. 5 d,e) could be identified more clearly in both the small and large liposomes, but between different sizes of proteoliposomes there was still no difference observed in the surface profile of the channel features. A line profile of the isosurface, tracing a path between two opposite VSDs, confirms a lack of height difference out to a distance of 6 nm from the channel axis (Supplementary Fig. 5g). Thus the effect of the membrane curvature on the BK structure at the current resolution appears to be small. All image processing and other numerical calculations were done in the Matlab programming environment (MathWorks, Natick, MA). The docking of density maps derived from the crystal structures of MlotiK, Kv1.2, and the MthK gating ring (PDB codes 3BEH, 2A79, 2FY8) was performed manually using Chimera29.
Supplementary Material
Acknowledgments
We thank Drs. Nikolaus Grigorieff and Chen Xu (Brandeis University) for their help with data collection. We also thank Dr. Shumin Bian for sharing his BK purification methods and Ms. Yangyang Yan for cell culture. Image processing and reconstruction made use of the Yale Biomedical High-Performance Computing Center.
Footnotes
The density maps have been deposited in the Electron Microscopy Data Bank with accession numbers EMD-5114 and EMD-5121.
References
- 1.Raunser S, Walz T. Electron crystallography as a technique to study the structure on membrane proteins in a lipidic environment. Annual review of biophysics. 2009;38:89. doi: 10.1146/annurev.biophys.050708.133649. [DOI] [PubMed] [Google Scholar]
- 2.Frank J. Three-Dimensional Electron Microscopy of Macromolecular Assemblies. Oxford University Press; Oxford: 2006. [Google Scholar]
- 3.Cui J, Yang H, Lee US. Molecular mechanisms of BK channel activation. Cellular and Molecular Life Sciences (CMLS) 2009;66(5):852. doi: 10.1007/s00018-008-8609-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neyton J, Miller C. Discrete Ba2+ block as a probe of ion occupancy and pore structure in the high-conductance Ca2+-activated K+ channel. Journal of General Physiology. 1988;92(5):569. doi: 10.1085/jgp.92.5.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horrigan FT, Aldrich RW. Coupling between voltage sensor activation, Ca2+ binding and channel opening in large conductance (BK) potassium channels. Journal of General Physiology. 2002;120(3):267. doi: 10.1085/jgp.20028605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salkoff L, Butler A, Ferreira G, et al. High-conductance potassium channels of the SLO family. Nature Reviews Neuroscience. 2006;7(12):921. doi: 10.1038/nrn1992. [DOI] [PubMed] [Google Scholar]
- 7.Fodor Anthony A., Aldrich Richard W. Convergent Evolution of Alternative Splices at Domain Boundaries of the BK Channel. Annual Review of Physiology. 2009;71(1):19. doi: 10.1146/annurev.physiol.010908.163124. [DOI] [PubMed] [Google Scholar]
- 8.Jiang YX, Lee A, Chen JY, et al. Crystal structure and mechanism of a calcium-gated potassium channel. Nature. 2002;417(6888):515. doi: 10.1038/417515a. [DOI] [PubMed] [Google Scholar]
- 9.Chanda B, Mathew MK. Functional reconstitution of bacterially expressed human potassium channels in proteoliposomes: Membrane potential measurements with JC-1 to assay ion channel activity. Biochimica et Biophysica Acta - Biomembranes. 1999;1416(1-2):92. doi: 10.1016/s0005-2736(98)00217-x. [DOI] [PubMed] [Google Scholar]
- 10.Reers M. J-aggregate formation of a carbocyanine as a quantitative fluorescent indicator of membrane potential. Biochemistry. 1991;30(18):4480. doi: 10.1021/bi00232a015. [DOI] [PubMed] [Google Scholar]
- 11.Galvez A, Gimenez-Gallego G, Reuben JP, et al. Purification and characterization of a unique, potent, peptidyl probe for the high conductance calcium-activated potassium channel from venom of the scorpion Buthus tamulus. Journal of Biological Chemistry. 1990;265(19):11083. [PubMed] [Google Scholar]
