Abstract
To determine human Ig heavy chain variable region (VH) gene segment organization on individual homologous chromosomes, an efficient approach has been developed. Single spermatozoa were used as subjects for the study. Upon sperm lysis, VH regions in each sperm were randomly sheared into fragments by the random Brownian force. The fragments were separated from each other by aliquoting the lysate into a certain number of tubes. The gene segments in the VH1 and VH4 families in each tube were identified by denaturing gradient gel electrophoresis after PCR amplification. The polymorphic VH sequences were used to determine the parental origins of the analyzed sperm. VH segment organization in the parental haplotypes was determined by aligning the overlapping fragments from the spermatozoa with the corresponding haplotypes. Based on this comparison between the resulting haplotype maps and the composite map reported previously, the VH region on chromosome 14 could be subdivided into four portions. The numbers and compositions of the VH gene segments differ considerably among the maps in two portions, but are highly conserved in the other two. The data also indicate that the VH region on chromosome 15 may contain a large duplicated block with copy number varying among haplotypes. The approach used in the present study may be used to construct high-resolution haplotype maps without molecular cloning.
For many human multigene families, determination of gene organization on individual homologous chromosomes is a challenging issue for several reasons: (i) the diploidy of the human genome, (ii) the genes sharing a high degree of sequence identity in each family, (iii) variation in DNA sequences and gene compositions among the haplotypes, (iv) different chromosomal locations, (v) occupation of large chromosomal regions, and (vi) recent duplications. In this paper, we show that all of these issues can be addressed by analyzing single DNA fragments from the haploid genomes of individual spermatozoa.
Two human Ig heavy chain variable region (VH) families, VH1 and VH4, were used for the analysis. Human VH gene segments are located on three chromosomes, 14q32, 15q11, and 16p11 (1–6). However, only about half of the VH segments on chromosome 14 are functional (7, 8). Each haploid human genome contains ≈120 distinct VH gene segments (5, 7, 8). The VH segments are classified into seven families (VH1–VH7) with segments sharing >80% sequence identity in each family (9–15). Extensive polymorphisms for the VH region on chromosome 14 have been reported (16–22). A composite map for this region was constructed recently (8). Several studies showed that the number and composition of the VH gene segments in some portions of the VH region on chromosome 14 varied among the haplotypes (23–30). However, the extent of variation in the entire region remains unclear because no haplotype maps for this region have been constructed. The VH segment organization on the other two chromosomes has not been determined.
MATERIALS AND METHODS
Preparation of Single VH Segment-Containing DNA Fragments from Individual Spermatozoa.
The procedure by Lien et al. (31) was modified and used for single sperm preparation. Briefly, sperm (≈500) were suspended in 200 μl of 0.5% melted low melting point agarose in H2O at 37°C. About 100 μl of the suspension was pipetted onto a microscopic slide (at a 45° angle) across the width and was allowed to flow to form a thin layer. After 10 min at room temperature, single sperm were scraped up with a syringe needle (27G½) and each was placed into a 0.5-ml microtube containing 4 μl of H2O. The tubes were incubated at 72°C for 1 min to melt the agarose. Each sperm was lysed by incubating at 37°C for 15 min in a 5-μl solution containing 10 mM Tris⋅HCl (pH 8.3), 10 mM EDTA, 0.1% SDS, 40 mM DTT, and 50 μg/ml of proteinase K. After adding 27 μl of H2O and flushing through a pipette tip once, each lysate was subdivided into a desired number of tubes (eight or 12 tubes for 10 sperm analyzed in the early stage of the study, and 16 tubes for 37 sperm analyzed later). To inactivate proteinase K, the tubes were incubated at 85°C for 10 min after adding a drop of mineral oil.
PCR Amplification.
Family-specific primers were designed for the VH1 and VH4 families according to the sequences conserved among the sequences in each family. The primers used for the first PCR step were V1M2 (5′-CCTCAGTGAAGGTCTCCTGCAAG-3′) and V1M4 (5′-cgccgcccccgccccCTGCTCAGCTCCATGTAGGCTGTG-3′) for the VH1 family, and V4M1 (5′-aatgagctccACACATTTCCTTAAA-TTC -3′) and V4M4 (5′-cgccgcccccgccccGAAGGCTTCACCAGTCCTG-3′) for the VH4 sequences. In the second step, VHGC (5′- cgcccgccgcgccccgcgcccgtcccgccgcccccgcccc-3′), as part of the GC clamp, was used for both families to replace V1M4 and V4M4 and two internal (nested) primers V1M3 (5′-GGACAAGGGCTTGAGTGGATGGGA-3′) and V4M2 (5′-GAACATGAAACACCTGTGGTTCT-3′) were used to replace V1M2 and V4M1 for the two families, respectively. Letters in lowercase in the primer sequences represent nongenomic sequences used either for enhancing amplification efficiency or for attaching the GC clamp to facilitate denaturing gradient gel electrophoresis (DGGE) separation. All PCR amplifications were performed with a DNA Thermal Cycler 480 (Perkin–Elmer). In the first step, each sample contained 1× PCR buffer (100 mM Tris⋅HCl, pH 8.3/50 mM KCl/1.5 MgCl2/100 μg/ml gelatin), the four dNTPs each at 100 μM, primers each at 50 nM for the VH4 family and 20 nM for the VH1, and 0.5 units of the Taq DNA polymerase. The final volume was 25 μl. Each PCR cycle consisted of a denaturation step at 95°C for 30 sec and an extension step at 72°C for 30 sec. The annealing step was at 50°C for 5 min for the first three cycles, 55°C for 3 min for seven cycles, and 60°C for 2 min for the last 30 cycles. In the second PCR step, the VH1 and VH4 segments were amplified separately with 1.5-μl aliquots from the first-step products. Each sample contained 1× PCR buffer, the four dNTPs each at 100 μM, the corresponding primers each at 0.2 μM, and 0.5 units of the enzyme in a final volume of 25 μl. Each PCR cycle was 95°C for 30 sec for denaturation and 72°C for 30 sec for extension. The annealing step was 55°C for 1 min for the first three cycles and 60°C for 1 min for 25 cycles. An additional PCR cycle was performed at 95°C for 2 min, 60°C for 1 min, and 72°C for 10 min after adding 5 μl of solution containing 0.5 units of the Taq polymerase and adjusting the concentration of each primer to 0.5 μM to minimize DNA heteroduplexes that may form DGGE bands.
