SUMMARY
During brain development, proper neuronal migration and morphogenesis is critical for the establishment of functional neural circuits. Here we report that srGAP2 negatively regulates neuronal migration and induces neurite outgrowth and branching through the ability of its F-BAR domain to induce filopodia-like membrane protrusions resembling those induced by I-BAR domains in vivo and in vitro. Previous work has suggested that in non-neuronal cells, filopodia dynamics decreases the rate of cell migration and the persistence of leading edge protrusions. srGAP2 knockdown reduces leading process branching and increases the rate of neuronal migration in vivo. Overexpression of srGAP2 or its F-BAR domain has the opposite effects, increasing leading process branching and decreasing migration. These results (1) suggest that F-BAR domains are functionally diverse and (2) highlight the functional importance of proteins directly regulating membrane deformation for proper neuronal migration and morphogenesis.
INTRODUCTION
During brain development, neural progenitor proliferation, neuronal migration and differentiation require considerable changes in cell shape involving coordinated cytoskeletal and membrane remodeling (Ayala et al., 2007; Luo, 2002). Cortical neuron migration involves the coordinated extension and adhesion of the leading process (LP) along radial glial processes with the forward translocation of the nucleus which requires regulation of centrosome and microtubule dynamics by proteins such as Lis1, Doublecortin, and Nudel among others (Ayala et al., 2007; Higginbotham and Gleeson, 2007; Lambert de Rouvroit and Goffinet, 2001). Recent genetic studies have identified regulators of leading process morphology and neuronal migration such as cyclin dependent kinase 5 (Cdk5) (Ohshima et al., 2007) or its activator p35 (Gupta et al., 2003). However, little is known about the molecular mechanisms underlying membrane dynamics during neuronal neuronal migration and morphogenesis.
The basis of neurite initiation, outgrowth and branching is rooted in the ability of the actin and microtubule cytoskeleton to undergo dynamic changes (Gupton and Gertler, 2007; Luo, 2002; Mattila and Lappalainen, 2008). Filopodia have been shown to play a role in neurite initiation (Dent et al., 2007; Kwiatkowski et al., 2007), growth cone dynamics (Burnette et al., 2007; Gallo and Letourneau, 2004), neurite outgrowth (Luo, 2002) and branching (Dent et al., 2004; Gallo and Letourneau, 1998). Downregulation of the actin anti-cappers, ENA/VASP proteins, which are potent inducers of filopodia resulted in failed neurite initiation. Interestingly, loss of ENA/VASP proteins also resulted in defects in cortical lamination (Kwiatkowski et al., 2007) suggesting a functional relationship between filopodia formation, neurite initiation and neuronal migration.
Classically, filopodia formation is thought to be primarily dependent on proteins that regulate actin polymerization at the barbed end of actin filaments and proteins bundling F-actin (Gupton and Gertler, 2007). In particular, proteins containing Inverse-BAR (I-BAR) domain such as IRSp53, that induce membrane deformation have been shown to induce filopodia formation independent of its ability to bundle F-actin (Lim et al., 2008; Mattila et al., 2007; Saarikangas et al., 2009). The BAR-domain superfamily contains three main groups: the Bin/Amphiphysin/Rvs (BAR) domain subfamily (Itoh and De Camilli, 2006), the Fes-Cip4 Homology (FCH)-BAR (also called F-BAR or EFC) domain subfamily ((Itoh et al., 2005; Tsujita et al., 2006); reviewed in (Frost et al., 2009)) and the I-BAR subfamily (reviewed in (Scita et al., 2008)). Crystal structures of the F-BAR domains of FBP17, CIP4, and FCHo2 demonstrated that these domains are elongated homodimers characterized by a shallow curvature formed by the anti-parallel interaction of two alpha-helical coiled coils (Henne et al., 2007; Shimada et al., 2007). In addition to sharing the general fold and quaternary organization of the BAR domain superfamily as a whole, F-BAR domains share functional properties with ‘classical’ BAR domains, most notably the ability to bind and deform membranes in vitro and in living cells (Frost et al., 2008; Itoh et al., 2005; Kakimoto et al., 2006; Shimada et al., 2007). However, to date, the in vivo functions of F-BAR domain-containing proteins are largely unknown (Frost et al., 2009).
Here we identify slit-robo GTPase Activating Protein (srGAP2) as a regulator of neuronal migration and morphogenesis through the unexpected ability of its N-terminal F-BAR domain to induce filopodia-like membrane protrusions resembling those induced by I-BAR domains. Our results highlight the functional importance of proteins directly regulating membrane deformation for proper neuronal migration and axon-dendrite morphogenesis.
