Abstract
Purpose
The level of drug metabolism and drug transport is correlated with the sensitivity of cancer cells towards platinum-based chemotherapy. We hypothesize that genetic polymorphisms in metabolising enzymes gene GSTP1 (glutathione S-transferase P1), and MRP2 (multidrug resistance-associated protein 2) (ABCC2), which result in inter-individual differences in metabolism and drug disposition, may predict clinical response to platinum agents in advanced non-small cell lung cancer (NSCLC) patients.
Methods
Totally 113 patients with advanced NSCLC were routinely treated with platinum-based chemotherapy, and clinical response was evaluated after four cycles. MRP2 C-24T (−24C>T), MRP2 Val417Ile (1249G>A), MRP2 Ile1324Ile (3972C>T), and GSTP1 Ile105Val (342A>G) genotype were determined by gene-chip method (a 3-D (three dimensions) polyacrylamide gel-based DNA microarray method) using DNA samples isolated from peripheral blood collected before treatment. Pearson Chi-square test and Fisher’s exact test were performed to measure the differences of the chemotherapeutic efficacy among variant genotype. The odds ratios and 95% confidence intervals were computed by logistic regression.
Results
The C→T change of MRP2 C-24T and the A→G change of GSTP1 Ile105Val polymorphism significantly increased platinum-based chemotherapy response.
Conclusion
The polymorphic status of MRP2 C-24T and GSTP1 Ile105Val might be the predictive markers for the treatment response of advanced NSCLC patients. The DNA microarray-based method is accurate, high throughput and inexpensive, suitable for single-nucleotide polymorphism genotyping in a large number of individuals.
Keywords: Single-nucleotide polymorphism, Gene chip, MRP2, GSTP1, Non-small cell lung cancer (NSCLC), Chemotherapy
Introduction
Lung cancer is the most common cause of cancer death in many countries; more than a million people in the world die from the disease each year [1]. About 80% of lung cancer patients are diagnosed with non-small cell lung cancer (NSCLC), of which nearly two-thirds are detected at advanced stages [2]. Chemotherapy is the major treatment method currently for advanced NSCLC, but the rate of efficacy with standard cytotoxic chemotherapy is only 30–40%, and this level has reached a “therapeutic plateau” [3]. At present, platinum-based double-agent therapies are the major remedial measure of the advanced NSCLC and have been shown to improve the overall survival [4]. Cisplatin is still the scaffolding of combination chemotherapy. Resistance to chemotherapy is also a challenge. Cisplatin resistance can develop as a result of decreased influx or increased efflux of drug, glutathione or metallothionein conjugation, drug detoxification, DNA repair, or skipping lesions during DNA replication [5]. It is more likely that the combination of these mechanisms results in the cisplatin resistance. Because drug resistance varies from person to person, genetic factors are believed to influence the effectiveness of lung cancer treatment [6].
The finding of human genome project indicated that 99% of DNA within different individuals were identical, and only 1% was variant, of which the major one was single-nucleotide polymorphism (SNP). SNP is a point mutation carried by some individuals of a population. The study of pharmacogenetics indicated that such tiny diversity in sequence of genome significantly influenced on individual treatment response, toxicity, and survival in cancer patients. Because the inter-individual variation is necessary for the optimization of medication, the genetic polymorphisms have the potential significance in drug disposition and pharmacokinetics. In addition, SNP has greater clinical significance in terms of its ease of clinical application, rather than its mRNA, which present some clinical difficulties in terms of obtaining tissue samples from lung cancer patients.
As known, platinum-based drugs inhibit tumor growth mainly by the formation of bulky DNA adducts, and the latter are mainly removed by DNA repair mechanisms, especially by the nucleotide excision repair (NER) mechanisms. Also, our work team has already done some work about the association of DNA repair gene polymorphisms and platinum-based chemotherapy. Moreover, it should not been disregarded that drug metabolism and drug transport also play an important role in response to platinum-based chemotherapy.
Phase II metabolising enzymes take advantage of electrophilic groups intrinsically carried in a structure, or introduced by phase I metabolism, to conjugate xenobiotics with donor molecules, such as glutathione (GSH), UDP glucuronic acid, or 3′-phosphoadenosine-5′- phosphosulfate (PAPS). The glutathione S-transferase (GST) family of phase II metabolizing enzymes catalyzes detoxifying endogenous reactions with GSH and protect cellular macromolecules from damage caused by a wide variety of endogenous and exogenous molecules, including cytotoxic, mutagens, carcinogens, and chemotherapeutic agents [7]. The resulting glutathione adducts have increased the solubility and can then be either excreted or further metabolized. In the current lung cancer chemotherapy, many of the commonly used drugs are metabolized by the glutathione system, especially the platinum drugs [8].
The 17 human cytosolic GST subunits are classified as seven gene families according to their biochemical characteristics and amino acid sequence similarities: α (GSTA), μ (GSTM), θ (GSTT), π (GSTP), ω (GSTO), ζ (GSTZ), and σ (GSTS) [9]. Human GSTs are nearly ubiquitously expressed, and GSTP is the most abundant subunit in lung and brain. GSTP1 is widely expressed in different human epithelial tissue [10] and is the most abundant GST isoform in the lung [11]. GSTP1 conjugates and protects particularly against the cytotoxic effects of some chemotherapeutic agents, including anthracyclines, alkylating agents, and their metabolites [12]. The GST gene families, including glutathione S-transferase P1 (GSTP1), contain several polymorphic loci. At least three of the genes that code for GSTs, GSTM1, GSTT1, and GSTP1, have been found to have functional polymorphisms that are frequently present in general populations [13]. The role of GSTs in the detoxification of antitumor agents suggests the possible implication of GST polymorphisms to the chemotherapeutic response. Patients with a GSTs-null genotype are believed to exhibit impaired detoxification of environmental genotoxic agents and chemotherapeutic drugs, which prompts the hypothesis that allelic variants associated with less effective detoxification of potential antitumor drugs can confer an increased sensitivity to chemotherapy.
In addition to the enzyme, drug transporters are important in determining drug absorption, drug distribution to tissues, and drug excretion in the urine and bile. Some studies have shown that the conjugates formed by GSTs are transported by MRP2 (multidrug resistance protein 2, also designated canalicular multispecific organic anion transporter, cMOAT, or ABCC2 protein), which are the part of the phase III biotransformation system, in an ATP-dependent manner [14, 15]. Furthermore, GSTP1 activity may work synergistically with MRP transport as a tri-GSH conjugate [16].
