Abstract
Activation of the ubiquitin-proteasome system has been described in different models of cardiac hypertrophy. Cardiac cell growth in response to pressure or volume overload, as well as physiological adaptive hypertrophy, is accompanied by an increase in protein ubiquitination, proteasome subunit expression, and proteasome activity. Importantly, an inhibition of proteasome activity prevents and reverses cardiac hypertrophy and remodelling in vivo. The focus of this review is to provide an update about the mechanisms by which proteasome inhibitors affect cardiac cell growth in adaptive and maladaptive models of cardiac hypertrophy. In the first part, we summarize how the proteasome affects both proteolysis and protein synthesis in a context of cardiac cell growth. In the second part, we show how proteasome inhibition can prevent and reverse cardiac hypertrophy and remodelling in response to different conditions of overload.
Keywords: Cell growth, Hypertrophy, Proteasome
1. Introduction
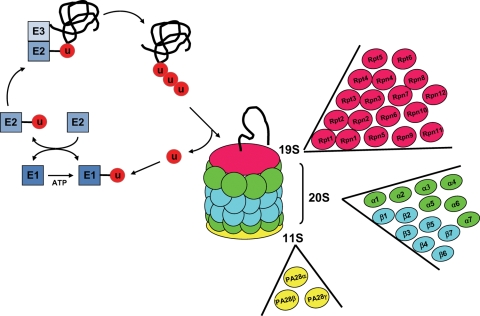
The ubiquitin-proteasome system (UPS) represents the main mechanism of degradation of intracellular cytosolic and nuclear proteins. The UPS has two major biological functions, i.e. ubiquitylation and proteasome-mediated proteolysis (Figure 1).1,2 Ubiquitylation involves specific enzymes. The ubiquitin-activating enzyme (E1) binds ubiquitin by a thioester bond in a reaction that requires ATP hydrolysis. The ubiquitin-conjugating enzyme (E2) transfers the ubiquitin moiety from E1 to the target protein, which is recognized by the ubiquitin–protein ligase (E3). E3 ubiquitin ligase catalyses the formation of the peptidyl bond between ubiquitin and the target protein. Once the first ubiquitin is bound to its target, the elongation of the poly-ubiquitin chain is performed by E2 and E3 (Figure 1).3–5 This poly-ubiquitin chain subsequently transfers the client protein to the proteasome for degradation through specific proteolytic activities.
Figure 1.
Structure of the ubiquitin-proteasome pathway. See text for details. U, ubiquitin. The figure shows the reactions catalysed by E1, E2, and E3 (blue). For the 26S proteasome, the 19S cap (red) is composed of six ATPase subunits (Rpt1–6) and 12 non-ATPase subunits (Rpn1–12), the 20S core is composed of two inner rings made of seven β subunits (blue) and two outer rings containing seven α subunits (green), and the 11S activator (yellow) is composed of three subunits, α, β, and γ.
The 26S proteasome is made of the catalytic 20S core particle and the regulatory 19S cap particle.6,7 The central 20S particle is made of four stacked rings, two rings of α subunits and two rings of β subunits, with the catalytic activity located inside the β rings (Figure 1), where the client protein is degraded by three main proteolytic activities: trypsin-like (located on the β2 subunit and cutting the petidyl bond involving basic amino acids), chymotrypsin-like (β5 subunit, for hydrophobic amino acids), and peptidylglutamyl hydrolase or caspase-like activity (β1 subunit, for acidic amino acids).8 The 19S cap (also named PA700) covering both ends of the 20S proteasome is composed of six distinct ATPases (Rpt1–6) providing an access to the catalytic core for the substrates to be degraded (Figure 1). The 19S particle also contains 12 non-ATPase (Rpn) subunits. In addition to the 19S cap, an ATP-independent activator of the 20S particle is PA28, or 11S particle, which is composed of three family members forming two complexes in vivo: a hetero-oligomer composed of α and β subunits and a homo-oligomer of γ subunits (Figure 1). The different PA28 complexes have different subcellular distributions and the γ oligomer is likely restricted to the nucleus.9,10 The PA28 proteasome is also called the immunoproteasome because it has been mainly characterized as producing the antigenic peptides presented by the major histocompatibility complex.11 However, we will see below that PA28 is also a component of the cardiac proteasome.
The clinical importance of the proteasome is rapidly expanding. For example, a dysfunction of the proteasome has been involved in neurological diseases, such as Huntington's and Alzheimer's diseases.12,13 Another potential clinical application is the regression of cancer growth by proteasome inhibition. The first proteasome inhibitor approved for use in clinical trials is bortezomib (PS-341, Velcade®), a specific and reversible inhibitor that shows promising results in both haematological and solid tumours.14–16 Our hypothesis is that the therapeutic potential of manipulating proteasome activity could be extended to heart disease. Cardiac cell growth represents the main mechanism of adaptation to an increase in workload, but it progressively leads to heart failure. Based on the findings in cancer, investigating the role of the proteasome during cardiac growth caused by volume or pressure overload might give new insights into how to rescue cardiac function. However, few studies addressed the role of the proteasome in the control of cardiac growth. Such a role is likely for the following reasons. First, increased cardiac wall stress from overload automatically creates an increase in protein synthesis. It is estimated that in normal conditions, up to 50% of nascent proteins will be degraded by the proteasome even before peptide elongation is achieved.17 Cellular stress increases the production of denatured proteins, which must be degraded to avoid the activation of pro-apoptotic signals. Secondly, as discussed further below, a qualitative regulation of E3 ubiquitin ligases during overload may lead to the degradation of specific substrates interfering with an increase in cell size. Thirdly, a role for the proteasome in the progression of heart disease is supported by the observation that proteasome activity decreases in different forms of severe cardiac disease, such as cardiomyopathies,18 heart failure,19 and ischaemia/reperfusion.20 Reactivation of the proteasome in these conditions might be part of the therapeutic approach.
In this review, we provide a brief overview of the pharmacological tools of proteasome inhibition, we describe how the proteasome can affect both cardiac proteolysis and protein translation, and we address three specific questions about the role of proteasome in cardiac hypertrophy. Does cardiac hypertrophy affect proteasome activity and proteasome subunit expression? Does a manipulation of proteasome activity affect cardiac hypertrophy? Can proteasome inhibition reverse cardiac remodelling after imposition of pressure or volume overload?
2. Pharmacological inhibition of proteasome activity
Proteasome inhibitors generally exert their effect by direct binding to the β subunits of the 20S particle.21,22 For instance, bortezomib is a dipeptide boronic acid, which reversibly inhibits the chymotryptic-like activity (β5) and the caspase-like activity (β1) of the proteasome (Table 1). Reciprocally, lactacystin and epoxyketones are irreversible proteasome inhibitors widely used in vitro and in vivo.23–25 Lactacystin inhibits proteasome activity by forming an ester bond preferentially with the N-terminus of the β5 subunit, but is not absolutely specific for the proteasome. The most specific inhibitors available are epoxyketones (Table 1), such as epoxomicin. Epoxomicin forms a cyclic morpholino structure with a Thr residue of the β5 subunit, a reaction that cannot take place with other proteases.4
Table 1.