DGGE.
The DGGE gels were prepared according to the manufacturer of the DGGE apparatus (C.B.S. Scientific, Del Mar, CA). Ten percent polyacrylamide was used for all gels. The denaturing gradients were 52–68% and 55–73% (100% denaturing strength defined as 7 M urea and 40% formamide) for the VH1 and VH4 families, respectively. Electrophoresis was performed at 60°C at 113 v for 15 h. Some VH1 bands were resolved with 50–65% denaturing range.
RESULTS
Identification of the VH Gene Segments in Single DNA Fragments from Individual Sperm.
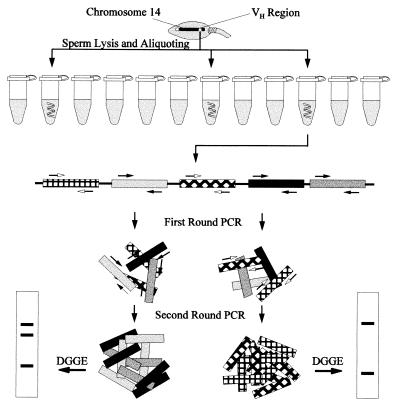
The procedure for identifying the VH gene segments in single DNA fragments from individual sperm is diagramed in Fig. 1. Single sperm cells were prepared from a healthy donor and lysed. Because the VH regions are relatively large (1,100 kb for the one on chromosome 14, ref. 8), these regions are unavoidably sheared by the random Brownian force when released from sperm. To separate the VH segment-containing fragments from each other, each sperm lysate was aliquoted into 16 tubes (into eight or 12 for a few analyzed at the early stage of the study). The VH1 and VH4 sequences in each tube were amplified to analyzable amounts with the two-round PCR procedure described in Materials and Methods. In the final PCR products, the sequences flanked by the primers were ≈83 bp for VH1 and ≈145 bp for VH4.
Figure 1.
Diagram for identification of VH gene segments in single DNA fragments from a single sperm. Only the VH region on chromosome 14 is shown. VH segments of two different families are shown as squares with different backgrounds and fillings. Family-specific PCR primers are represented by black and white arrows. The two vertical squares at the bottom represent DGGE gel strips with bands shown as bars.
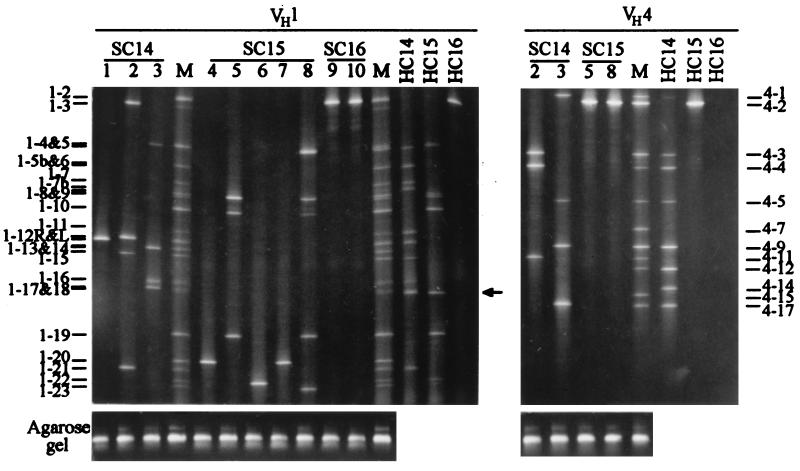
Because the lengths of the amplified VH sequences from each family were either the same or very similar, these sequences could not be separated by regular gel electrophoresis and were resolved by DGGE (32–36). DGGE was designed to separate the DNA fragments differing by as little as 1 bp. Because most VH-amplified sequences differed from each other by >1 bp, these sequences should be readily separated from each other by DGGE. The VH1 and VH4 sequences amplified from the diploid genome of the sperm donor were used as the molecular markers in DGGE. The DGGE bands for the VH sequences amplified from the tubes with VH segment-containing fragments from one sperm are shown in Fig. 2. The DGGE bands were numbered based on their positions on the DGGE gels (from top to bottom). The VH loci represented by the DGGE bands were named with prefixes 1- and 4-, respectively, followed by the band numbers.