Multidrug resistance-associated protein 2 are expressed in the outer plasma membrane as well as in intracellular vesicles and the Golgi apparatus. This indicates a role in the sequestration of drugs into vesicles and cellular drug export. The human MRP subfamily contains nine members. MRP2 is a 1,545 amino acid, 190–200 kDa protein having two ATP-binding domains, and 17 transmembrane regions in its sequence [17]. MRP2 belongs to the subfamily C of ABC (ATP-binding cassette) transporter proteins [18], the efflux transporters that have a role in limiting influx and facilitating efflux to prevent the intracellular accumulation of their own substrate compounds [17]. Drugs are detoxified and conjugated in vivo and then exported out of the cells. In tumor cells, such functions confer multidrug resistance for various anticancer agents. MRP2 has broad-substrate specificity and was identified to transport relatively hydrophilic compounds, including the glucuronide, glutathione, and sulfate conjugates of endogenous and exogenous compounds covering organic anions derived from phases I and II metabolism of xenobiotics and anionic anticancer drugs, such as cisplatin [19, 20]. The interindividual variations on activity and expression level of the transporter, due to SNP of MRP2 gene, might be a critical factor in selection of the anticancer drugs.
It has been speculated that SNPs in drug metabolizing enzymes genes and drug transporters genes may alter their expression or activity, affect drug disposition, and in turn influence the effects of cancer treatment [21]. Therefore, we could expect that the lower the levels of expression of GSTP1 and MRP2, the more susceptible the tumors to platinum therapies. Here, a study of 113 NSCLC patients was conducted to evaluate if functional polymorphisms in GSTP1 and MRP2 are associated with patient response to chemotherapy.
Patients and methods
Study subjects
All patients for the study were recruited from several hospitals in Nanjing of China between March 2006 and September 2007. To avoid the confounding effect of differences in outcome resulting from clinical stage, only advanced NSCLC patients were included in the analysis. Because SNPs evaluated in the present study are potentially relevant to therapies based on platinum, only the response to the first platinum-based regimen was assessed, and the patients who had received previous chemotherapy were excluded. There were 113 patients who were eligible in this study. These 113 eligible patients, all of Chinese Han people, were diagnosed with histologically confirmed advanced NSCLC (stages IIIA–IV), and had a measurable lesion by CT scan, a Karnofsky performance status of not less than 60. The status of all patients in electrocardiogram, blood chemistries, hepatic, and renal function at the beginning of treatment was normal. The study was approved by an Ethics Review Committee at the hospital and patients gave consent to participate. The main characteristics of patients were shown in Table 1.
Table 1.
Patient clinicopathologic characteristics and chemotherapy regimens
| Characteristics | Patient no. | (%) |
|---|---|---|
| Age (years) | ||
| Median (range) | 59.6 (34-84) | |
| Gender | ||
| Female | 37 | 32.7 |
| Male | 76 | 67.3 |
| Histology | ||
| Squamous cell carcinoma | 30 | 26.5 |
| Adenocarcinoma | 80 | 70.8 |
| Large cell and undifferentiated carcinoma | 3 | 2.7 |
| Chemotherapy regimens | ||
| DDP/CBP+TAX/TXT/DOC | 49 | 43.4 |
| DDP/CBP+GEM | 59 | 52.2 |
| DDP/CBP+NVB | 5 | 4.4 |
DDP cisplatin, CBP carboplatin, TAX taxol/paclitaxel, TXT taxetere, DOC docetaxel, GEM gemcitabine, NVB vinorelbine
Chemotherapy regimens and therapeutic effect evaluation
All patients had received platinum-based chemotherapy, 49 (43.4%) were given TP/TC/DP/DC regimens (DDP/CBP+TAX/TXT/DOC), 59 (52.2%) had GP/GC regimens (DDP/CBP+GEM), and 5 (4.4%) received NP/NC regimens (DDP/CBP+NVB) (Table 1). Concrete dosage: DDP (cisplatin) 30 mg/m2 on day 2–4; CBP (carboplatin) AUC = 4–5 g on day 1; TAX (taxol/paclitaxel) 175 mg/m2 on day 1 (kept for 3 h), TXT (taxetere)/DOC (docetaxel) 75 mg/m2 on day 1 (kept for 1 h); GEM (gemcitabine) 1 g/m2 on day 1 and day 8; NVB (vinorelbine) 25 mg/m2 on days 1 and 8. All chemotherapeutic drugs were administered intravenously, and the treatment cycles were repeated every 3–4 weeks. Patient responses to treatment were determined after four cycles by the WHO criteria [22], which classify the response into four categories: complete response (CR), partial response (PR), stable disease (SD), and progressive disease (PD). CR was defined as complete disappearance of all measurable lesions. PR required at least 50% reduction in measurable lesions. Patients with SD had less than a 50% decrease or no more than a 25% increase in the size of measurable lesions. PD was assigned to patients when measurable lesions increased by more than 25% or new lesions appeared. For data analysis, CR and PR were combined as responders, and SD and PD were grouped as non-responders.
DNA collection and genotyping
Each patient provided 5 ml pretreatment blood for the study. The blood samples were collected in citric acid/EDTA anticoagulation tubes and stored at −80°C until analysis. Genomic DNA was isolated from the blood samples using QIAGEN DNA mini Kit (China), and stored at 4°C until use.
Single-nucleotide polymorphisms were analyzed with a 3-D polyacrylamide gel-based DNA microarray genotyping method. This method was invented by researchers of State Key Laboratory of Bioelectronics, Southeast University in 2005 (Patent code: 200510040597.3) [23]. Probes and primers were designed by Primer Premier 5.0 Software. The sequences of primers and probes are shown in Table 2. One of each pair primers was modified with acrylamide phosphoramidite (Acrydite™; Matrix Technologies) at its 5′-terminal. Each couple of probes was labeled with Cy3 and Cy5 fluorescent dyes at 5′-terminal respectively. The PCR reactions were performed in 30 μl reaction solution containing 10 pmol primer and 50 ng genomic DNA. The PCR reaction consisted of an initial step at 95°C for 5 min, then 35–40 cycles of denaturing at 94°C for 30 s, annealing at 48–60°C (according to T m) for 30 s and extension at 72°C for 40 s, and a last extension at 72°C for 5 min.
Table 2.