Proteasome inhibitors: classes and targets
| Class | Compound | Inhibition | Proteasome target |
|---|---|---|---|
| Peptide aldehydes | MG-132, MG-115 | Reversible | β1, 2, and 5 subunits |
| Boronic acid peptides | MG-262, PS-341 | Reversible | β1 and 5 subunits |
| Lactacystins | β-Lactone, PS-519 | Irreversible | β5 subunit |
| Epoxyketones | Epoxomicin, eponemycin | Irreversible | β5 subunit |
Bortezomib is the first proteasome inhibitor used against cancer, particularly in malignancies from secretory cells.26 The accelerated rate of translation and secretion in such cells includes a large proportion of misfolded proteins in the endoplasmic reticulum (ER) and Golgi,27 which are shuttled back from the ER to the UPS, a process known as ER-associated degradation.28 In the presence of proteasome inhibitors, this retrograde process cannot take place. The accumulation of unfolded proteins in the ER results in the activation of the ‘unfolded protein response’, a process that activates pro-apoptotic cascades to destroy the tumour cell by apoptosis.28–30 It has been shown that proteasome inhibitors can challenge ER stress in the heart as well,31,32 and, accordingly, several clinical case reports have mentioned a potential cardiotoxicity of bortezomib in patients treated for malignancies.33,34 In a large series of patients treated for myeloma, however, the cardiotoxicity of bortezomib was barely different from that of high-dose dexamethasone.35 Reciprocally, other studies have shown a protective effect of proteasome inhibitors, diametrically opposite to the pro-apoptotic effects described in cancer cells. For example, proteasome inhibitors increase cell survival in animal models of ischaemia/reperfusion in the kidney, heart, and brain.36–38 Similarly, treatment of hypertensive rats with a proteasome inhibitor suppresses the development of cardiac fibrosis.39 In diseases associated with bone loss, treatment of mice with proteasome inhibitors increases osteoblast differentiation and bone formation.40
Therefore, it is possible that variable physiological consequences result from proteasome inhibition depending on the cell type that is targeted, the type of proteasome inhibitor, and the dose used. Another explanation relates to the fact that the proteasome cleaves the client protein with different proteolytic activities, as explained above. Epoxomicin, for example, is highly specific for the chymotryptic-like activity (β5)41 and therefore does not totally inhibit the proteasome. Epoxomicin can block proteasome activity by no more than 30–40%,42 which may explain its limited toxicity for normal cells with low secretory function in organs such as the heart, brain, and kidney. In fact, a 25% inhibition of the chymotryptic activity is sufficient to induce apoptosis in tumour cells, whereas an 80% inhibition of the chymotryptic activity in normal cells, such as blood, liver, and spleen, is well tolerated.4 In addition, proteasome composition, activity, and post-translational modifications vary depending on the tissues examined.43 Comparison of proteasomes from heart and liver shows that the liver carries a significantly higher amount of inducible subunits, such as β1i and β5i.43 Also, the liver proteasome contains higher amount of phosphorylated proteins and is less sensitive to proteasome inhibitors.43
3. The proteasome affects both proteolysis and protein synthesis
3.1. The proteasome and proteolysis
Cardiac myofibrillar proteins are in a constant state of degradation and resynthesis. The balance between these two processes in response to stress mechanisms determines the number of functional contractile units and myocardial mass. It has been shown that myosin heavy chain is degraded by the proteasome,44 suggesting that an impairment of the proteasome pathway might affect cardiac function and mass. Other contractile proteins, such as actin, troponin, and tropomyosin, are also targeted by the proteasome.45 It is possible that a dysfunction in proper ubiquitination and proteasomal degradation of proteins during chronic hypertrophy might be interpreted as a signal of decompensation that would precipitate heart failure.46 Moreover, specific ubiquitin ligases (E3), such as muscle-atrophy F-box (atrogin-1) and muscle-specific ring finger-1 (MuRF1), which were initially only associated with skeletal muscle atrophy,47,48 are now implicated in the pathophysiology of cardiac hypertrophy.49–51 Atrogin-1 interacts with the calcium-activated serine/threonine phosphatase calcineurin and causes its degradation by the proteasome, leading to the inhibition of calcineurin-induced cardiac hypertrophy in response to pathological stimuli.50
The targeted proteolysis of specific substrates also involves transcription factors, such as NF-κB, which participates in the inflammatory reaction triggered by necrosis following ischaemia.52 NF-κB is a heterodimer sequestered in the cytoplasm by the inhibitory-binding protein, IκB. Upon stimulation by stress factors (hypoxia, reactive oxygen species, lipopolysaccharide, TNFα, etc.), IκB is phosphorylated by specific IκB kinases, which targets IκB for ubiquitylation and proteolysis.53 As a result, NF-κB translocates into the nucleus and activates the transcription of genes encoding cytokines, adhesion molecules, interleukins, the inducible isoform of nitric oxide synthase, and matrix metalloproteases (MMPs) among others, which altogether promote the inflammatory reaction characterizing the acute phase of myocardial infarction (MI).54 Blocking NF-κB activity with proteasome inhibitors limits the infiltration of ischaemic myocardium by leucocytes and decreases infarct size by ∼50% in vivo.55
3.2. The proteasome and protein synthesis
The proteasome not only regulates proteolysis, but it may also control protein translation. We describe below three potential mechanisms by which proteasome inhibition could block translation initiation and elongation, but only the first one has been demonstrated in the heart.
The first potential mechanism is the degradation of repressors of hypertrophy, such as the inducible cyclic AMP early repressor (ICER).56 Upon stimulation of β-adrenergic receptors, increased cAMP production leads to an activation of the cAMP response element-binding protein (CREB), a transcription factor activating the expression of genes involved in cardiac cell growth, contractility, and protection against apoptosis.57 By binding the cAMP response element, ICER prevents the transactivation of these genes by CREB, which is followed by an inhibition of hypertrophy and increased apoptosis.56 It was shown in cardiac myocytes that the UPS controls the degradation rate of ICER and thereby its activity,58 by the binding of specific E3 ligases that are themselves transcriptionally regulated.59
The second mechanism relates to ribosome assembly. Ubiquitin, a very abundant protein in the cell, is first translated as a precursor, or hybrid protein, from which the ubiquitin moiety is subsequently freed by cleavage. The proteins to which ubiquitin is initially fused have been identified as two specific subunits of the ribosome, the proteins L28 and L60.60 These ubiquitin tails act as chaperones to facilitate ribosome assembly, after which they are cleaved.60 A saturation of the UPS upon proteasome inhibition may therefore interfere with that process. That theory is supported by the observation that short-term proteasome inhibition interferes with ribosome maturation in the nucleolus.61
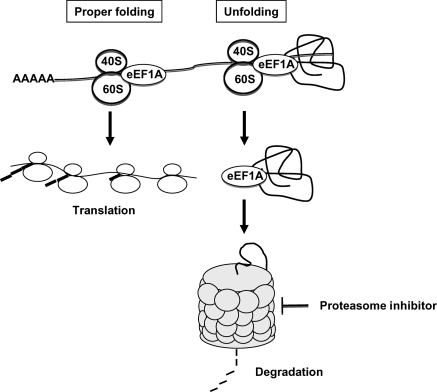
The third potential mechanism is by modifying the activity of the elongation factor eEF1A, which participates in co-translational protein degradation, a mechanism by which nascent but denatured proteins can be rapidly degraded by the proteasome.62 Specifically, eEF1A binds the denatured client protein, but not a correctly folded peptide,63 and subsequently delivers it to the 19S proteasome.64 eEF1A is indispensable for protein elongation because it binds aminoacyl tRNAs to the A-site of the ribosomes. It is therefore possible that upon proteasome inhibition, eEF1A becomes saturated with damaged peptides and can no longer play a role in translation (Figure 2).
Figure 2.
