Abstract
Methicillin-resistant Staphylococcus aureus (MRSA) from Malaysia were shown to possess staphylococcal cassette chromosome mec (SCCmec)-III and IIIA. Spa sequencing and multi-locus sequence typing (MLST) documented t037 and ST 239 (CC8) for 83.3% of the isolates. This confirms observations in several other Far Eastern countries and corroborates the epidemicity of this clone.
The emergence and global spread of methicillin-resistant Staphylococcus aureus (MRSA) over the past five decades constitutes one of the most serious contemporary challenges to the successful treatment of hospital-acquired infections. The enhanced success of MRSA in acquiring resistance to additional groups of antimicrobial agents renders infections caused by these pathogens very difficult to manage and expensive to treat [1]. The prevalence rate of MRSA varies widely among cities, countries and continents because of the clonal dissemination of locally prevalent MRSA clones. The genotypic characteristics of MRSA strains from 12 Asian countries assessed recently by multi-locus sequence typing (MLST) and staphylococcal cassette chromosome mec (SCCmec) typing identified clonal cluster 5 (CC5) MRSA of SCCmec type II (CC5-MRSA-II) as a prototype clone in Korea and Japan while CC239-MRSA-III (or IIIA) appeared to be the major clone in other Asian countries [2]. The above study did not include isolates from Malaysia, another Asian country where the prevalence of MRSA in several hospitals is over 40% [3], but information on the molecular characteristics and clonal distribution of Malaysian MRSA is still limited. Therefore, the aim of the current study was to determine the SCCmec and sequence types of the predominant clones circulating in a large public hospital in Malaysia.
Thirty-six non-repetitive invasive MRSA strains isolated from different clinical sources (blood, pus, urine and tracheal aspirates) collected from a public hospital in Klang Valley (Malaysia) from September 2006 to February 2007 were examined. Isolates were classified as HA and CA-MRSA based on the date of admission, specimen collection date, the number of previous admissions and hospital exposures, and prolonged hospital stays. All isolates were reconfirmed as MRSA by standard laboratory procedures and subjected to SCCmec and Spa typing [4, 5]. Selected isolates from the dominant Spa types were further typed by MLST [6]. Pulsed field gel electrophoresis (PFGE) typing was performed to define the precise relatedness among the strains [7].
Among the 36 isolates (Fig. 1), 15 (41.6%) and 19 (52.7%) possessed SCCmec type III and IIIA respectively, while 2 isolates (5.5%) were SCCmec type V. The SCCmec V isolates were also Panton-Valentine leukocidin (PVL)-positive as was reported previously [8]. Molecular typing by Spa sequencing and MLST identified t037 and ST 239 belonging to CC8 as the most predominant MRSA, covering 83.3% of the isolates studied. Other Spa and sequence types such as t074, t421, t3103, t127 (ST1) and t657 (ST 772) were found incidentally. Spa t074 and t421 are closely related to t037 and arose from the same Spa ancestor (CC-37), while t657 and t3013 are singletons. Strain relatedness within the hospital as defined by PFGE typing grouped all the isolates from the t037, t074 and t421 into one major cluster at approximately 75% similarity, with t657 and t127 in the sub-cluster at 50% similarity, while t3013 was distantly related (less than 40%) and segregated separately (bottom part of Fig. 1). Although the PFGE pattern showed a 40–100% variation among the strains, no clones specific to clinical source, ward or the period of collection were detected. The combined result of PFGE, Spa, MLST and SCCmec typing clearly shows that ST 239 III (or IIIA)-Spa t037 was predominantly circulating in the hospital during the study period (Fig. 1).
Fig. 1.
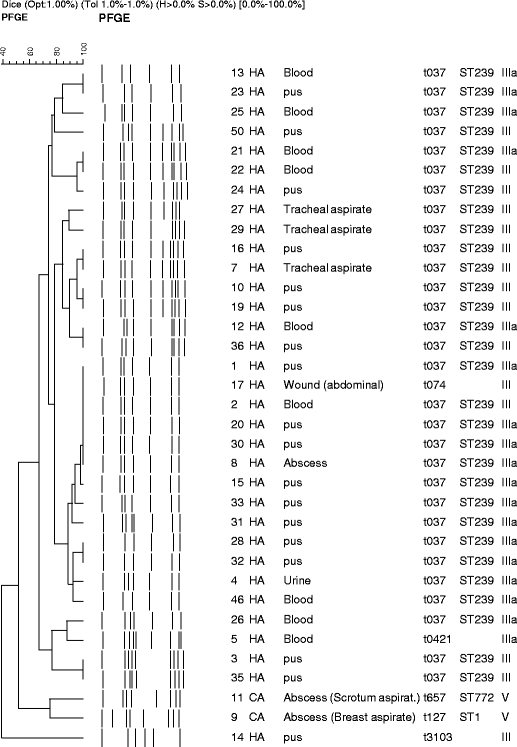
Dendrogram calculated on the basis of pulsed field gel electrophoresis data obtained for Malaysian methicillin-resistant Staphylococcus aureus (MRSA) strains. Next to the dendrogram the banding patterns generated by pulsed field gel electrophoresis (PFGE) are shown. In the subsequent text columns the strains code, community- (CA) or hospital-acquisition (HA) source, clinical material and Spa-MLST-SCCmec types (multi-locus sequence typing-staphylococcal cassette chromosome mec) are identified. Note that one isolate was untypeable by PFGE and is not included in this figure. Strains 11 and 9 are Panton-Valentine leukocidin (PVL)-positive, as was shown before
To date, five major pandemic MRSA clones have been reported. These include the Iberian or ST 247-IA clone, the Brazilian or ST 239-IIIA clone, the Hungarian or ST 239-III clone, the New York/Japan or ST 5-II clone, and the paediatric or ST 5-IV clone. Much of the old nomenclature reflects the geographic area in which these MRSA clones were first identified [1]. In Malaysia we have now also identified ST 239, a clone that was first identified in Brazil in 1995 [9] and which was later reported from Hungary in 1997 [10]. Interestingly, the 239 clone disappeared from Hungarian institutions between 1998 and 2003 where it was largely replaced by ST 228-I and ST 5-II [11]. In addition, the ST 239 clone was also identified in Saudi Arabia and Romania [2]. A recent paper from 11 Asian countries reported that ST 239 accounts for at least 90% of the HA-MRSA in the greater Asian region, which comprises over 60% of the world’s population [2].