Sequences of primers and probes
| Locus | Primers and probes |
|---|---|
| MRP2 | Forward primer: 5′-CCTTTACGGAGAACATCAGA-3′ |
| (C-24T, rs717620) | Reverse primer: 5′-Acrydite™-TTTGCATTACATTTCCCAGA-3′ |
| Probe: 5′-Cy3-AGTCTTCGTTCCA-3′ | |
| 5′-Cy5-AGTCTTTGTTCCA-3′ | |
| MRP2 (Val 417 Ile) | Forward primer: 5′-TGGAGGCAAGAAGTCACAGT-3′ |
| (G1249A, rs2273697) | Reverse primer: 5′-Acrydite™-GATTACAAGCACCATCACCC-3′ |
| Probe: 5′-Cy3-TACACCGTTGGAG-3′ | |
| 5′-Cy5-TACACCATTGGAG-3′ | |
| MRP2 (Ile 1,324 Ile) | Forward primer: 5′-Acrydite™-CACTGCTACCCTTCTCCTGTTC-3′ |
| (C3972T, rs3740066) | Reverse primer: 5′-CTGACCCTTTCCCTCCATCC-3′ |
| Probe: 5′-Cy3-GCTACCGATGTCA-3′ | |
| 5′-Cy5-GCTACCAATGTCA-3′ | |
| GSTP1 (Ile 105 Val) | Forward primer: 5′-CAGGGCTCTATGGGAAGGAC-3′ |
| (A342G, rs1695) | Reverse primer: 5′-Acrydite™-CAGGAGATCAGAAACCACCAGTT-3′ |
| Probe: 5′-Cy3-AAATACATCTCCC-3′ | |
| 5′-Cy5-AAATACGTCTCCC-3′ |
After PCR amplification and gel electrophoresis test, PCR products were processed by ethanol precipitation, evaporation, or left untreated. Solutions containing acrylamide-modified PCR products, glycerol, ammonium persulfate (APS), and acrylamide monomers were prepared, spotted, and polymerized onto the acryl-modified slide. In the process, TEMED is introduced onto the spotted microarray to immobilize the modified nucleic acids. Following the attachment to obtain ssDNA for hybridization analysis, dsDNA on the slides was denatured in 0.1 M NaOH for 10 min. After hybridization, the slide was subjected to electrophoresis under 5–30 V/cm for 5–20 min in 1× Tris–Borate–EDTA (TBE) buffer at 4°C. Images of the slides were captured by a scanner (LuxScan™-10 K Confocal Scanner, Packard BioScience Company, USA) and were analyzed with Genepix Pro 3.0 Software. Sequencing of 10% samples was performed to validate the results.
Statistical analysis
Statistical analysis was performed using SPSS Software Package Version 13.0 (SPSS Inc., Chicago, IL, USA). The significance of differences in frequencies and genotypes between good and poor responders was calculated using the χ2 test. χ2 test was also performed to test for Hardy–Weinberg equilibrium (HWE), haplotype frequencies and haplotype-trait association. Continuity correction test or Fisher’s exact test was performed when >20% cells have expected count <5, but not <1, and n > 40, or when theoretical frequency had expected count <1 or n < 40. Multivariate analyses were performed with the use of unconditional logistic regression analysis to assess the association between treatment response and each genetic polymorphism while adjusting for patient gender, age at diagnosis, tumor histology, disease stage, and chemotherapy regimens. The logistic regression model was used to calculate the OR and their 95% CI. In the regression analysis, the outcome variable was patient response to treatment; patients who had poor respond to treatment (SD+PD) were compared with good responders (CR+PR). All P values reported were two-sided, and P values <0.05 were considered statistically significant.
Results
Images of DNA-microarray hybridization for SNPs genotyping
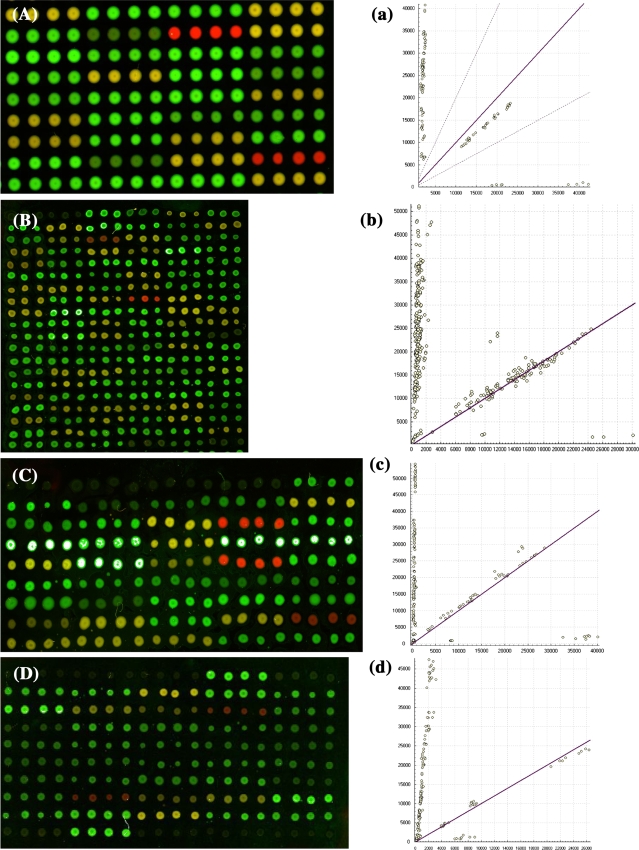
On the basis of the immobilization efficiency, acryl-modified glass slides were selected to fabricate DNA microarrays. By allele-specific oligonucleotide dual-color fluorescence hybridization, homozygous wild type, homozygous mutant type, and heterozygote type yielded green, red, and yellow fluorescence, respectively. Figure 1 showed the microarray images.
Fig. 1.
Microarray hybridization scanning patterns of SNPs genotyping. A, B, C and D the microarray images of locus MRP2 C-24T, MRP2 Val417Ile, MRP2 Ile1324Ile and GSTP1 Ile105Val; green, yellow and red represent wild, hybrid and mutation type, respectively. a, b, c and d the corresponding scatter plots of A, B, C and D showing the genotype assignment. Each scatter spot shows the signal intensities from each sample without correction for the average local background signal from the microarrays. The scatter spots close to the longitudinal (y) axis indicate the wild homozygote and those close to the lateral (x) axis indicate the mutational homozygote. The spots far from the x axis and the y axis indicated the heterozygote
Sequencing result
Sequencing of 10% samples randomly selected was performed. The result was 100% concordance to that of the genotyping suggesting that the 3-D DNA microarray method is reliable.
Treatment response and genotype
Of 113 patients, 30 (26.5%) had some responses (CR+PR) and 83 (73.5%) showed no response (SD+PD).