Transport of denatured proteins to the proteasome by eEF1A. The elongation factor eEF1A binds aminoacyl tRNAs to the A-site of the ribosomes, and it participates in co-translational protein degradation by delivering denatured client proteins to the 19S proteasome. If eEF1A becomes saturated with damaged peptides, it can no longer promote translation. 40S, small ribosome subunit; 60S, large ribosome subunit; AAAAA, poly A tail.
4. Proteasome and cardiac cell growth
Cardiac hypertrophy represents a stimulation of cardiac cell growth in response to increased workload, which can be physiological (e.g. exercise) or pathological (e.g. pressure overload due to systemic hypertension or aortic valve stenosis, or volume overload due to MI or valvular regurgitation). The hypertrophied heart is characterized by an increase in wall thickness, which counteracts the increased wall stress caused by the overload. During cardiac hypertrophy, the increase in protein synthesis rate in the heart results in an increase in abundance of sarcomeric proteins.
4.1. Cardiac workload affects proteasome expression and activity
An increase in proteasome subunit protein expression and activity is well documented in several models of cardiac hypertrophy. For instance, chronic (2 years) pressure overload induced by thoracic aortic banding in dogs caused an up-regulation of transcripts encoding poly-ubiquitin, ubiquitin-processing enzymes, as well as proteasome subunits.23 This genomic response was accompanied by an increase in proteasome protein expression, an increase in chymotryptic- and tryptic-like activities of the proteasome, and a decrease in phosphorylation of the α7 protein.23 In an additional study, an increase in transcripts encoding proteasome subunits and ubiquitinating enzymes was found in the left atrium of a dog model with terminal congestive heart failure caused by pacing-induced ventricular hypertrophy.65
Furthermore, data collected in patients offer concrete evidence for a regulation of the proteasome in chronic conditions of left ventricular hypertrophy and heart failure. In patients with aortic valve stenosis, for example, ubiquitination was increased by two-fold compared with control during the stage of compensated hypertrophy and was further increased by 12-fold after the transition into heart failure.66 Increased expression of regulatory proteins of the proteasome was further confirmed by functional proteomics in myocardial samples from patients with idiopathic, dilated cardiomyopathy.67
Activation of the proteasome occurs as well in response to pharmacologically induced cardiomyopathy. Rats made diabetic upon treatment with streptozotocin show a two-fold increase in cardiac proteasome activity and a marked increase in expression of proteasome subunits from the 19S, 20S, and 11S particles.68 Such increase is accompanied by a decrease in ejection fraction and fractional shortening, and an increase in chamber diameter, all indicators of a deterioration of cardiac function.68 Of interest, that model showed an increase in expression of the 11S particle (PA28 or immunoproteasome), demonstrating that this specific particle can be involved in removing damaged or senescent proteins in the heart, such as the carbonyl proteins that accumulate in response to diabetes. This novel observation extends the physiological function of PA28 beyond its participation in antigen presentation.11 A pharmacologically induced activation of cardiac proteasome has also been demonstrated upon treatment with doxorubicin, a potent anthracycline used for the treatment of cancer and which induces irreversible cardiomyopathy leading to heart failure.69 Doxorubicin increases the expression of the carboxy terminus of Hsp70-interacting protein, an ubiquitin ligase playing a central role in protein quality control.69
4.2. Inhibition of the proteasome prevents and reverses cardiac hypertrophy
We will address first the development and regression of maladaptive hypertrophy, followed by an example of adaptive hypertrophy.
4.2.1. Proteasome inhibition prevents maladaptive hypertrophy
The afterload is defined as the pressure against which the ventricles must contract to eject blood. In case of increased afterload, the left ventricle must generate more pressure to open the aortic valve and to eject blood into the systemic circulation, which is defined as pressure overload, such as typically found in patients with systemic hypertension. This condition is reproduced in animal models by aortic banding to generate a pressure gradient that reproduces an increased afterload. Because pressure overload ultimately results in heart failure, exploring pharmacological agents that can regress hypertrophy, preserve contractile function, and/or minimize the remodelling are directly relevant on a clinical perspective. For example, treatment of mice with rapamycin, an inhibitor of the mammalian target of rapamycin (mTOR), can both prevent and reverse hypertrophy in a model of pressure overload, while preserving contractile function.70,71 A similar role was described recently for proteasome inhibitors. In our study, treatment with epoxomicin started before aortic banding in mice completely prevented the development of cardiac hypertrophy, although no deterioration in cardiac function was observed.23 Similar results were found in Dahl salt-sensitive rats treated simultaneously with a high salt diet and with bortezomib.72 The same group showed that suppression of cardiac hypertrophy by proteasome inhibitors is a reversible process, suggesting that sustained cardiac proteasome inhibition is required to prevent the development of cardiac hypertrophy.72 Altogether, these studies show that proteasome activation is required for the development of cardiac hypertrophy by pressure overload. The mechanism by which proteasome inhibition prevents the development of cardiac hypertrophy is multifactorial. We explained above the role of specific E3 ligases in the turnover of sarcomeric proteins, as well as the specific function of the proteasome in the degradation of inhibitors of hypertrophy. In addition, several pro-hypertrophic signalling pathways can be down-regulated in response to proteasome inhibition, such as Akt, ERK1/2, calcineurin, and cyclins.72,73
4.2.2. Proteasome inhibition reverses maladaptive hypertrophy
Although the studies summarized above show that proteasome inhibition can prevent hypertrophy, it would be much more clinically relevant if proteasome inhibitors, when administered after the onset of pressure overload, could improve contractile function by preventing further development of cardiac cell hypertrophy. To address that possibility, animals were submitted to 3 weeks of aortic banding to induce cardiac hypertrophy. Treatment with epoxomicin was started 2 weeks after banding, at which time point significant hypertrophy had developed and cardiac function deteriorated. Blocking proteasome activity in that model reversed pre-existing hypertrophy and stabilized cardiac function.25 Unlike the effects of proteasome inhibitors on cancer cells, no increase in apoptosis was found at the dose used in that study.25 Comparable results were found in a murine model of isoproterenol-induced cardiac hypertrophy in the presence of the proteasome inhibitor PS-519.74 Although proteasome activity was not measured in that study, addition of a proteasome inhibitor also caused a regression of hypertrophy and a decrease in myocyte cross-sectional area.74
Because cardiac hypertrophy is considered an adaptive response, preventing hypertrophy with proteasome inhibitors in the face of overload may seem counterintuitive. However, several recent studies have challenged this concept. In one example, increased wall stress upon banding in mice with genetic deletions preventing the development of hypertrophy was not accompanied by functional deterioration of the heart, at the opposite of the corresponding wild-type mice.75 A second example, mentioned above, is illustrated by the mTOR inhibitor rapamycin, which limits hypertrophy in a context of pressure overload by two mechanisms that are very similar to our observations with proteasome inhibitors. First, the administration of rapamycin before the induction of pressure overload blocks the development of hypertrophy without impairing contractile function,70 which reproduces the prophylactic effects of epoxomicin that we described.23 Secondly, the administration of rapamycin after induction of overload reverses hypertrophy and improves contractility,71 again reproducing similar effects of epoxomicin.25
4.2.3. Proteasome inhibition reverses adaptive hypertrophy
Adaptive hypertrophy is defined as an increase in cardiac cell size resulting from increased cardiac workload without increased wall stress, as typically found in the athlete's heart. We studied adaptive hypertrophy in a model of cardiac-specific over-expression of H11 kinase (H11K), also denoted Hsp22 or HspB8, which is a member of the family of small heat shock proteins.24 H11K is expressed predominantly in the heart and skeletal muscle, and plays a critical role in the maintenance of cardiac cell survival and in promoting cell growth.24 A transgenic (TG) mouse with cardiac-specific over-expression of H11K is characterized by a 30% increase in the heart weight/body weight ratio, by the re-expression of a foetal gene programme, and by concentric hypertrophy with preserved contractile function at echocardiography.76 The mechanism of hypertrophy in that model critically involves the activation of the bone morphogenetic protein receptor and the subsequent stimulation of the Akt signalling pathway.76–78 In that model, over-expression of H11K in the heart resulted in a significant increase in proteasome subunit protein expression and activity, as well as a subcellular redistribution of the proteasome towards the nuclear compartment.24 Interestingly, an age-dependent increase in proteasome activity was found in the H11K TG which parallels the corresponding increase in heart weight.24 A 1-week daily treatment with the proteasome inhibitor epoxomicin decreased the chymotryptic activity of the proteasome and significantly reduced parameters of cardiac hypertrophy, such as heart weight/body weight ratio, heart weight/tibial length ratio, and cardiac cell size, without affecting cardiac function.24 Over-expression of H11K in cardiac myocytes was sufficient to activate the proteasome and increase protein synthesis, which was blocked by epoxomicin.24 Addition of epoxomicin to cardiac myocytes also blocked the stimulation of protein synthesis by pro-hypertrophic stimuli, such as insulin and angiotensin-II.24 This finding further demonstrates that proteasome activity and protein translation are closely associated, which extends the role of the proteasome beyond protein degradation only.