We identify ST 239-III (or IIIa) as the predominant clone in a single hospital over a 5-month period. Although we analysed a limited number of isolates only, this still indicates that this clone is a major one in Malaysia, which is consistent with the previous reports on the dominance of ST 239 in Asia. However, investigations with a large number of strains from different geographical regions in Malaysia need to be carried out to confirm the predominance of ST 239-III (or IIIa) in Malaysia.
Acknowledgement
This work was supported by Escience fund 02-01-04-SF0853 from the Ministry of Science, Technology and Innovation (MOSTI), Malaysia.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Aries De Sousa M, de Lencastre H. Bridges from hospitals to the laboratory: genetic portraits of methicillin resistant Staphylococcus aureus clones. FEMS Immunol Med Microbiol. 2004;40(2):101–111. doi: 10.1016/S0928-8244(03)00370-5. [DOI] [PubMed] [Google Scholar]
- 2.Chongtrakool P, Ito T, Ma XX, Kondo Y, Trakulsomboon S, Tiensasitorn C, et al. Staphylococcal cassette chromosome mec (SCCmec) typing of methicillin-resistant Staphylococcus aureus strains isolated in 11 Asian countries: a proposal for a new nomenclature for SCCmec elements. Antimicrob Agents Chemother. 2006;50(3):1001–1012. doi: 10.1128/AAC.50.3.1001-1012.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rohani MY, Raudzah A, Lau MG, Zaidatul AA, Salbiah MN, Keah KC, et al. Susceptibility pattern of Staphylococcus aureus isolated in Malaysian Hospitals. Int J Antimicrob Agents. 2000;13(3):209–213. doi: 10.1016/S0924-8579(99)00129-6. [DOI] [PubMed] [Google Scholar]
- 4.Zhang K, McClure JA, Elsayed S, Louie T, Conly JM. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2005;43(10):5026–5033. doi: 10.1128/JCM.43.10.5026-5033.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harmsen D, Claus H, Witte W, Rothgänger J, Claus H, Turnwald D, et al. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol. 2003;41(12):5442–5448. doi: 10.1128/JCM.41.12.5442-5448.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38(3):1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murchan S, Kaufmann ME, Deplano A, de Ryck R, Struelens M, Zinn CE, et al. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J Clin Microbiol. 2003;41(4):1574–1585. doi: 10.1128/JCM.41.4.1574-1585.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neela V, Ehsanollah GR, Zamberi S, Van Belkum A, Mariana NS. Prevalence of Panton–Valentine leukocidin genes among carriage and invasive Staphylococcus aureus isolates in Malaysia. Int J Infect Dis. 2008;13:e131–e132. doi: 10.1016/j.ijid.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 9.Teixeira LA, Resende CA, Ormonde LR, Rosenbaum R, Am F, de Lencastre H, et al. Geographic spread of epidemic multiresistant Staphylococcus aureus clone in Brazil. J Clin Microbiol. 1995;33(9):2400–2404. doi: 10.1128/jcm.33.9.2400-2404.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Lencastre H, Severina EP, Milch H, Thege MK, Tomasz A. Wide geographic distribution of a unique methicillin-resistant Staphylococcus aureus clone in Hungarian hospitals. Clin Microbiol Infect. 1997;3(3):289–296. doi: 10.1111/j.1469-0691.1997.tb00616.x. [DOI] [PubMed] [Google Scholar]
- 11.Conceicao T, Aries De Sousa M, Fuzi M, Toth A, Paszti J, Ungvari E, et al. Replacement of methicillin resistant Staphylococcus aureus clones in Hungary over time: a ten year surveillance study. Clin Microbiol Infect. 2007;13(10):971–979. doi: 10.1111/j.1469-0691.2007.01794.x. [DOI] [PubMed] [Google Scholar]