Table 3 shows the frequencies of genotypes in different response patients, and the association of genotypes with the treatment response. (1) Genotype frequencies for both MRP2 and GSTP1 polymorphisms were found to be in Hardy–Weinberg equilibrium (HWE). Allele frequencies and HWE of each locus were shown in Tables 4 and 5. (2) The polymorphic genotypes of MRP2 (C-24T) and GSTP1 (Ile105Val) were significantly different between patients who responded and did not respond to the platinum-based treatment. After combining the heterozygous and homozygous variant genotypes, the difference remained statistically significant, suggesting that MRP2 (C-24T) and GSTP1 (Ile105Val) genotype differed between the two groups. (3) Genotype influences the treatment response. Patients carrying at least one variant allele (MRP2-C-24T C/T+T/T and GSTP1 A/G+G/G) were more likely to be responders compared with those who did not carry the variant allele; after adjusting for patient gender, age at diagnosis, tumor histology, disease stage, and chemotherapy regimens, the OR for response were 4.493 and 2.881, and the 95% CI were between 1.728 and 11.682 (P = 0.002), and between 1.167 and 7.113 (P = 0.022), respectively. For other SNPs locus, however, the genotypes were not substantially different between the groups. (4) Table 6 displays statistics from case–control tests performed on each individual haplotype of MRP2 (C-24T) and GSTP1 (Ile105Val). A significant P value indicates that there is an association between the haplotype C-A (MRP2-24C and GSTP1105A) and the treatment response. The patients carrying MRP2-24C and GSTP1105A simultaneously were more to be non-responders.
Table 3.
Genotype and response to chemotherapy among NSCLC patients (n = 113)
| Genotype | Cases | Response to chemotherapy | OR (95% CI) | P value | Adjusted OR (95% CI) | Adjusted P value | |
|---|---|---|---|---|---|---|---|
| CR + PR (%) n = 30 | SD + PD (%) n = 83 | ||||||
| MRP2 (C-24T) | |||||||
| C/C | 66 | 11 (36.7) | 55 (66.3) | 1 | 0.015 | 4.069 (1.518–10.910) | 0.005 |
| C/T | 43 | 16 (53.3) | 27 (32.5) | 2.959 (1.211–7.246) | 0.023 | 10.514 (0.842–131.319) | 0.068 |
| T/T | 4 | 3 (10.0) | 1 (1.2) | 14.925 (1.425–166.667) | 0.005 | 4.493 (1.728–11.682) | 0.002 |
| C/T+T/T | 47 | 19 (63.3) | 28 (33.7) | 3.390 (1.420–8.130) | |||
| MRP2 (Val417Ile) | |||||||
| G/G | 84 | 20 (66.7) | 64 (77.1) | 1 | |||
| G/A | 26 | 9 (30.0) | 17 (20.5) | 1.695 (0.654–4.386) | 0.274 | 1.910 (0.697–5.229) | 0.208 |
| A/A | 3 | 1 (3.3) | 2 (2.4) | 1.600 (0.138–18.519) | 0.568 | 1.616 (0.115–22.779) | 0.722 |
| G/A+A/A | 29 | 10 (33.3) | 19 (22.9) | 1.684 (0.674–4.202) | 0.262 | 1.879 (0.710–4.968) | 0.204 |
| MRP2 (Ile1324Ile) | |||||||
| C/C | 74 | 20 (66.7) | 54 (65.1) | 1 | |||
| C/T | 33 | 8 (26.7) | 25 (30.1) | 0.864 (0.335–2.227) | 0.762 | 1.066 (0.385–2.951) | 0.901 |
| T/T | 6 | 2 (6.7) | 4 (4.8) | 1.350 (0.229–7.937) | 0.665 | 1.508 (0.210–10.830) | 0.683 |
| C/T+T/T | 39 | 10 (33.3) | 29 (34.9) | 0.931 (0.385–2.252) | 0.874 | 1.133 (0.441–2.911) | 0.796 |
| GSTP1 (Ile105Val) | |||||||
| A/A | 71 | 13 (43.3) | 58 (69.9) | 1 | |||
| A/G | 38 | 15 (50.0) | 23 (27.7) | 2.907 (1.200–7.042) | 0.016 | 2.788 (1.106–7.029) | 0.030 |
| G/G | 4 | 2 (6.7) | 2 (2.4) | 4.464 (0.574–34.483) | 0.176 | 4.083 (0.457–36.463) | 0.208 |
| A/G+G/G | 42 | 17 (56.7) | 25 (30.1) | 3.030 (1.282–7.194) | 0.010 | 2.881 (1.167–7.113) | 0.022 |
Adjusted OR (95% CI): OR (95% CI) after adjusting for patient gender, age at diagnosis, tumor histology, disease stage, and chemotherapy regimens
Table 4.
Allele frequencies
| Locus | Allele | Frequency | Standard Error | 95% CI |
|---|---|---|---|---|
| MRP2 (C-24T) | C | 0.7743 | 0.0265 | 0.7168–0.8230 |
| T | 0.2257 | 0.0265 | 0.1770–0.2832 | |
| MRP2 (Val417Ile) | A | 0.1416 | 0.0238 | 0.0973–0.1903 |
| G | 0.8584 | 0.0238 | 0.8097–0.9027 | |
| MRP2 (Ile1324Ile) | C | 0.8009 | 0.0277 | 0.7434–0.8540 |
| T | 0.1991 | 0.0277 | 0.1460–0.2566 | |
| GSTP1 (Ile105Val) | A | 0.7965 | 0.0263 | 0.7434–0.8451 |
| G | 0.2035 | 0.0263 | 0.1549–0.2566 |
Table 5.
Genotype frequencies
| Locus | Genotype | Frequency | HWD coeff | Standard error | 95% CI |
|---|---|---|---|---|---|
| MRP2 (C-24T) | C/C | 0.5841 | −0.0155 | 0.0151 | −0.0459–0.0149 |
| C/T | 0.3805 | −0.0155 | 0.0151 | −0.0459–0.0149 | |
| T/T | 0.0354 | −0.0155 | 0.0151 | −0.0459–0.0149 | |
| MRP2 (Val417Ile) | A/A | 0.0265 | 0.0065 | 0.0126 | −0.0165–0.0318 |
| A/G | 0.2301 | 0.0065 | 0.0126 | −0.0165–0.0318 | |
| G/G | 0.7434 | 0.0065 | 0.0126 | −0.0165–0.0318 | |
| MRP2 (Ile1324Ile) | C/C | 0.6549 | 0.0135 | 0.0163 | −0.0185–0.0484 |
| C/T | 0.2920 | 0.0135 | 0.0163 | −0.0185–0.0484 | |
| T/T | 0.0531 | 0.0135 | 0.0163 | −0.0185–0.0484 | |
| GSTP1 (Ile105Val) | A/A | 0.6283 | −0.0060 | 0.0146 | −0.0313–0.0218 |
| A/G | 0.3363 | −0.0060 | 0.0146 | −0.0313–0.0218 | |
| G/G | 0.0354 | −0.0060 | 0.0146 | −0.0313–0.0218 |
Table 6.