4.3. Proteasome inhibition reverses myocardial remodelling
A major consequence of stress-induced pressure and volume overload is ventricular remodelling, which consists in ventricular dilation and in the accumulation of extracellular matrix, in particular of collagen.79 Collagen accumulation in the failing heart affects cardiac geometry and wall stress, thereby altering myocyte function.80,81 Collagen exists in several distinct types, of which type I and type III predominate in the myocardium. Collagen type I is the most abundant form and it has the highest tensile strength.82 Increase in collagen formation occurs in response to growth signals (especially angiotensin-II and the transforming growth factor β) activating the fibroblasts. Although collagen provides strength, it also increases myocyte stress. Collagen is degraded by MMPs. The gelatinases MMP-2 and MMP-9 are expressed both in cardiac fibroblasts and in myocytes, and degrade several types of collagen (including the types I and III).83 Disruption of the collagen matrix by MMPs increases the stress for the cardiomyocytes, leading to further hypertrophy.80 Also, this dissociation of collagen bundles by MMPs automatically leads to additional collagen synthesis through transcriptional activation of the corresponding genes.80 There is therefore a direct relationship between extracellular matrix turnover and cardiac cell size. It has been shown in rat cardiac fibroblasts that proteasome inhibitors decrease the transcriptional activity of genes encoding MMP-2 and MMP-9, as well as collagen types I and III.39
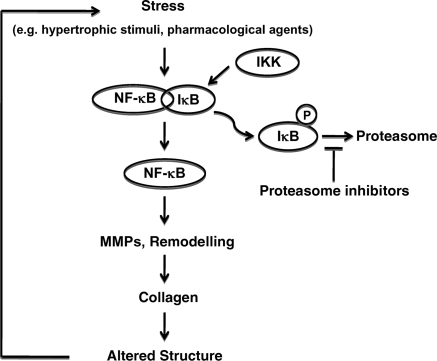
The mechanism of regression of collagen accumulation upon proteasome inhibition relies on the fact that proteasome inhibitors prevent the activation of the transcription factor NF-κB, which is activated upon cardiac stress, such as overload or ischaemia, by the proteasomal degradation of its specific inhibitor, IκB.84 Blocking the proteasome prevents the degradation of IκB and the activation of NF-κB, which results in decreased accumulation of extracellular matrix in the myocardium (Figure 3). Inhibition of NF-κB is also achieved in cardiac tissue by over-expression of PR39,85 a peptide binding reversibly to the α7 subunit of the proteasome and which selectively prevents the degradation of IκB without affecting the overall proteasome activity.86 Mice over-expressing PR39 in the heart are characterized by a decrease in infarct size after coronary occlusion,85 increased angiogenesis,87 and up-regulation of markers of hypertrophy.88 Unlike the chemical inhibitors targeting the β subunits of the proteasome, the biological inhibition by PR39 demonstrates that the α subunits also modulate the activity and substrate specificity of the proteasome.
Figure 3.
Interference of proteasome inhibitors with cardiac remodelling. During cardiac stress, NF-κB is activated upon removal of IκB, which is phosphorylated by IKK, and degraded by the proteasome. NF-κB activation stimulates the expression of MMPs, leading to cardiac remodelling, increased expression of collagen, alteration in cardiac structure, and increased stress. Inhibition of the proteasome prevents IκB degradation, thereby restraining NF-κB activity.
Although we insisted above on the importance of proteasome inhibition in preventing tissue remodelling that leads to heart failure, the failing heart is paradoxically characterized by a dramatic down-regulation of proteasome activity.19 It is not clear whether such down-regulation is causally related to the progression of the disease or whether it represents an attempt for a compensatory mechanism. Also, it remains unclear whether the pharmacological treatment of heart failure affects proteasome activity. The decrease in cardiac output that accompanies any form of heart failure triggers a neurohormonal response from both the renin–angiotensin and the sympathetic systems. Therefore, the main therapeutic avenues to blunt the functional degradation of the failing heart include inhibitors of angiotensin-converting enzyme (ACE), blocking the renin–angiotensin axis, and β-blockers, attenuating the sympathetic stimulation. The protective mechanisms following ACE inhibition involve a decrease in afterload, a decrease in scar tissue, an inhibition of apoptosis, and a decrease in ventricular hypertrophy.89 Low-dose β-blockers improve the contractile performance of the failing heart.90 Proteasome activity might participate in the therapeutic effects of ACE-inhibitors and β-blockers, although this has not been explored in the heart. It is known that in skeletal muscle, for example, angiotensin-II stimulates protein degradation through a stimulation of the UPS leading to muscle wasting, which is inhibited by proteasome inhibitors.91 This is reversed by insulin-like growth factor-I, which down-regulates UPS activity92 and restores muscle mass.93 However, this stimulation of the UPS by angiotensin-II is opposite to the down-regulation found in the failing heart. As for the sympathetic system, the cyclic AMP-dependent protein kinase (PKA), which lies downstream the β-adrenergic receptors, activates proteasome assembly and activity.94 The desensitization of the failing heart to catecholaminergic stimulation might therefore participate in the down-regulation of proteasome activity in that condition. The therapeutic goal of β-blockers in heart failure is to resensitize the sympathetic response, which could be accompanied by a reactivation of the UPS through PKA. It is therefore possible that proteasome inhibition could be beneficial in a context of hypertrophy and remodelling, whereas proteasome reactivation might be necessary in the failing heart.