- 12.Miller C, Latorre R, Reisin I. Coupling of voltage-dependent gating and Ba++ block in the high-conductance, Ca++-activated K+ channel. Journal of General Physiology. 1987;90(3):427. doi: 10.1085/jgp.90.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L, Ounjai P, Sigworth FJ. Streptavidin crystals as nanostructured supports and image-calibration references for cryo-EM data collection. Journal of Structural Biology. 2008;164:190. doi: 10.1016/j.jsb.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Liguo, Bose Pulkit S., Sigworth Fred J. Using cryo-EM to measure the dipole potential of a lipid membrane. PNAS. 2006;103(49):18528. doi: 10.1073/pnas.0608714103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang QX, Chester DW, Sigworth FJ. Spherical reconstruction: A method for structure determination of membrane proteins from cryo-EM images. Journal Of Structural Biology. 2001;133119(2-3) doi: 10.1006/jsbi.2001.4376. [DOI] [PubMed] [Google Scholar]
- 16.Grigorieff N. FREALIGN: High-resolution refinement of single particle structures. Journal of Structural Biology. 2007;157(1):117. doi: 10.1016/j.jsb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 17.Long SB, Campbell EB, MacKinnon R. Voltage sensor of kv1.2: Structural basis of electromechanical coupling. Science. 2005;309(5736):903. doi: 10.1126/science.1116270. [DOI] [PubMed] [Google Scholar]; Long Stephen B., Tao Xiao, Campbell Ernest B., et al. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450(7168):376. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 18.Clayton GM, Altieri S, Heginbotham L, et al. Structure of the transmembrane regions of a bacterial cyclic nucleotide-regulated channel. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(5):1511. doi: 10.1073/pnas.0711533105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu G, Zakharov SI, Yang L, et al. Position and role of the BK channel alpha subunit s0 helix inferred from disulfide crosslinking. Journal of General Physiology. 2008;131(6):537. doi: 10.1085/jgp.200809968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang YX, Lee A, Chen JY, et al. X-ray structure of a voltage-dependent K+ channel. Nature. 2003;423(6935):33. doi: 10.1038/nature01580. [DOI] [PubMed] [Google Scholar]
- 21.Fodor AA, Aldrich RW. Statistical limits to the identification of ion channel domains by sequence similarity. Journal of General Physiology. 2006;127(6):755. doi: 10.1085/jgp.200509419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang HH, Shi JY, Zhang GH, et al. Activation of Slo1 BK channels by Mg2+ coordinated between the voltage sensor and RCK1 domains. Nat. Struct. Mol. Biol. 2008;15(11):1152. doi: 10.1038/nsmb.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye S, Li Y, Chen L, et al. Crystal Structures of a Ligand-free MthK Gating Ring: Insights into the Ligand Gating Mechanism of K+ Channels. Cell. 2006;126(6):1161. doi: 10.1016/j.cell.2006.08.029. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Z, Zhou Y, Ding JP, et al. A limited access compartment between the pore domain and cytosolic domain of the BK channel. Journal of Neuroscience. 2006;26(46):11833. doi: 10.1523/JNEUROSCI.3812-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bingham JP, Bian S, Tan ZY, et al. Synthesis of a biotin derivative of iberiotoxin: Binding interactions with streptavidin and the BK Ca2+-activated K+ channel expressed in a human cell line. Bioconjugate Chemistry. 2006;17(3):689. doi: 10.1021/bc060002u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semiautomated software for high-resolution single-particle reconstructions. Journal of Structural Biology. 1999;128(1):82. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 27.Stewart A, Grigorieff N. Noise bias in the refinement of structures derived from single particles. Ultramicroscopy. 2004;102(1):67. doi: 10.1016/j.ultramic.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 28.Grigorieff N, Beckmann E, Zemlin F. Lipid location in deoxycholate-treated purple membrane at 2.6 angstrom. Journal of Molecular Biology. 1995;254(3):404. doi: 10.1006/jmbi.1995.0627. [DOI] [PubMed] [Google Scholar]
- 29.Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera - A visualization system for exploratory research and analysis. Journal of Computational Chemistry. 2004;25(13):1605. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.