Figure 2.
DGGE separation and chromosomal location of the VH sequences amplified from single DNA fragments generated from a single sperm. Results for the VH1 and VH4 families are shown separately. The chromosomal origin of the VH sequences in each lane is indicated by the chromosome number after SC for a sperm chromosome and HC for a human chromosome in the somatic cell hybrids. M, VH sequences amplified from the sperm donor’s genomic DNA and used as molecular markers. Lanes 1–10, VH sequences amplified from single DNA fragments and resolved by DGGE. VH1 and VH4 sequences in the lanes sharing the same numbers were those in the same tubes before the two families were amplified separately in the second round of PCR. Some bands such as 1–6 and 1–5b could not be separated well in the gel shown here but were well resolved in a gel with a slightly changed denaturing range. A band amplified from the Chinese hamster genome background in lanes HC14 and HC15 is indicated by an arrow (lane HC16 was from a human-mouse hybrid). Results from agarose gel (1%) electrophoresis for the same set of sperm samples are aligned with the DGGE results for illustrating the size uniformity of the amplified VH sequences.
Determination of the VH Gene Segment Organization in Individual Haplotypes.
VH segment-containing fragments from 47 single sperm were analyzed with the procedure described above. The results were used to determine the VH gene segment organization in individual haplotypes through three steps.
Determining the chromosomal locations of the VH gene segments. To determine the VH segment organization in individual haplotypes, it is necessary to learn the chromosomal locations of these segments. Human-rodent somatic cell hybrids containing single human chromosomes 14, 15, or 16, respectively, in mapping panel 2 of the National Institute of General Medical Sciences repository were used as standards for the analysis. By comparing the DGGE bands (Fig. 2, lanes M) from the sperm donor’s diploid genome with those from the somatic hybrids (Fig. 2, lanes HC14, HC15, and HC16), the chromosomal locations of most VH sequences in the sperm donor were determined. To confirm that comigration of the DGGE bands from the two sources was because of their sequences identity rather than comigration of different sequences, the DGGE bands were excised from the gel, reamplified with PCR, and subjected to sequence analysis. The results showed that all comigrating bands from these two sources contained identical sequences. With the known chromosomal locations, the VH sequences amplified from the sperm donor’s genome were used as molecular markers to determine the chromosomal locations of the VH sequences identified from the sperm samples.
Several VH sequences detected from the sperm donor’s genomic DNA were not detected from the somatic hybrids. The chromosomal locations of these sequences were readily determined because in the analysis of single VH segment-containing fragments from single sperm these sequences frequently were codetected with certain sequences whose chromosomal locations were known through the somatic hybrid assay.
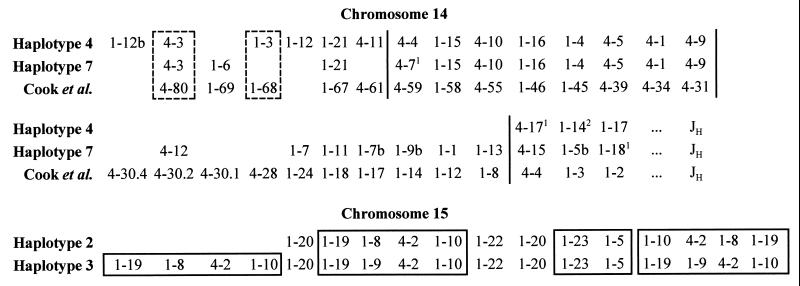
Determination of the VH segment compositions in the parental haplotypes. The parental haplotypes for chromosome 14 were named as haplotypes 4 and 7, respectively, based on the initial observation that two allelic VH4 sequences, 4–4 and 4–7, segregated among the sperm analyzed. Of the 47 sperm analyzed, 28 contained haplotype 4, and 19 contained haplotype 7. As shown in Fig. 3 (Upper), 17 VH segments (nine VH1 and eight VH4) were detected from sperm with haplotype 4, and 21 (13 VH1 and eight VH4) from those with haplotype 7. Eight VH segments (Fig. 3, 1–12L, 1–3, 1–12R, 4–11, 4–4, 4–17, 1–14, and 1–17) were specific for haplotype 4, and 12 VH segments (Fig. 3, 1–6, 4–7, 4–12, 1–7, 1–11, 1–7b, 1–9b, 1–1, 1–13, 4–15, 1–5b, and 1–18) were for haplotype 7. These haplotype-specific sequences were, in turn, used to confirm the haplotypes in the sperm analyzed.
Figure 3.