Tests for haplotype-trait association between MRP2 (C-24T) and GSTP1 (Ile105Val)
| Haplotype | Frequencies | 95% CI | Chi-square | P value | Prob Exact | ||
|---|---|---|---|---|---|---|---|
| Trait 1 | Trait 2 | Combined | |||||
| C–A | 0.71074 | 0.43370 | 0.64446 | 0.58192–0.70701 | 14.8140 | 0.0001 | <0.0001 |
| C–G | 0.11456 | 0.19963 | 0.12987 | 0.08595–0.17380 | 2.9280 | 0.0871 | 0.1600 |
| T–A | 0.12661 | 0.24963 | 0.15200 | 0.10509–0.19891 | 5.2672 | 0.0217 | 0.0500 |
| T–G | 0.04809 | 0.11703 | 0.07367 | 0.03953–0.10780 | 3.2455 | 0.0716 | 0.1400 |
Trait response to chemotherapy, Trait 1 SD+PD n = 83, Trait 2 CR+PR n = 30
Discussion
One of the major obstacles of cancer chemotherapy is the development of drug resistance, which prevents the application of sufficient high doses to eradicate less-sensitive tumor cell populations. Interindividual differences in response to xenobiotics, which include many clinically used drugs, are extensive and represent a major problem in rational therapeutics. Such differences in many cases may be caused by inherited differences in enzymes and transporters, which function in drug elimination [24]. Owing to its possible effect on gene expression, we anticipated that polymorphisms of drug metabolizing enzymes genes and drug transporters genes may influence tumor response to platinum-based chemotherapy. The identification of molecular variables that predict either sensitivity or resistance to chemotherapy is of major interest in selecting the most likely effective first-line treatment.
Some studies suggested that there is no difference whether cisplatin- or carboplatin-based chemotherapy regimens in the clinical efficacy [25, 26]. Moreover, the results tend to be similar whether the partner drug is paclitaxel, docetaxel, or gemcitabine. Similar results are generally obtained with carboplatin [25–27]. Hence, the data of these 113 patients were analyzed as a combination group.
Glutathione S-transferases are crucial for the cell defence system. These phase II detoxification enzymes are involved in the detoxification of a variety of chemotherapeutics including platinum. In vitro analyses revealed a significant association between high GSTP1 expression of tumor cells and decreased sensitivity to platinum agents [28]. Cisplatin is detoxified by glutathione through adduct formation [29]. GSTP1 interacts with platinum-based compounds [30], and glutathione-conjugated platinum can be quickly effluxed from cells [31]. Thus, it is plausible that high GST activity may result in more rapid drug metabolism that diminishes the cytotoxic effects of chemotherapy on tumor cells [32].
Multidrug resistance-associated protein 2 is responsible for the intracellularly formed glucuronide and GSH conjugates of clinically important drugs. MRP2 is expressed in many tumor tissues, and the tumor cells overexpressing MRP2 might acquire the multidrug resistance [18]. A significant correlation has been observed between MRP2 mRNA levels and cisplatin resistance in colorectal carcinoma [33] suggesting that MRP2 contributes to resistance against treatment with the chemotherapeutic drugs.
Because the important factors influencing interindividual differences in the drug disposition, many analyses of SNPs of drug metabolizing enzymes and drug transporters have been performed.
Board et al. [34] first identified the GSTP1 polymorphisms. Alterations in the structure, function, or expression levels of GSTP1 due to genetic polymorphisms could alter the ability to detoxify chemotherapeutic agents and modulate drug response. The Ile105Val GSTP1 polymorphism, an A/G SNP located within the substrate binding domain of GSTP1 at position +313 within exon 5 results in an amino acid substitution of isoleucine by valine at codon 105 of the enzyme. This substitution has been shown to significantly influence the catalytic activity and thermal stability of the enzyme and to affect the conjugation capacity of GSTP1 for certain substrates, including platinum agents [35, 36]. Rather than being present or absent, the GSTP1 gene has alleles that encode enzymes with different activities. The highest level of GSTP1 activity is seen in individuals with homozygous wild genotype (Ile/Ile). The activity is somewhat reduced in heterozygotes (Ile/Val) and further diminished for those with homozygous mutant genotype (Val/Val) [35]. It is estimated that approximately 50% of people have either one or two valine alleles (Ile/Val or Val/Val) [37]. Thus, this polymorphism is linked to clinical outcome of patients who received platinum-based chemotherapy [38, 39]. Individuals with these variant GSTP1 genotypes that result in reduced GST enzymatic activity may be good responders due to decreased detoxification of chemotherapeutic agents.
Based on this, we hypothesize that lung cancer patients with GSTP1 Ile105Val polymorphisms conferring low activity may have a more favorable prognosis. Our result confirmed this hypothesis. We suggest that this might occur due to the reduced metabolism and slower removal of chemotherapeutic agents, which would yield a prolonged cytotoxic effect; this could lead to a better treatment response and subsequently improved patient survival. Homozygous mutants of GSTP1 (Ile105Val) showed no effect on treatment response, probably the numbers were too small to identify such an effect. Therefore, we are enlarging the samples to confirm the results.