Although preventing collagen build-up is beneficial for cardiac remodelling in the chronic setting, it could increase the risk of cardiac rupture at the acute phase following MI. Cardiac rupture results from an imbalance between mechanical stretch and tensile resistance of the necrotic tissue, due to an imbalance between the enzymatic activity of MMPs and the rate of collagen deposition.95 Necrotic myocardium subject to rupture shows increased expression of different forms of MMPs,96 whereas decreasing MMP activity reciprocally limits the risk of rupture.97 Blocking collagen build-up and deposition upon proteasome inhibition might increase the risk of myocardial rupture, although this remains to be demonstrated. Such possibility is supported by a study showing that over-expression of the β2-adrenergic receptor decreases the risk of myocardial rupture after MI by increasing collagen deposition.98
5. Conclusions
Proteasome activity and proteasome protein subunit expression increase in different conditions of cardiac hypertrophy, both in animal models and in patients. Reciprocally, pharmacological inhibition of proteasome activity prevents and reverses hypertrophy. Proteasome inhibitors also decrease ventricular remodelling in a context of cardiac overload, at least in part by preventing the activation of the transcription factor NF-κB, thereby limiting collagen deposition. A partial inhibition of proteasome activity is sufficient to observe these beneficial effects, at concentrations that do not induce cell toxicity. Several lines of evidence point to the possibility that proteasome inhibition interferes not only with protein degradation, but also with protein synthesis. These observations further support the concept of manipulating proteasome activity for the treatment of hypertrophic heart disease.
Conflict of interest: none declared.
Funding
NIH grant HL 072863 and AHA grant 0230017N (C.D.).
References
- 1.Etlinger JD, Goldberg AL. A soluble ATP-dependent proteolytic system responsible for the degradation of abnormal proteins in reticulocytes. Proc Natl Acad Sci USA. 1977;74:54–58. doi: 10.1073/pnas.74.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ciechanover A, Hod Y, Hershko A. A heat-stable polypeptide component of an ATP-dependent proteolytic system from reticulocytes. Biochem Biophys Res Commun. 1978;81:1100–1105. doi: 10.1016/0006-291x(78)91249-4. [DOI] [PubMed] [Google Scholar]
- 3.Hershko A, Heller H, Elias S, Ciechanover A. Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J Biol Chem. 1983;258:8206–8214. [PubMed] [Google Scholar]
- 4.Meiners S, Ludwig A, Stangl V, Stangl K. Proteasome inhibitors: poisons and remedies. Med Res Rev. 2008;28:309–327. doi: 10.1002/med.20111. [DOI] [PubMed] [Google Scholar]
- 5.Powell SR. The ubiquitin-proteasome system in cardiac physiology and pathology. Am J Physiol Heart Circ Physiol. 2006;291:H1–H19. doi: 10.1152/ajpheart.00062.2006. [DOI] [PubMed] [Google Scholar]
- 6.Hough R, Pratt G, Rechsteiner M. Purification of two high molecular weight proteases from rabbit reticulocyte lysate. J Biol Chem. 1987;262:8303–8313. [PubMed] [Google Scholar]
- 7.Drews O, Zong C, Ping P. Exploring proteasome complexes by proteomic approaches. Proteomics. 2007;7:1047–1058. doi: 10.1002/pmic.200600574. [DOI] [PubMed] [Google Scholar]
- 8.Tanaka K, Yoshimura T, Kumatori A, Ichihara A, Ikai A, Nishigai M, et al. Proteasomes (multi-protease complexes) as 20S ring-shaped particles in a variety of eukaryotic cells. J Biol Chem. 1988;263:16209–16217. [PubMed] [Google Scholar]
- 9.Rechsteiner M, Hill C. Mobilizing the proteolytic machine: cell biological roles of proteasome activators and inhibitors. Trends Cell Biol. 2005;15:27–33. doi: 10.1016/j.tcb.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 10.Ahn K, Erlander M, Leturcq D, Peterson PA, Fruh K, Yang Y. In vivo characterization of the proteasome regulator PA28. J Biol Chem. 1996;271:18237–18242. doi: 10.1074/jbc.271.30.18237. [DOI] [PubMed] [Google Scholar]
- 11.Rechsteiner M, Realini C, Ustrell V. The proteasome activator 11S REG (PA28) and class I antigen presentation. Biochem J. 2000;345(Pt. 1):1–15. [PMC free article] [PubMed] [Google Scholar]
- 12.Layfield R, Lowe J, Bedford L. The ubiquitin-proteasome system and neurodegenerative disorders. Essays Biochem. 2005;41:157–171. doi: 10.1042/EB0410157. [DOI] [PubMed] [Google Scholar]
- 13.Bennett EJ, Shaler TA, Woodman B, Ryu KY, Zaitseva TS, Becker CH, et al. Global changes to the ubiquitin system in Huntington's disease. Nature. 2007;448:704–708. doi: 10.1038/nature06022. [DOI] [PubMed] [Google Scholar]
- 14.Orlowski RZ, Stinchcombe TE, Mitchell BS, Shea TC, Baldwin AS, Stahl S, et al. Phase I trial of the proteasome inhibitor PS-341 in patients with refractory hematologic malignancies. J Clin Oncol. 2002;20:4420–4427. doi: 10.1200/JCO.2002.01.133. [DOI] [PubMed] [Google Scholar]
- 15.Zavrski I, Jakob C, Kaiser M, Fleissner C, Heider U, Sezer O. Molecular and clinical aspects of proteasome inhibition in the treatment of cancer. Recent Results Cancer Res. 2007;176:165–176. doi: 10.1007/978-3-540-46091-6_14. [DOI] [PubMed] [Google Scholar]
- 16.Aghajanian C, Soignet S, Dizon DS, Pien CS, Adams J, Elliott PJ, et al. A phase I trial of the novel proteasome inhibitor PS341 in advanced solid tumor malignancies. Clin Cancer Res. 2002;8:2505–2511. [PubMed] [Google Scholar]
- 17.Chuang SM, Madura K. Saccharomyces cerevisiae Ub-conjugating enzyme Ubc4 binds the proteasome in the presence of translationally damaged proteins. Genetics. 2005;171:1477–1484. doi: 10.1534/genetics.105.046888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, Robbins J. Heart failure and protein quality control. Circ Res. 2006;99:1315–1328. doi: 10.1161/01.RES.0000252342.61447.a2. [DOI] [PubMed] [Google Scholar]
- 19.Tsukamoto O, Minamino T, Okada K, Shintani Y, Takashima S, Kato H, et al. Depression of proteasome activities during the progression of cardiac dysfunction in pressure-overloaded heart of mice. Biochem Biophys Res Com. 2006;340:1125–1133. doi: 10.1016/j.bbrc.2005.12.120. [DOI] [PubMed] [Google Scholar]
- 20.Bulteau A-L, Lundberg KC, Humphries KM, Sadek HA, Szweda PA, Friguet B, et al. Oxidative modification and inactivation of the proteasome during coronary occlusion/reperfusion. J Biol Chem. 2001;276:30057–30063. doi: 10.1074/jbc.M100142200. [DOI] [PubMed] [Google Scholar]