Maps for the VH1 and VH4 segments on chromosomes 14 and 15. The alignment of the maps for chromosome 14 is read from left to right and then from top to bottom, and is subdivided into four portions by vertical bars (see text). Loci in the dashed-lined boxes are aligned putatively because sequence data in the map by Cook et al. (8) are not available. The superscript letters indicate the numbers of base pairs different between the allelic sequences within the amplified regions. The unresolved pairs, 4–9 and 4–1 in both haplotypes 4 and 7, 1–17 and 1–14, and 1–16 and 1–4 in haplotype 4 are placed in the alignment according to the order information in the map by Cook et al. (8). 1–9b and 1–1 in haplotype 7 could not be amplified by the family-specific primers and were placed on the map by analyzing additional sperm with both gene segment-specific and family-specific primers. VH segments that are closely located on chromosome 15 are boxed. The CEs on the right of the maps may be oriented differently as indicated by the observation that segments 1–23 and 1–5 were codetected with 1–10 and 4–2 from a sperm with haplotype 2 but with 1–19 and 1–9 from a sperm with haplotype 3. The sequences with no counterpart sequences in the maps and the accession numbers of their counterpart sequences in the GenBank are: 1–3, Z12305 (10); 1–5, Z29632 (5); 1–8, Z29632 (5); 1–9, Z17390 (5); 1–10, Z29596 (5); 1–12, Z12312 (10); 1–12b, AF030491; 1–19, Z29631 (5); 1–20, AF030492; 1–22, Z29633 (5); 1–23, L25542 (6); 4–2, X92231 (52); and 4–3, U23548.
Because the two parental haplotypes of chromosome 15 contained either two or three copies of a duplicated VH segment-containing block, these haplotypes were called haplotypes 2 and 3 (Fig. 3, Lower). Of the 47 sperm analyzed, 17 apparently contained haplotype 2, and 28 were with haplotype 3. The remaining two sperm seemed to be recombinants. Thirteen VH segments (11 VH1 and two VH4) were detected from haplotype 2 and 17 (14 VH1 and three VH4) from haplotype 3.
The sequence, 1–2 (GenBank accession no. AF030490), was the only VH1 sequence detected from chromosome 16. The copy number of this sequence detected from each sperm ranged from zero to three. Because of the small number of the detected VH1 sequences and the fact that there is no VH4 sequence on chromosome 16 (4–6), no further analysis was performed for the VH segments on this chromosome.
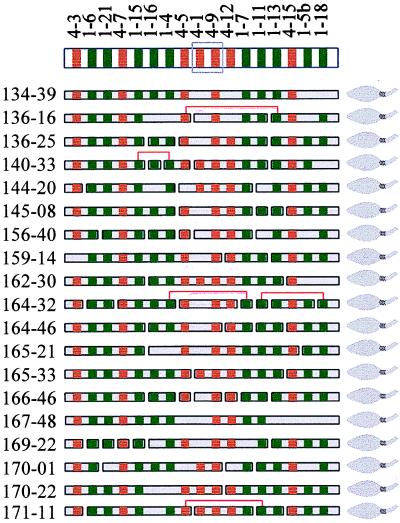
Determination of the VH segment organization in individual haplotypes. By treating the VH segments as restriction enzyme sites, the order of the VH segments in each haplotype was determined by a procedure similar to restriction enzyme mapping. Practically, each haplotype was deduced by aligning the overlapping VH segment-containing fragments from the sperm cells with the corresponding haplotype. The deduced order of the VH segments in haplotype 7 and the aligned VH segment-containing fragments from the 19 sperm is shown in Fig. 4. A small fraction (eight of 90 for haplotype 4 and five of 79 for haplotype 7) of tubes may have received two VH segment-containing fragments in each. This assumption is consistent with the expectations based on the Poisson distribution. These tubes were easily identified because if they were not considered as the tubes containing two fragments in each, the gene order deduced based on the information from most tubes would have to be seriously disrupted. The orders of 4–9 and 4–1 in both haplotypes 4 and 7, 1–17 and 1–14, and 1–16 and 1–4 in haplotype 4 could not be resolved because of either their relatively close physical locations (for example, the distance between 1–16 and 1–4 is only ≈4 kb; ref. 7), or the low detection rate (for 4–1).
Figure 4.
Diagram of the VH segment-containing fragments in the 19 sperm with haplotype 7. The deduced order of the VH segments in this haplotype is shown at the top. Sperm numbers are on the left. VH1 (green) and VH4 (red) segments are represented by squares. Undetected segments are left blank at the corresponding positions. Codetected but apparently nonadjacent fragments are connected with brackets. Unresolved segments, 4–1 and 4–9, because of the low detection rate of 4–1 are boxed.
The VH Gene Segment Organization in the Haplotypes of Chromosome 14.