The MRP2 gene is located on chromosomal locus 10q24 and consists of 32 exons (31 coding exons) and spans 69 kb. SNP analysis of MRP2 has been performed, and numerous SNPs have been identified [40, 41]. A recent study provided detailed information on ABCC2 variations and haplotype structures in Japanese [42]. Because of its importance in determining the disposition of anionic drugs, and as a consequence, in determining the pharmacological and/or adverse effects of substrate drugs, correlations between genotype and phenotype need to be established [40]. However, although several studies have suggested the association of SNPs with altered MRP2 expression or function, knowledge about biochemical consequences is still very poor and their association with clinical phenotype remains to be clarified. The functional analysis of these SNPs mutants of MRP2 still remains to be performed. Among several kinds of MRP2 SNPs, C-24T (promoter), G1249A (exon 10) and C3972T (exon 28) are frequently observed [41, 43]. Their allele frequency is 18.8, 12.5, and 21.9%, respectively. In this paper, the allele frequency found in this population is 22.6, 14.2, and 19.9%, respectively, which is similar to the literature. G1249A polymorphism is a G→A base change that results in amino acid alterations from Val to Ile at 417, and 1,249 AA is associated with decreased mRNA [44]; whereas C3972T is the ‘silent’ mutation at 1,324 (Ile1324Ile). several studies have suggested their association with altered MRP2 expression or function [45, 46]. Besides being able to quickly export glutathione-conjugated platinum, upregulated expression of MRP2 is also associated with decreased formation of platinum-DNA adducts and decreased G2-arrest in the cisplatin-resistant cell lines [47]. Amino acid substitution due to SNPs might weaken this effectiveness, and therefore, might enhance the sensitivity to platinating agents. Yang et al. [48] reported that variant in exon 10 (G1249A) was significantly correlated with response to chemotherapy. We also have analyzed these three frequent SNPs in MRP2 gene to investigate their significance in chemosensitivity to platinum-based chemotherapy. Different from Yang’s result, however, our results indicate that the mutant alleles in promoter (C-24T) was significantly related to sensitiveness of NSCLC to therapeutic agents, while the SNPs in exon 10 (G1249A) and exon 28 (C3972T) were not associated with response to chemotherapy. In this study, the presence of the A allele in exon 10 showed a similar trend of better response. However, it is statistically impossible to find the differences of the chemotherapeutic efficacy among variant genotype. We think that there should exist other mechanisms that contribute to the complicacy of the result. For example, most MRP2 substrates are also transported by OATP2 (organic anion transporting polypeptide), so the SNPs and/or inter-individual differences in the expression level of OATP2 maybe also need to be analyzed simultaneously.
Our observation suggests that the polymorphic status of GSTP1 Ile105Val and MRP2 C-24T might predict treatment response of advanced stage NSCLC patients. Moreover, there is an association between the haplotype C-A (MRP2 -24C and GSTP1 105A) and the response to chemotherapy. However, the limitation of our study must be acknowledged. Usually, in the retrospective study, it may be more precise and objective to evaluate the overall survival and progression-free survival as prognostic factors. Considering the difficulty in clinical practice, we chose chemotherapy response as end point of prediction, which was also critical to illuminate the mechanism affected outcome. Ideally, validation studies should be carried out to measure such parameters as predictive and prognostic factors which is also the expected aim of our next research. Therefore, larger sample size and prospective studies, the independent collection of clinical outcomes data and genotyping, and in vivo functional studies are needed to confirm the results and identify the clear biological basis of these findings.
Here, we used a new gel-based DNA microarray genotyping method, reported by Xiao et al. [23], for gene mutation analysis and functional SNP screening. This method based on Rehman’s idea [49]was improved and gave a uniform sample concentration and viscosity for all of the spots on microarrays during spotting, and had a much lower fluorescent background as well as higher S/N (signal/noise). According to the report, by modifying the process for immobilization of nucleic acids, a high quantity microarray with a uniform sample concentration of spots is fabricated. The gel immobilization of nucleic acids has a high immobilization yield and good hybridization efficiency. PCR products modified with acrylamide groups can be used in parallel for gel immobilization to prepare the microarray, in which PCR products are purified only by ethanol precipitation or even untreated. As an alterative to conventional washing, electrophoresis effectively removed the non-specifically bound targets in hybridization and therefore reduce the high background after hybridization, so that the power for discriminating single-nucleotide mismatch is significantly enhanced. Different genotypes of an SNP have successfully been determined by the ratio of signal intensities from two allele-specific oligonucleotides labeled with different dyes. When compared with the common used genotyping methods, such as TagMan assay, PCR–RFLP (restriction fragment length polymorphism), this method is simple, rapid, robust, efficient, and cost-effective for the detection of single-nucleotide mismatch, extremely powerful in genotyping of large-scale samples, and could be used with high reproducibility [23, 50, 51].
Acknowledgments
This work was financially assisted by the Prophase Force-Study program of Jiangsu Province Nature Fund (grant BK2005203), by the Medicine Science Technology Research “Eleventh Five-Year” Program of PLA (No. 06MA111) and by the Focal Project of Nanjing Medicine Technology Development (grant ZKX05030)
Conflict of interest statement
The authors indicated no potential conflicts of interest.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Contributor Information
Xinchen Sun, Phone: +86-25-83275408, FAX: +86-25-83272011, Email: sunxch505@yahoo.com.cn.