- 21.Sin N, Kim KB, Elofsson M, Meng L, Auth H, Kwok BH, et al. Total synthesis of the potent proteasome inhibitor epoxomicin: a useful tool for understanding proteasome biology. Bioorg Med Chem Lett. 1999;9:2283–2288. doi: 10.1016/s0960-894x(99)00376-5. [DOI] [PubMed] [Google Scholar]
- 22.Craiu A, Gaczynska M, Akopian T, Gramm CF, Fenteany G, Goldberg AL, et al. Lactacystin and clasto-lactacystin β-lactone modify multiple proteasome β-subunits and inhibit intracellular protein degradation and major histocompatibility complex class I antigen presentation. J Biol Chem. 1997;272:13437–13445. doi: 10.1074/jbc.272.20.13437. [DOI] [PubMed] [Google Scholar]
- 23.Depre C, Wang Q, Yan L, Hedhli N, Peter P, Chen L, et al. Activation of the cardiac proteasome during pressure overload promotes ventricular hypertrophy. Circulation. 2006;114:1821–1828. doi: 10.1161/CIRCULATIONAHA.106.637827. [DOI] [PubMed] [Google Scholar]
- 24.Hedhli N, Wang L, Wang Q, Rashed E, Tian Y, Sui X, et al. Proteasome activation during cardiac hypertrophy by the chaperone H11 Kinase/Hsp22. Cardiovasc Res. 2008;77:497–505. doi: 10.1093/cvr/cvm054. [DOI] [PubMed] [Google Scholar]
- 25.Hedhli N, Lizano P, Hong C, Fritzky LF, Dhar SK, Liu H, et al. Proteasome inhibition decreases cardiac remodeling after initiation of pressure overload. Am J Physiol Heart Circ Physiol. 2008;295:H1385–H1393. doi: 10.1152/ajpheart.00532.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee AH, Iwakoshi NN, Anderson KC, Glimcher LH. Proteasome inhibitors disrupt the unfolded protein response in myeloma cells. Proc Natl Acad Sci USA. 2003;100:9946–9951. doi: 10.1073/pnas.1334037100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oyadomari S, Yun C, Fisher EA, Kreglinger N, Kreibich G, Oyadomari M, et al. Cotranslocational degradation protects the stressed endoplasmic reticulum from protein overload. Cell. 2006;126:727–739. doi: 10.1016/j.cell.2006.06.051. [DOI] [PubMed] [Google Scholar]
- 28.Marciniak S, Ron D. Endoplasmic reticulum stress signaling in disease. Physiol Rev. 2006;86:1133–1149. doi: 10.1152/physrev.00015.2006. [DOI] [PubMed] [Google Scholar]
- 29.Nawrocki S, Carew J, Pino M, Highshaw R, Dunner K, Huang P, et al. Bortezomib sensitizes pancreatic cancer cells to endoplasmic reticulum stress-mediated apoptosis. Cancer Res. 2005;65:11658–11666. doi: 10.1158/0008-5472.CAN-05-2370. [DOI] [PubMed] [Google Scholar]
- 30.Obeng E, Carlson L, Gutman D, Harrington W, Lee K, Boise L. Proteasome inhibitors induce a terminal unfolded protein response in multiple myeloma cells. Blood. 2006;107:4907–4916. doi: 10.1182/blood-2005-08-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Appelman YE, Doevendans PA. Proteasome inhibition and stress compromise the heart in chemotherapy. Cardiovasc Res. 2008;79:547–548. doi: 10.1093/cvr/cvn191. [DOI] [PubMed] [Google Scholar]
- 32.Fu H, Minamino T, Tsukamoto O, Sawada T, Asai M, Kato H, et al. Overexpression of endoplasmic reticulum-resident chaperone attenuates cardiomyocyte death induced by proteasome inhibition. Cardiovasc Res. 2008;79:600–610. doi: 10.1093/cvr/cvn128. [DOI] [PubMed] [Google Scholar]
- 33.Voortman J, Giaccone G. Severe reversible cardiac failure after bortezomib treatment combined with chemotherapy in a non-small cell lung cancer patient: a case report. BMC Cancer. 2006;6:129. doi: 10.1186/1471-2407-6-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hacihanefioglu A, Tarkun P, Gonullu E. Acute severe cardiac failure in a myeloma patient due to proteasome inhibitor bortezomib. Int J Hematol. 2008;88:219–222. doi: 10.1007/s12185-008-0139-7. [DOI] [PubMed] [Google Scholar]
- 35.Yeh ET, Bickford CL. Cardiovascular complications of cancer therapy: incidence, pathogenesis, diagnosis, and management. J Am Coll Cardiol. 2009;53:2231–2247. doi: 10.1016/j.jacc.2009.02.050. [DOI] [PubMed] [Google Scholar]
- 36.Itoh M, Takaoka M, Shibata A, Ohkita M, Matsumura Y. Preventive effect of lactacystin, a selective proteasome inhibitor, on ischemic acute renal failure in rats. J Pharmacol Exp Ther. 2001;298:501–507. [PubMed] [Google Scholar]
- 37.Pye J, Ardeshirpour F, McCain A, Bellinger DA, Merricks E, Adams J, et al. Proteasome inhibition ablates activation of NF-κB in myocardial reperfusion and reduces reperfusion injury. Am J Physiol Heart Circ Physiol. 2003;284:H919–H926. doi: 10.1152/ajpheart.00851.2002. [DOI] [PubMed] [Google Scholar]
- 38.Zhang L, Zhang ZG, Liu X, Hozeska A, Stagliano N, Riordan W, et al. Treatment of embolic stroke in rats with bortezomib and recombinant human tissue plasminogen activator. Thromb Haemost. 2006;95:166–173. [PubMed] [Google Scholar]
- 39.Meiners S, Hocher B, Weller A, Laule M, Stangl V, Guenther C, et al. Downregulation of matrix metalloproteinases and collagens and suppression of cardiac fibrosis by inhibition of the proteasome. Hypertension. 2004;44:471–477. doi: 10.1161/01.HYP.0000142772.71367.65. [DOI] [PubMed] [Google Scholar]
- 40.Roodman GD. Bone building with bortezomib. J Clin Invest. 2008;118:462–464. doi: 10.1172/JCI34734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meng L, Mohan R, Kwok BH, Elofsson M, Sin N, Crews CM. Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo antiinflammatory activity. Proc Natl Acad Sci USA. 1999;96:10403–10408. doi: 10.1073/pnas.96.18.10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kisselev AF, Callard A, Goldberg AL. Importance of the different proteolytic sites of the proteasome and the efficacy of inhibitors varies with the protein substrate. J Biol Chem. 2006;281:8582–8590. doi: 10.1074/jbc.M509043200. [DOI] [PubMed] [Google Scholar]
- 43.Gomes AV, Young GW, Wang Y, Zong C, Eghbali M, Drews O, et al. Contrasting proteome biology and functional heterogeneity of the 20S proteasome complexes in mammalian tissues. Mol Cell Proteomics. 2009;8:302–315. doi: 10.1074/mcp.M800058-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eble DM, Spragia ML, Ferguson AG, Samarel AM. Sarcomeric myosin heavy chain is degraded by the proteasome. Cell Tissue Res. 1999;296:541–548. doi: 10.1007/s004410051315. [DOI] [PubMed] [Google Scholar]
- 45.Taylor RG, Tassy C, Briand M, Robert N, Briand Y, Ouali A. Proteolytic activity of proteasome on myofibrillar structures. Mol Biol Rep. 1995;21:71–73. doi: 10.1007/BF00990974. [DOI] [PubMed] [Google Scholar]
- 46.Field ML, Clark JF. Inappropriate ubiquitin conjugation: a proposed mechanism contributing to heart failure. Cardiovasc Res. 1997;33:8–12. doi: 10.1016/s0008-6363(96)00141-1. [DOI] [PubMed] [Google Scholar]