Based on the order of the VH segments and sequence comparison, the two haplotype maps for chromosome 14 generated in the present study were aligned with the composite map by Cook et al. (8) (Fig. 3, Upper). The orders of the VH segments in these maps are all consistent, indicating that the method used in the current study is highly reliable. The alignment for the three maps could be subdivided into four portions (indicated by the vertical bars): (i) the most JH-distal portion (upper left). This portion is highly polymorphic in VH segment composition. Of the seven loci, five (1–12b, 1–6/1–69, 1–3/1–68, 1–12, and 4–11/4–61) contained null alleles. It should be pointed out that two VH1 segments, YAC7 and DP-10, both were placed to the 1–69 locus in the map by Cook et al. (8). These two sequences differ by 7 bp within 294 bp. Our sequence data indicate that 1–6 in haplotype 7 is identical to YAC7 and 1–12 in haplotype 4 is identical to DP-10. Our map data also showed that 1–12 is between 1–3 and 1–21 in haplotype 4. Therefore, 1–6 and 1–12 are likely two different loci as shown in Fig. 3 rather than two allelic sequences of one locus. Sasso et al. (30) showed that the VH1 gene segment, 1–69 (VH26), was present in a very complicated fashion. It may have been duplicated into two loci. This observation is consistent with our results, (ii) the upper-right portion of the alignment. This portion is surprisingly conserved. Not only are the VH segment number in all three maps the same, but the nucleotide sequences of the eight segments are all identical in the amplified regions, except for a 1-bp difference between 4–7 and its allelic counterparts, (iii) the lower left portion. This portion features large insertion/deletion polymorphisms. No VH segment in this portion was detected from haplotype 4. For the four VH4 segments, 4–30.4, 4–30.2, 4–30.1, and 4–28 on the map of Cook et al. (8), only the counterparts for 4–30.2 were detected in haplotype 7. VH4 segments in this portion also were shown to be polymorphic in the previous studies (8, 24–26), (iv) the most JH-proximal portion (lower right). This portion includes three VH segments with no variation in VH segment composition although allelic differences at the nucleotide sequences level were detected at each locus.
The size of the VH region covered by the composite map by Cook et al. (8) was estimated to be ≈1,100 kb. Based on this estimate, the differences between the VH segment numbers in the maps, and the average distance between adjacent VH segments in the map by Cook et al., the VH region is estimated to be ≈800 kb in haplotype 4 and ≈970 kb in haplotype 7. Because a large portion of the composite map was generated with diploid materials (7), the estimated size by Cook et al. may be greater than the actual sizes of the VH region in the haplotypes. In other words, the estimated sizes of haplotypes 4 and 7 could be closer to the actual sizes. The mean numbers of VH segment-containing fragments detected from each sperm were 3.21 and 4.16 for haplotypes 4 and 7, respectively. Therefore, the average size of these fragments could be 230–250 kb with a range from several kb to >1,000 kb (Fig. 4). Based on the previous map information (8), the distances between the genes resolved in the current study ranged from 9 kb (between 4–15 and 1–5b) to 115 kb (between 1–15 and 1–16).
The VH Gene Segment Organization in the Haplotypes of Chromosome 15.
The order of the VH gene segments and the map alignment for the haplotypes 2 and 3 of chromosome 15 are shown Fig. 3 (Lower). More than one copy was detected for the four VH segments, 1–10, 4–2, 1–9 (or 1–8), and 1–19, from most single sperm. These segments must be closely located because they were codetected very frequently (106 of 119). Because these duplicated blocks are the major components on the map for chromosome 15, they are called the core elements (CEs). The two haplotypes of chromosome 15, haplotypes 2 and 3, were named based on the number of CEs in these haplotypes. Other than the CEs, two copies of 1–20 and one copy of 1–5, 1–22, and 1–23 were detected from both haplotypes. Segments 1–5 and 1–23 also may be very closely located because they were codetected in 30 of 31 sperm from which both segments were detected.
The two VH segments 1–8 and 1–9 differing by 1 bp can be used to distinguish the CEs. CEs containing 1–8 are called CE-8 and those contained 1–9 are called CE-9. As shown in Fig. 3 (Lower), haplotype 2 contained only two CE-8s, and haplotype 3 contained one CE-8 and two CE-9s. The haplotypes in two sperm could not be placed to any of these parental types. One of these sperm contained three CE-8s, and the other contained two CE-8s and one CE of unknown type because experimentally neither 1–8 nor 1–9 was detected from this CE. These sperm may have resulted either from duplication of a CE-8 in haplotype 2 or from genetic recombination between the two different parental haplotypes. In any case, the genetic event responsible for the occurrence of these recombinant sperm should be on the order of 2/47 = 0.04.
The means numbers of the VH segment-containing fragments detected from chromosome 15 were 4.18 and 5.07 per sperm for haplotypes 2 and 3, respectively. Based on these numbers and the estimated size of the VH segment-containing fragments from chromosome 14, the sizes of the VH regions on chromosome 15 were estimated as ≈1,000 kb for haplotype 2 and 1,200 kb for haplotype 3. Because fewer VH segments were detected from this chromosome, the density of the VH segments on chromosome 15 could be lower than those on chromosome 14. Other than 1–12b and 1–20, all VH segments on chromosome 15 identified in the present study were reported previously (5, 6). VH segment duplication on this chromosome was implicated by analyzing the DH segments (6). However, no analysis on VH segment organization on this chromosome has been reported previously. Difficulty caused by the large distances between the VH loci may be one of the reasons.