Baoan Chen, Phone: +86-25-83272006, FAX: +86-25-83272011, Email: cba8888@hotmail.com.
References
- 1.Guilbert JJ. The world health report 2002: reducing risks, promoting healthy life. Educ Health (Abingdon) 2003;16(2):230. doi: 10.1080/1357628031000116808. [DOI] [PubMed] [Google Scholar]
- 2.Spiro SG, Silvestri GA. The treatment of advanced non-small cell lung cancer. Curr Opin Pulm Med. 2005;11:287–291. doi: 10.1097/01.mcp.0000166590.03042.56. [DOI] [PubMed] [Google Scholar]
- 3.Breathnach O, Freidlin B, Conley B, et al. Twenty-two years of phase III trials for patients with advanced non-small cell lung cancer: sobering results. J Clin Oncol. 2001;19:1734–1742. doi: 10.1200/JCO.2001.19.6.1734. [DOI] [PubMed] [Google Scholar]
- 4.Stephens RJ, Fairlamb D, Gower N, et al. (2002) The Big Lung Trial (BLT): determining the value of cisplatin-based chemotherapy for all patients with non-small cell lung cancer (NSCLC)—Preliminary results in the supportive care setting. Proc Am Soc Clin Oncol 21 (Abstract 1161)
- 5.Rabik CA, Dolan ME. Molecular mechanisms of resistance and toxicity associated with platinating agents. Cancer Treat Rev. 2007;33(1):9–23. doi: 10.1016/j.ctrv.2006.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Viktorsson K, De Petris L, Lewensohn R. The role of p53 in treatment responses of lung cancer. Biochem Biophys Res Commun. 2005;331:868–880. doi: 10.1016/j.bbrc.2005.03.192. [DOI] [PubMed] [Google Scholar]
- 7.Strange RC, Spiteri MA, Ramachandran S, Fryer AA. Glutathione-S transferase family of enzymes. Mutat Res. 2001;482(1–2):21–26. doi: 10.1016/s0027-5107(01)00206-8. [DOI] [PubMed] [Google Scholar]
- 8.Fuertes MA, Castilla J, Alonso C, et al. Cisplatin biochemical mechanism of action: from cytotoxicity to induction of cell death through interconnections between apoptotic and necrotic pathways. Curr Med Chem. 2003;10:257–266. doi: 10.2174/0929867033368484. [DOI] [PubMed] [Google Scholar]
- 9.Mannervik B, Board PG, Hayes JD, Listowsky I, Pearson WR. Nomenclature for mammalian soluble glutathione transferases. Methods Enzymol. 2005;401:1–8. doi: 10.1016/S0076-6879(05)01001-3. [DOI] [PubMed] [Google Scholar]
- 10.Terrier P, Townsend AJ, Coindre JM, Triche TJ, Cowan KH. An immunohistochemical study of pi class glutathione S-transferase expression in normal human tissue. Am J Pathol. 1990;137:845–853. [PMC free article] [PubMed] [Google Scholar]
- 11.Anttila S, Hirvonen A, Vainio H, Husgafvel-Pursiainen K, Hayes JD, Ketterer B. Immunohistochemical localization of glutathione S-transferases in human lung cancer. Cancer Res. 1993;53:5643–5648. [PubMed] [Google Scholar]
- 12.D’Al`o F, Voso MT, Guidi F, et al. Polymorphisms of CYP1A1 and glutathione S-transferase and susceptibility to adult acute myeloid leukemia. Haematologica. 2004;89(6):664–670. [PubMed] [Google Scholar]
- 13.Rebbeck TR. Molecular epidemiology of the human glutathione S transferase genotypes GSTM1 and GSTT1 in cancer susceptibility. Cancer Epidemiol Biomarkers Prev. 1997;6(9):733–743. [PubMed] [Google Scholar]
- 14.Haimeur A, Conseil G, Deeley RG, Cole SP. The MRP-related and BCRP/ABCG2 multidrug resistance proteins: biology, substrate specificity and regulation. Curr Drug Metab. 2004;5:21–53. doi: 10.2174/1389200043489199. [DOI] [PubMed] [Google Scholar]
- 15.Rebbeor JF, Connolly GC, Ballatori N. Inhibition of Mrp2- and Ycf1p-mediated transport by reducing agents: evidence for GSH transport on rat MRP2. Biochim Biophys Acta. 2002;1559:171–178. doi: 10.1016/S0005-2736(01)00454-0. [DOI] [PubMed] [Google Scholar]
- 16.Leslie EM, Haimeur A, Waalkes MP. Arsenic transport by the human multidrug resistance protein 1 (MRP1/ABCC1): evidence that a tri-glutathione conjugate is required. J Biol Chem. 2004;279(31):32700–32708. doi: 10.1074/jbc.M404912200. [DOI] [PubMed] [Google Scholar]
- 17.Takano M, Yumoto R, Murakami T. Expression and function of efflux drug transporters in the intestine. Pharmacol Ther. 2006;109:137–161. doi: 10.1016/j.pharmthera.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 18.Borst P, Evers R, Kool M, Wijnholds J. A family of drug transporters: the multidrug resistance-associated proteins. J Natl Cancer Inst. 2000;92:1295–1302. doi: 10.1093/jnci/92.16.1295. [DOI] [PubMed] [Google Scholar]
- 19.Taniguchi K, Wada M, Kohno K, et al. A human canalicular multispecific organic anion transporter (cMOAT) gene is overexpressed in cisplatin resistant human cancer cell lines with decreased drug accumulation. Cancer Res. 1996;56:4124–4129. [PubMed] [Google Scholar]
- 20.Bakos E, Evers R, Sinkule JA, Varadi A, Borst P, Sarkadi B. Interactions of the human multidrug resistance proteins MRP1 and MRP2 with organic anions. Mol Pharmacol. 2000;57:760–768. doi: 10.1124/mol.57.4.760. [DOI] [PubMed] [Google Scholar]
- 21.Bernig T, Chanock SJ. Challenges of SNP genotyping and genetic variation: its future role in diagnosis and treatment of cancer. Expert Rev Mol Diagn. 2006;6:319–331. doi: 10.1586/14737159.6.3.319. [DOI] [PubMed] [Google Scholar]
- 22.Miller AB, Hoogstraten B, Staquet M, Winkler A. Reporting results of cancer treatment. Cancer. 1981;47:207–214. doi: 10.1002/1097-0142(19810101)47:1<207::AID-CNCR2820470134>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 23.Xiao PF, Cheng L, Wan Y, et al. An improved gel-based DNA microarray method for detecting single nucleotide mismatch. Electrophoresis. 2006;27:3904–3915. doi: 10.1002/elps.200500918. [DOI] [PubMed] [Google Scholar]
- 24.Evans WE, Relling MV. Pharmacogenomics: translating functional genomics into rational therapeutics. Science. 1999;286:487–491. doi: 10.1126/science.286.5439.487. [DOI] [PubMed] [Google Scholar]
- 25.Schiller JH, Harrington D, Belani CP, et al. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. New Engl J Med. 2002;346:92–98. doi: 10.1056/NEJMoa011954. [DOI] [PubMed] [Google Scholar]
- 26.Zatloukal P, Petruzelka L, Zemanova M, et al. Gemcitabine plus cisplatin vs. gemcitabine plus carboplatin in stage IIIb and IV non-small cell lung cancer: a phase III randomized trial. Lung Cancer. 2003;41:321–331. doi: 10.1016/S0169-5002(03)00233-2. [DOI] [PubMed] [Google Scholar]
- 27.Scagliotti GV, De Marinis F, Rinaldi M, et al. Phase III randomized trial comparing three platinum-based doublets in advanced non-small-cell lung cancer. J Clin Oncol. 2002;20(21):4285–4291. doi: 10.1200/JCO.2002.02.068. [DOI] [PubMed] [Google Scholar]