- 47.Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 48.Gomes MD, Lecker SH, Jagoe RT, Navon A, Goldberg AL. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc Natl Acad Sci USA. 2001;98:14440–14445. doi: 10.1073/pnas.251541198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gaussin V, Tomlinson JE, Depre C, Engelhardt S, Antos CL, Takagi G, et al. Common genomic response in different mouse models of β adrenergic-induced cardiomyopathy. Circulation. 2003;108:2926–2933. doi: 10.1161/01.CIR.0000101922.18151.7B. [DOI] [PubMed] [Google Scholar]
- 50.Li HH, Kedar V, Zhang C, McDonough H, Arya R, Wang DZ, et al. Atrogin-1/muscle atrophy F-box inhibits calcineurin-dependent cardiac hypertrophy by participating in an SCF ubiquitin ligase complex. J Clin Invest. 2004;114:1058–1071. doi: 10.1172/JCI22220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kedar V, McDonough H, Arya R, Li H-H, Rockman HA, Patterson C. Muscle-specific RING finger 1 is a bona fide ubiquitin ligase that degrades cardiac troponin I. Proc Natl Acad Sci USA. 2004;101:18135–18140. doi: 10.1073/pnas.0404341102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li C, Browder W, Kao RL. Early activation of transcription factor NF-κB during ischemia in perfused rat heart. Am J Physiol. 1999;276:H543–H552. doi: 10.1152/ajpheart.1999.276.2.H543. [DOI] [PubMed] [Google Scholar]
- 53.Scheidereit C. IκB kinase complexes: gateways to NF-κB activation and transcription. Oncogene. 2006;25:6685–6705. doi: 10.1038/sj.onc.1209934. [DOI] [PubMed] [Google Scholar]
- 54.Frangogiannis NG. The immune system and cardiac repair. Pharmacol Res. 2008;58:88–111. doi: 10.1016/j.phrs.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stansfield WE, Moss NC, Willis MS, Tang R, Selzman CH. Proteasome inhibition attenuates infarct size and preserves cardiac function in a murine model of myocardial ischemia-reperfusion injury. Ann Thorac Surg. 2007;84:120–125. doi: 10.1016/j.athoracsur.2007.02.049. [DOI] [PubMed] [Google Scholar]
- 56.Tomita H, Nazmy M, Kajimoto K, Yehia G, Molina CA, Sadoshima J. Inducible cAMP early repressor (ICER) is a negative-feedback regulator of cardiac hypertrophy and an important mediator of cardiac myocyte apoptosis in response to β-adrenergic receptor stimulation. Circ Res. 2003;93:12–22. doi: 10.1161/01.RES.0000079794.57578.F1. [DOI] [PubMed] [Google Scholar]
- 57.Muller FU, Lewin G, Baba HA, Boknik P, Fabritz L, Kirchhefer U, et al. Heart-directed expression of a human cardiac isoform of cAMP-response element modulator in transgenic mice. J Biol Chem. 2005;280:6906–6914. doi: 10.1074/jbc.M407864200. [DOI] [PubMed] [Google Scholar]
- 58.Folco E, Koren G. Degradation of the inducible cAMP early repressor (ICER) by the ubiquitin-proteasome pathway. Biochem J. 1997;328:37–43. doi: 10.1042/bj3280037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pati D, Meistrich ML, Plon SE. Human Cdc34 and Rad6B ubiquitin-conjugating enzymes target repressors of cyclic AMP-induced transcription for proteolysis. Mol Cell Biol. 1999;19:5001–5013. doi: 10.1128/mcb.19.7.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Spence J, Gali RR, Dittmar G, Sherman F, Karin M, Finley D. Cell cycle-regulated modification of the ribosome by a variant multiubiquitin chain. Cell. 2000;102:67–76. doi: 10.1016/s0092-8674(00)00011-8. [DOI] [PubMed] [Google Scholar]
- 61.Stavreva DA, Kawasaki M, Dundr M, Koberna K, Muller WG, Tsujimura-Takahashi T, et al. Potential roles for ubiquitin and the proteasome during ribosome biogenesis. Mol Cell Biol. 2006;26:5131–5145. doi: 10.1128/MCB.02227-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schubert U, Anton L, Gibbs J, Norbury C, Yewdell J, Bennink J. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–774. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- 63.Hotokezaka Y, Tobben U, Hotokezaka H, Van Leyen K, Beatrix B, Smith DH, et al. Interaction of the eukaryotic elongation factor 1A with newly synthesized polypeptides. J Biol Chem. 2002;277:18545–18551. doi: 10.1074/jbc.M201022200. [DOI] [PubMed] [Google Scholar]
- 64.Chuang SM, Chen L, Lambertson D, Anand M, Kinzy TG, Madura K. Proteasome-mediated degradation of cotranslationally damaged proteins involves translation elongation factor 1A. Mol Cell Biol. 2005;25:403–413. doi: 10.1128/MCB.25.1.403-413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cardin S, Pelletier P, Libby E, Le Bouter S, Xiao L, Kaab S, et al. Marked differences between atrial and ventricular gene-expression remodeling in dogs with experimental heart failure. J Mol Cell Cardiol. 2008;45:821–831. doi: 10.1016/j.yjmcc.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 66.Hein S, Arnon E, Kostin S, Schonburg M, Elsasser A, Polyakova V, et al. Progression from compensated hypertrophy to failure in the pressure-overloaded human heart: structural deterioration and compensatory mechanisms. Circulation. 2003;107:984–991. doi: 10.1161/01.cir.0000051865.66123.b7. [DOI] [PubMed] [Google Scholar]
- 67.Weekes J, Morrison K, Mullen A, Wait R, Barton P, Dunn M. Hyperubiquitination of proteins in dilated cardiomyopathy. Proteomics. 2003;3:208–216. doi: 10.1002/pmic.200390029. [DOI] [PubMed] [Google Scholar]
- 68.Powell SR, Samuel SM, Wang P, Divald A, Thirunavukkarasu M, Koneru S, et al. Upregulation of myocardial 11S-activated proteasome in experimental hyperglycemia. J Mol Cell Cardiol. 2008;44:618–621. doi: 10.1016/j.yjmcc.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 69.Liu J, Zheng H, Tang M, Ryu YC, Wang X. A therapeutic dose of doxorubicin activates ubiquitin-proteasome system-mediated proteolysis by acting on both the ubiquitination apparatus and proteasome. Am J Physiol Heart Circ Physiol. 2008;295:H2541–H2550. doi: 10.1152/ajpheart.01052.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shioi T, McMullen JR, Tarnavski O, Converso K, Sherwood MC, Manning WJ, et al. Rapamycin attenuates load-induced cardiac hypertrophy in mice. Circulation. 2003;107:1664–1670. doi: 10.1161/01.CIR.0000057979.36322.88. [DOI] [PubMed] [Google Scholar]
- 71.McMullen JR, Sherwood MC, Tarnavski O, Zhang L, Dorfman AL, Shioi T, et al. Inhibition of mTOR signaling with rapamycin regresses established cardiac hypertrophy induced by pressure overload. Circulation. 2004;109:3050–3055. doi: 10.1161/01.CIR.0000130641.08705.45. [DOI] [PubMed] [Google Scholar]
- 72.Meiners S, Dreger H, Fechner M, Bieler S, Rother W, Gunther C, et al. Suppression of cardiomyocyte hypertrophy by inhibition of the ubiquitin-proteasome system. Hypertension. 2008;51:302–308. doi: 10.1161/HYPERTENSIONAHA.107.097816. [DOI] [PubMed] [Google Scholar]