DISCUSSION
Determination of gene organization by sperm analysis has many advantages over the conventional methods. Because each sperm contains only one set of chromosomes, the complications involved in using diploid materials are removed. With the molecular cloning method, the sizes of the cloned DNA fragments are restricted by the upper and lower limits of the vector capacity. With sperm analysis, it is possible to generate DNA fragments of several hundred to >1,000 kb so that the order of the genes separated by relatively large distances can be determined and a large amount of work involved in cloning can be avoided. On the other hand, any closely located duplicated genes also may be resolved if the force applied to shearing sperm DNA is strong enough, or if the sperm DNA is digested with one or more restriction enzymes that do not cut the amplified sequences. Our group has developed a high-capacity multiplex PCR protocol (37) with which a large number of single-copy sequences can be amplified simultaneously by PCR and resolved either by regular gel electrophoresis or by DGGE (unpublished data). By combining this method with sperm analysis, the organization of a large number of single-copy sequences also could be examined. With the conventional approaches, resolving recently duplicated genes is a serious issue because the duplicates are, or almost are, identical. With sperm analysis, although it may require thousands of tubes to separate the copies from each other for the genes coding for ribosomal RNAs, for many duplicated genes with relatively small copy numbers, their copy numbers could be readily estimated as long as each sperm lysate is aliquoted into a sufficient number of tubes.
When a large number of genes with different chromosomal locations are studied, the analysis could be very complex. This complexity can be significantly reduced by first determining the chromosomal locations of the genes by using the sequences amplified from the human-rodent somatic hybrids containing single human chromosomes as molecular markers as shown in the present study. With known chromosomal locations of the genes, the number of tubes to which each sperm lysate is aliquoted to can be significantly reduced because genes on different chromosomes can be analyzed separately even if they are codetected from the same tubes.
A method called radiation hybrid (RH) mapping was first described by Cox et al. (38) and has been used by many laboratories for constructing physical maps. With this method, human cells containing chromosomes sheared by radiation are fused with rodent cells. Because closely located markers tend to retain together in the hybrid cells, the order of the markers can be determined by examining the codetection rates of the markers. This method is powerful when the markers are separated by large distances (several megabases). Compared with the RH method, our approach is used for the genes separated by smaller distances (from several kb to several hundred kb) and for determining gene organization in greater detail. Experimentally, because no viable cells are required, our experimental procedure is much simpler. With our method, the gene order in individual haplotypes can be determined.
Another method called “Happy Mapping” was used to order DNA sequences (27, 39, 40) by aliquoting sheared genomic DNA to one haploid equivalent per tube and by analyzing the codetection rate of the markers. Both Happy Mapping and the sperm analysis used in the present study are based on limiting dilution without involving molecular cloning for map construction. These two approaches are different in the following aspects: (i) Haplotype map construction. Because each sperm contains precisely one haploid genome, sperm analysis can be used to construct haplotype maps readily. A few polymorphic markers are needed only for the purposes of distinguishing the sperm of different parental origins and the recombinants from nonrecombinants. Happy Mapping starts with a DNA solution containing DNA fragments from many copies of two mixed haplotypes. With this approach, haplotype maps can be constructed only if most, if not all, DNA fragments contain one or more known polymorphic markers. Practically, identification of these markers requires DNA blocks larger than 1 kb at a density comparable to or higher than the marker density and determination of the sequences in these blocks. (ii) Background noise and map resolution. For both approaches, the basic information “units” are the DNA fragments containing the markers rather than the haploid genomes. Sperm analysis pursues separation of these basic units and therefore generates data with very low background noise, i.e., the fraction of tubes receiving >1 marker. With sperm analysis, the distribution of two markers in a lysate is completely independent from those in others. When the two markers are linked, it is impossible to generate any “background noise” because each lysate contains only one DNA fragment with the two markers. Background noise can be generated only when the two markers are separated (unlinked) in a lysate and can be expressed as 1/n2, where n is the tube number for each sperm lysate, or the dilution factor. According to this correlation, the background noise generated from the unlinked markers decreases exponentially as the dilution factor increases linearly. It also indicates that the effect on reducing the fraction of informative tubes caused by increasing the dilution factor may not “cancel out” that caused by reducing background noise. In contrast, when two markers are aliquoted from a DNA solution with the Happy Mapping approach, the background noise may be generated from a number of possible combinations of the markers and of different copies of the markers by the Poisson distribution. The background noise also may be generated from the intermixing of the linked and unlinked markers. Our simulation analysis indicated that with such a complex distribution pattern, the background noise cannot be reduced significantly by increasing the dilution factor at least in a practical range (1 to 1/20 haploid genome equivalent per tube) while this can be achieved by sperm analysis. To achieve the same degree of statistic confidence for linkage detection, 40–50 times more samples are required by Happy Mapping if the DNA sample is aliquoted into one haploid genome equivalent per tube compared with sperm analysis when each sperm lysate is aliquoted into 16 tubes.