- 28.Oguri T, Fujiwara Y, Katoh O, et al. Glutathione S transferase-pi gene expression and platinum drug exposure in human lung cancer. Cancer Lett. 2000;156:93–99. doi: 10.1016/S0304-3835(00)00447-X. [DOI] [PubMed] [Google Scholar]
- 29.Rudin CM, Yang ZJ, Schumaker LM, et al. Inhibition of glutathione synthesis reverses Bcl-2-mediated cisplatin resistance. Cancer Res. 2003;63(2):312–318. [PubMed] [Google Scholar]
- 30.Goto S, Iida T, Cho S, Oka M, Kohno S, Kondo T. Overexpression of glutathione S-transferase pi enhances the adduct formation of cisplatin with glutathione in human cancer cells. Free Radic Res. 1999;31(6):549–558. doi: 10.1080/10715769900301121. [DOI] [PubMed] [Google Scholar]
- 31.Harpole DH, Jr, Moore MB, Herndon JE, II, Aloia T, D’Amico TA, Sporn T. The prognostic value of molecular marker analysis in patients treated with trimodality therapy for esophageal cancer. Clin Cancer Res. 2001;7(3):562–569. [PubMed] [Google Scholar]
- 32.Beeghly A, Katsaros D, Chen H, et al. Glutathione S-transferase polymorphisms and ovarian cancer treatment and survival. Gynecol Oncol. 2006;100(2):330–337. doi: 10.1016/j.ygyno.2005.08.035. [DOI] [PubMed] [Google Scholar]
- 33.Hinoshita E, Uchiumi T, Taguchi K, et al. Increased expression of an ATP-binding cassette superfamily transporter, multidrug resistance protein 2, in human colorectal carcinomas. Clin Cancer Res. 2000;6:2401–2407. [PubMed] [Google Scholar]
- 34.Board PG, Weber GC, Coggan M. Isolation of a cDNA clone and localization of the human glutathione S-transferase 3 genes to chromosome bands 11q13 and 12q13-14. Ann Hum Genet. 1989;53:205–213. doi: 10.1111/j.1469-1809.1989.tb01786.x. [DOI] [PubMed] [Google Scholar]
- 35.Watson MA, Stewart RK, Smith GB, Massey TE, Bell DA. Human glutathione S-transferase P1 polymorphisms: relationship to lung tissue enzyme activity and population frequency distribution. Carcinogenesis. 1998;19(2):275–280. doi: 10.1093/carcin/19.2.275. [DOI] [PubMed] [Google Scholar]
- 36.Allan JM, ChP Wild, Rollinson S, et al. Polymorphism in glutathione S-transferase P1 is associated with susceptibility to chemotherapy induced leukaemia. Proc Natl Acad Sci USA. 2001;98:11592–11597. doi: 10.1073/pnas.191211198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garte S, Gaspari L, Alexandrie AK, et al. Metabolic gene polymorphism frequencies in control populations. Cancer Epidemiol Biomarkers Prev. 2001;10(12):1239–1248. [PubMed] [Google Scholar]
- 38.Stoehlmacher J, Park DJ, Zhang W, et al. Association between glutathione S-transferase P1, T1, and M1 genetic polymorphism and survival of patients with metastatic colorectal cancer. J Natl Cancer Inst. 2002;94:936–942. doi: 10.1093/jnci/94.12.936. [DOI] [PubMed] [Google Scholar]
- 39.Stoehlmacher J, Park DJ, Zhang W, et al. A multivariate analysis of genomic polymorphisms: prediction of clinical outcome to 5-FU/oxaliplatin combination chemotherapy in refractory colorectal cancer. Br J Cancer. 2004;91:344–354. doi: 10.1038/sj.bjc.6601975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzuki H, Sugiyama Y. Single nucleotide polymorphisms in multidrug resistance associated protein 2 (MRP2/ABCC2): its impact on drug disposition. Adv Drug Deliv Rev. 2002;54(10):1311–1331. doi: 10.1016/S0169-409X(02)00075-3. [DOI] [PubMed] [Google Scholar]
- 41.Ito S, Ieiri I, Tanabe M, Suzuki A, Higuchi S, Otsubo K. Polymorphism of the ABC transporter genes, MDR1, MRP1 and MRP2/cMOAT, in healthy Japanese subjects. Pharmacogenetics. 2001;11:175–184. doi: 10.1097/00008571-200103000-00008. [DOI] [PubMed] [Google Scholar]
- 42.Sai K, Saito Y, Itoda M, et al. Genetic variations and haplotypes of ABCC2 encoding MRP2 in a Japanese population. Drug Metab Pharmacokinet. 2008;23(2):139–147. doi: 10.2133/dmpk.23.139. [DOI] [PubMed] [Google Scholar]
- 43.Itoda M, Saito Y, Soyama A, et al. Polymorphisms in the ABCC2 (cMOAT/MRP2) gene found in 72 established cell lines derived from Japanese individuals: an association between single nucleotide polymorphisms in the 59-untranslated region and exon 28. Drug Metab Dispos. 2002;30:363–364. doi: 10.1124/dmd.30.4.363. [DOI] [PubMed] [Google Scholar]
- 44.Meyer zu Schwabedissen HE, Jedlitschky G, Gratz M, et al. Variable expression of MRP2 (ABCC2) in human placenta: influence of gestational age and cellular differentiation. Drug Metab Dispos. 2005;33:896–904. doi: 10.1124/dmd.104.003335. [DOI] [PubMed] [Google Scholar]
- 45.Haenisch S, Zimmermann U, Dazert E, et al. Influence of polymorphisms of ABCB1 and ABCC2 on mRNA and protein expression in normal and cancerous kidney cortex. Pharmacogenomics J. 2007;7:56–65. doi: 10.1038/sj.tpj.6500403. [DOI] [PubMed] [Google Scholar]
- 46.Han JY, Lim HS, Yoo YK, et al. Associations of ABCB1, ABCC2, and ABCG2 polymorphisms with irinotecan-pharmacokinetics and clinical outcome in patients with advanced non-small cell lung cancer. Cancer. 2007;110:138–147. doi: 10.1002/cncr.22760. [DOI] [PubMed] [Google Scholar]
- 47.Liedert B, Materna V, Schadendorf D, Thomale J, Lage H. Overexpression of cMOAT(MRP2/ABCC2) is associated with decreased formation of platinum-DNA adducts and decreased G2-arrest in melanoma cells resistant to cisplatin. J Invest Dermatol. 2003;121(1):172–176. doi: 10.1046/j.1523-1747.2003.12313.x. [DOI] [PubMed] [Google Scholar]
- 48.Yang XS, Masayuki S, Hitoshi K, et al. Single nucleotide polymorphisms in MRP2 gene and their significance on chemosensitivity of advanced ovarian cancer. J Pract Obstet Gynecol. 2004;20(3):153–155. [Google Scholar]
- 49.Rehman FN, Audeh M, Abrams ES, Hammond PW, Kenney M, Boles TC. Immobilization of acrylamide-modified oligonucleotides by co-polymerization. Nucleic Acids Res. 1999;27:649–655. doi: 10.1093/nar/27.2.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y, Zhang DD, Zheng WL, et al. Multiple gene methylation of nonsmall cell lung cancers evaluated with 3-dimensional microarray. Cancer. 2008;112:61325–61336. doi: 10.1002/cncr.23312. [DOI] [PubMed] [Google Scholar]
- 51.Wan Y, Wang Y, Luo JF, et al. Bisulfite modification of immobilized DNAs for methylation detection. Biosens Bioelectron. 2007;22:2415–2421. doi: 10.1016/j.bios.2006.08.033. [DOI] [PubMed] [Google Scholar]