- 73.Fang CX, Dong F, Thomas DP, Ma H, He L, Ren J. Hypertrophic cardiomyopathy in high-fat diet-induced obesity: role of suppression of forkhead transcription factor and atrophy gene transcription. Am J Physiol Heart Circ Physiol. 2008;295:H1206–H1215. doi: 10.1152/ajpheart.00319.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stansfield WE, Tang R-H, Moss NC, Baldwin AS, Willis MS, Selzman CH. Proteasome inhibition promotes regression of left ventricular hypertrophy. Am J Physiol Heart Circ Physiol. 2008;294:H645–H650. doi: 10.1152/ajpheart.00196.2007. [DOI] [PubMed] [Google Scholar]
- 75.Esposito G, Rapacciuolo A, Naga Prasad SV, Takaoka H, Thomas SA, Koch WJ, et al. Genetic alterations that inhibit in vivo pressure-overload hypertrophy prevent cardiac dysfunction despite increased wall stress. Circulation. 2002;105:85–92. doi: 10.1161/hc0102.101365. [DOI] [PubMed] [Google Scholar]
- 76.Depre C, Hase M, Gaussin V, Zajac A, Wang L, Hittinger L, et al. H11 kinase is a novel mediator of myocardial hypertrophy in vivo. Circ Res. 2002;91:1007–1014. doi: 10.1161/01.res.0000044380.54893.4b. [DOI] [PubMed] [Google Scholar]
- 77.Depre C, Wang L, Sui X, Qiu H, Hong C, Hedhli N, et al. H11 kinase prevents myocardial infarction by preemptive preconditioning of the heart. Circ Res. 2006;98:280–288. doi: 10.1161/01.RES.0000201284.45482.e8. [DOI] [PubMed] [Google Scholar]
- 78.Sui X, Li D, Qiu H, Gaussin V, Depre C. Activation of the bone morphogenetic protein receptor by H11Kinase/Hsp22 promotes cardiac cell growth and survival. Circ Res. 2009;104:887–895. doi: 10.1161/CIRCRESAHA.108.192328. [DOI] [PubMed] [Google Scholar]
- 79.Gallagher GL, Jackson CJ, Hunyor SN. Myocardial extracellular matrix remodeling in ischemic heart failure. Front Biosci. 2007;12:1410–1419. doi: 10.2741/2157. [DOI] [PubMed] [Google Scholar]
- 80.Weber K, Brilla C. Pathological hypertrophy and cardiac interstitium. Circulation. 1991;83:1849–1865. doi: 10.1161/01.cir.83.6.1849. [DOI] [PubMed] [Google Scholar]
- 81.Iimoto D, Covell J, Harper E. Increase in cross-linking of type 1 and type 3 collagens associated with volume overload hypertrophy. Circ Res. 1988;63:399–408. doi: 10.1161/01.res.63.2.399. [DOI] [PubMed] [Google Scholar]
- 82.Gonzalez A, Lopez B, Querejeta R, Diez J. Regulation of myocardial fibrillar collagen by angiotensin II. A role in hypertensive heart disease? J Mol Cell Cardiol. 2002;34:1585–1593. doi: 10.1006/jmcc.2002.2081. [DOI] [PubMed] [Google Scholar]
- 83.Funck RC, Wilke A, Rupp H, Brilla CG. Regulation and role of myocardial collagen matrix remodeling in hypertensive heart disease. Adv Exp Med Biol. 1997;432:35–44. doi: 10.1007/978-1-4615-5385-4_4. [DOI] [PubMed] [Google Scholar]
- 84.Hayden MS, Ghosh S. Signaling to NF-κB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 85.Gao Y, Lecker S, Post MJ, Hietaranta AJ, Li J, Volk R, et al. Inhibition of ubiquitin-proteasome pathway-mediated IκB α degradation by a naturally occurring antibacterial peptide. J Clin Invest. 2000;106:439–448. doi: 10.1172/JCI9826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gaczynska M, Osmulski PA, Gao Y, Post MJ, Simons M. Proline- and arginine-rich peptides constitute a novel class of allosteric inhibitors of proteasome activity. Biochemistry. 2003;42:8663–8670. doi: 10.1021/bi034784f. [DOI] [PubMed] [Google Scholar]
- 87.Li J, Post M, Volk R, Gao Y, Li M, Metais C, et al. PR39, a peptide regulator of angiogenesis. Nat Med. 2000;6:49–55. doi: 10.1038/71527. [DOI] [PubMed] [Google Scholar]
- 88.Post MJ, Sato K, Murakami M, Bao J, Tirziu D, Pearlman JD, et al. Adenoviral PR39 improves blood flow and myocardial function in a pig model of chronic myocardial ischemia by enhancing collateral formation. Am J Physiol Regul Integr Comp Physiol. 2006;290:R494–R500. doi: 10.1152/ajpregu.00460.2005. [DOI] [PubMed] [Google Scholar]
- 89.Weber K. Extracellular matrix remodeling in heart failure. Circulation. 1997;96:4065–4082. doi: 10.1161/01.cir.96.11.4065. [DOI] [PubMed] [Google Scholar]
- 90.Foody J, Farrell M, Krumholz H. β-Blocker therapy in heart failure. JAMA. 2002;287:883–889. doi: 10.1001/jama.287.7.883. [DOI] [PubMed] [Google Scholar]
- 91.Sanders PM, Russell ST, Tisdale MJ. Angiotensin II directly induces muscle protein catabolism through the ubiquitin-proteasome proteolytic pathway and may play a role in cancer cachexia. Br J Cancer. 2005;93:425–434. doi: 10.1038/sj.bjc.6602725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Russell ST, Eley HL, Tisdale M. Mechanism of attenuation of angiotensin-II-induced protein degradation by insulin-like growth factor-I (IGF-I) Cell Signal. 2007;19:1583–1595. doi: 10.1016/j.cellsig.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 93.Song Y-H, Li Y, Du J, Mitch WE, Rosenthal N, Delafontaine P. Muscle-specific expression of IGF-1 blocks angiotensin II-induced skeletal muscle wasting. J Clin Invest. 2005;115:451–458. doi: 10.1172/JCI22324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Asai M, Tsukamoto O, Minamino T, Asanuma H, Fujita M, Asano Y, et al. PKA rapidly enhances proteasome assembly and activity in in vivo canine hearts. J Mol Cell Cardiol. 2009;46:452–462. doi: 10.1016/j.yjmcc.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 95.Hayashidani S, Tsutsui H, Ikeuchi M, Shiomi T, Matsusaka H, Kubota T, et al. Targeted deletion of MMP-2 attenuates early LV rupture and late remodeling after experimental myocardial infarction. Am J Physiol Heart Circ Physiol. 2003;285:H1229–H1235. doi: 10.1152/ajpheart.00207.2003. [DOI] [PubMed] [Google Scholar]
- 96.van den Borne S, Cleutjens J, Hanemaaijer R, Creemers E, Smits J, Daemen M, et al. Increased matrix metalloproteinase-8 and -9 activity in patients with infarct rupture after myocardial infarction. Cardiovasc Pathol. 2008;Feb 21 doi: 10.1016/j.carpath.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 97.Matsumura S, Iwanaga S, Mochizuki S, Okamoto H, Ogawa S, Okada Y. Targeted deletion or pharmacological inhibition of MMP-2 prevents cardiac rupture after myocardial infarction in mice. J Clin Invest. 2005;115:599–609. doi: 10.1172/JCI22304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gao X, Dilley R, Samuel C, Percy E, Fullerton M, Dart A, et al. Lower risk of postinfarct rupture in mouse heart overexpressing β2-adrenergic receptors: importance of collagen content. J Cardiovasc Pharmacol. 2002;40:632–640. doi: 10.1097/00005344-200210000-00018. [DOI] [PubMed] [Google Scholar]