Two VH4 segments on chromosome 14, 4–61 (4–11 in haplotype 4) and 4–59 (4–4 and 4–7 in the haplotypes) were found at the border between a diversified and a conserved portion (Fig. 3). This finding indicates that the relative locations of these two genes may vary considerably among the haplotypes because gene composition in the adjacent diversified portion may vary considerably. Results from our recent study (41) indicate that the 4–61 locus is highly polymorphic with at least four alleles including a null allele at a frequency of 23%. The presence or absence of the 4–61 segment also could affect the relative location of 4–51. On the other hand, these two segments were shown to be involved in generating anti-DNA autoantibodies in the patients with systemic lupus erythematosus (42–44). The usage of the VH segments could be considerably affected by their organization. For example, when a frequently used VH segment is absent, other VH segment(s) may be used more frequently. With sperm analysis, it is possible to compare the VH gene organization in the haplotypes of normal individuals with that of patients with autoimmune diseases. The study could provide important information about the effect of VH segment organization on autoimmune diseases.
Acknowledgments
We thank Drs. Norman Arnheim, Patrick K. Bender, Patrick Charmley, Kai-Mon Lee, Robert Nagele, and W. Steven Ward for comments on the manuscript. This work was supported in part by an institutional grant from the Coriell Institute and the Emlen Stokes Chair in Genetics awarded to H.L.
ABBREVIATIONS
- DGGE
denaturing gradient gel electrophoresis
- VH
heavy chain variable region
- CE
core element
Footnotes
References
- 1.Croce C M, Shander M, Martinis J, Cicurel L, D’Ancona G G, Dolby T W, Koprowski H. Proc Natl Acad Sci USA. 1979;76:3416–3419. doi: 10.1073/pnas.76.7.3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cox D W, Markovic V D, Teshima I E. Nature (London) 1982;297:428–430. doi: 10.1038/297428a0. [DOI] [PubMed] [Google Scholar]
- 3.Kirsch I R, Morton C C, Nakahara K, Leder P. Science. 1982;216:301–303. doi: 10.1126/science.6801764. [DOI] [PubMed] [Google Scholar]
- 4.Cherif D, Berger R. Genes Chromosomes Cancer. 1990;2:103–108. doi: 10.1002/gcc.2870020205. [DOI] [PubMed] [Google Scholar]
- 5.Tomlinson I M, Cook G P, Carter N P, Elaswarapu R, Smith S, Walter G, Buluwela L, Rabbitts T H, Winter G. Hum Mol Genet. 1994;3:853–860. doi: 10.1093/hmg/3.6.853. [DOI] [PubMed] [Google Scholar]
- 6.Nagaoka H, Ozawa K, Matsuda F, Hayashida H, Matsumura R, Haino M, Shin E K, Fukita Y, Imai T, Anand R, et al. Genomics. 1994;22:189–197. doi: 10.1006/geno.1994.1360. [DOI] [PubMed] [Google Scholar]
- 7.Matsuda F, Shin E K, Nagaoka H, Matsumura R, Haino M, Fukita Y, Taka-ishi S, Imai T, Riley J H, Anand R, et al. Nat Genet. 1993;3:88–94. doi: 10.1038/ng0193-88. [DOI] [PubMed] [Google Scholar]
- 8.Cook G P, Tomlinson I M, Walter G, Riethman H, Carter N P, Buluwela L, Winter G, Rabbitts T H. Nat Genet. 1994;7:162–168. doi: 10.1038/ng0694-162. [DOI] [PubMed] [Google Scholar]
- 9.Lee K H, Matsuda F, Kinashi T, Kodaira M, Honjo T. J Mol Biol. 1987;195:761–768. doi: 10.1016/0022-2836(87)90482-7. [DOI] [PubMed] [Google Scholar]
- 10.Tomlinson I M, Walter G, Marks J D, Llewelyn M B, Winter G. J Mol Biol. 1992;227:776–798. doi: 10.1016/0022-2836(92)90223-7. [DOI] [PubMed] [Google Scholar]
- 11.Schroeder H W, Jr, Hillson J L, Perlmutter R M. Science. 1987;238:791–793. doi: 10.1126/science.3118465. [DOI] [PubMed] [Google Scholar]
- 12.Humphries C G, Shen A, Kuziel W A, Capra J D, Blattner F R, Tucker P W. Nature (London) 1988;331:446–449. doi: 10.1038/331446a0. [DOI] [PubMed] [Google Scholar]
- 13.Buluwela L, Rabbitts T H. Eur J Immunol. 1988;18:1843–1845. doi: 10.1002/eji.1830181130. [DOI] [PubMed] [Google Scholar]
- 14.Willems van Dijk K, Mortari F, Kirkham P M, Schroeder H W, Jr, Milner E C. Eur J Immunol. 1993;23:832–839. doi: 10.1002/eji.1830230410. [DOI] [PubMed] [Google Scholar]
- 15.Berman J E, Mellis S J, Pollock R, Smith C L, Suh H, Heinke B, Kowal C, Surti U, Chess L, Cantor C R, et al. EMBO J. 1988;7:727–738. doi: 10.1002/j.1460-2075.1988.tb02869.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Souroujon M C, Rubinstein D B, Schwartz R S, Barrett K J. J Immunol. 1989;143:706–711. [PubMed] [Google Scholar]
- 17.Willems van Dijk K, Schroeder H W, Jr, Perlmutter R M, Milner E C. J Immunol. 1989;142:2547–2554. [PubMed] [Google Scholar]
- 18.Willems van Dijk K, Sasso E H, Milner E C. J Immunol. 1991;146:3646–3651. [PubMed] [Google Scholar]
- 19.Guillaume T, Rubinstein D B, Young F, Tucker L, Logtenberg T, Schwartz R S, Barrett K J. J Immunol. 1990;145:1934–1945. [PubMed] [Google Scholar]
- 20.Sasso E H, Willems Van Dijk K, Milner E C. J Immunol. 1990;145:2751–2757. [PubMed] [Google Scholar]
- 21.Rubinstein D B, Symann M, Guillaume T. Scand J Immunol. 1993;37:33–38. doi: 10.1111/j.1365-3083.1993.tb01661.x. [DOI] [PubMed] [Google Scholar]
- 22.Walter M A, Surti U, Hofker M H, Cox D W. EMBO J. 1990;9:3303–3313. doi: 10.1002/j.1460-2075.1990.tb07530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shin E K, Matsuda F, Nagaoka H, Fukita Y, Imai T, Yokoyama K, Soeda E, Honjo T. EMBO J. 1991;10:3641–3645. doi: 10.1002/j.1460-2075.1991.tb04930.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen P P, Yang P M. Scand J Immunol. 1990;31:593–599. doi: 10.1111/j.1365-3083.1990.tb02810.x. [DOI] [PubMed] [Google Scholar]
- 25.Sasso E H, Willems van Dijk K, Bull A, van der Maarel S M, Milner E C. J Immunol. 1992;149:1230–1236. [PubMed] [Google Scholar]
- 26.Milner E C, Hufnagle W O, Glas A M, Suzuki I, Alexander C. Ann NY Acad Sci. 1995;764:50–61. doi: 10.1111/j.1749-6632.1995.tb55806.x. [DOI] [PubMed] [Google Scholar]
- 27.Walter G, Tomlinson I M, Cook G P, Winter G, Rabbitts T H, Dear P H. Nucleic Acids Res. 1993;21:4524–4529. doi: 10.1093/nar/21.19.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rubinstein D B, Symann M, Stewart A K, Guillaume T. Mol Immunol. 1993;30:403–412. doi: 10.1016/0161-5890(93)90070-r. [DOI] [PubMed] [Google Scholar]
- 29.Sasso E H, Willems van Dijk K, Bull A P, Milner E C. J Clin Invest. 1993;91:2358–2367. doi: 10.1172/JCI116468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sasso E H, Buckner J H, Suzuki L A. J Clin Invest. 1995;96:1591–1600. doi: 10.1172/JCI118198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lien S, Kaminski S, Alestrom P, Rogne S. Genomics. 1993;16:41–44. doi: 10.1006/geno.1993.1137. [DOI] [PubMed] [Google Scholar]
- 32.Fischer S G, Lerman L S. Cell. 1979;16:191–200. doi: 10.1016/0092-8674(79)90200-9. [DOI] [PubMed] [Google Scholar]
- 33.Fischer S G, Lerman L S. Proc Natl Acad Sci USA. 1983;80:1579–1583. doi: 10.1073/pnas.80.6.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Myers R M, Lumelsky N, Lerman L S, Maniatis T. Nature (London) 1985;313:495–498. doi: 10.1038/313495a0. [DOI] [PubMed] [Google Scholar]
- 35.Sheffield V C, Cox D R, Lerman L S, Myers R M. Proc Natl Acad Sci USA. 1989;86:232–236. doi: 10.1073/pnas.86.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abrams E S, Murdaugh S E, Lerman L S. Genomics. 1990;7:463–475. doi: 10.1016/0888-7543(90)90188-z. [DOI] [PubMed] [Google Scholar]
- 37.Lin Z, Cui X, Li H. Proc Natl Acad Sci USA. 1996;93:2582–2587. doi: 10.1073/pnas.93.6.2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cox D R, Burmeister M, Price E R, Kim S, Myers R M. Science. 1990;250:245–250. doi: 10.1126/science.2218528. [DOI] [PubMed] [Google Scholar]
- 39.Dear P H, Cook P R. Nucleic Acids Res. 1989;17:6795–6807. doi: 10.1093/nar/17.17.6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dear P H, Cook P R. Nucleic Acids Res. 1993;21:13–20. doi: 10.1093/nar/21.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cui X, Li H. Hum Genet. 1997;100:96–100. doi: 10.1007/s004390050472. [DOI] [PubMed] [Google Scholar]
- 42.Demaison C, Chastagner P, Theze J, Zouali M. Proc Natl Acad Sci USA. 1994;91:514–518. doi: 10.1073/pnas.91.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Manheimer-Lory A, Katz J B, Pillinger M, Ghossein C, Smith A, Diamond B. J Exp Med. 1991;174:1639–1652. doi: 10.1084/jem.174.6.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kasaian M T, Ikematsu H, Balow J E, Casali P. J Immunol. 1994;152:3137–3151. [PMC free article] [PubMed] [Google Scholar]