RESULTS
Expression of srGAP2 in the Developing Cortex
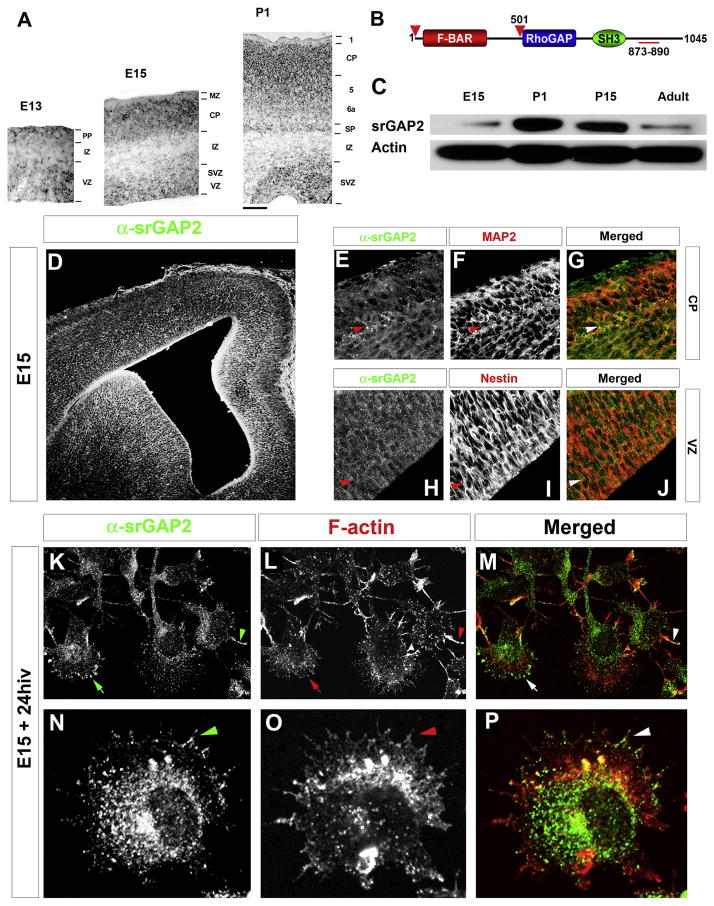
To begin our study of the role of srGAP2 in cortical development, we first examined its pattern of expression. srGAP1-3 have recently been reportred to be expressed throughout the cortex during and after radial migration (Bacon et al., 2009; Mattar et al., 2004; Yao et al., 2008). Our analysis confirmed that srGAP2 mRNA is expressed throughout the developing cortex and is found both in proliferative zones (ventricular and subventricular zones, VZ and SVZ respectively) at E13 and E15 and in postmitotic regions (cortical plate, CP) at E15 and P1 (Fig. 1A). In order to determine the pattern of srGAP2 protein expression, we used a previously characterized polyclonal antibody raised against the C-terminus of srGAP2 (Fig. 1B–C; (Yao et al., 2008)). srGAP2 protein is expressed throughout cortical development culminating at postnatal day 1 (P1) corresponding to the peak of neuronal migration in the cortex. Its expression is maintained at P15 and reduced, but still present, in adult cortex (Fig. 1C).
Figure 1. srGAP2 is expressed in neuronal progenitors and post-mitotic neurons and localizes to sites of membrane protrusion.
(A) In situ hybridization for srGAP2 in developing cortex at embryonic day (E)13 and E15 and postnatal day (P)1.
(B) Domain organization of srGAP2 which contains an F-BAR domain, a RhoGAP and a SH3 domain from N-to C-terminal ends (1–1045 aa, predicted MW 118kDa). The black bar indicates the localization of the antigen (A2, aa 873–890) used to affinity purify the srGAP2-specific polyclonal antibody to the C-terminus of srGAP2 (Yao et al., 2008).
(C) Western blot for srGAP2 protein levels during cortical development at the indicated time points (E15, P1, P15 and Adult) obtained by SDS-PAGE and immunoblotting with A2-rabbit polyclonal antibody (Yao et al, 2008).
(D–J) Immunofluorescence staining of srGAP2 protein expression on fixed coronal sections of E15 mouse cortex. srGAP2 protein colocalizes (arrowheads) with MAP2 (postmitotic neuron marker) in the CP (D–F) and also colocalizes with Nestin (arrowheads) (neuronal precursor marker) in the VZ (G–I).
(K–P) Immunofluorescence staining of srGAP2 protein in early dissociated cortical neuron cultures (E15+ 24 hours in vitro –hiv). srGAP2 protein is found close to the plasma membrane of immature cortical neurons (arrow in K-M) and to F-actin-rich filopodia (stained with Alexa546-phalloidin; arrowheads in K–M and N–P).
Immunofluorescent staining for srGAP2 shows that it is ubiquitously expressed in the cortical wall (Fig. 1D) being found both in Nestin-positive neuronal progenitors in the VZ (Fig. 1H–J) and MAP2-positive post-mitotic neurons in the CP (Fig. 1E–G). At the subcellular level, endogenous srGAP2 is found at the cell periphery (Fig. 1K–M arrows) and was often localized along F-actin-rich filopodia-like protrusions (arrowhead in Fig. 1K–M and Fig. 1N–P) in acutely dissociated E15 cortical neurons.
Full-length srGAP2 and its F-BAR Domain Induce Filopodia Formation
Over-expression of F-BAR domain-containing proteins such as FBP17 or CIP4 have been shown to cause membrane invagination and tubulation in cell lines (Itoh et al., 2005; Tsujita et al., 2006). Surprisingly, expression of srGAP2 did not induce any membrane invaginations but instead, induced filopodia formation (Fig. S1D–F, S1P). This effect requires its F-BAR domain since deletion of the F-BAR domain (srGAP2ΔF-BAR-EGFP) does not induce filopodia formation in COS7 cells (Fig. S1G–I, S1P).
Interestingly, expression of the F-BAR domain of srGAP2 did not inhibit endocytosis, assessed using Alexa546-Transferrin uptake assay (Fig. S2), as do the F-BAR domains of FBP17 and CIP4 (Itoh et al., 2005). Furthermore, expression of the isolated F-BAR domain fused to EGFP induced filopodia formation just as full-length srGAP2 (Fig. S1J–K, S1P). Of note, the F-BAR domain is a potent membrane-targeting motif (Fig. S1J). These data suggest that the F-BAR domain of srGAP2 is necessary and sufficient for membrane localization and the induction of filopodia-like membrane protrusions.
In order to distinguish the membrane targeting function of the F-BAR domain from its membrane deformation activity, we identified a small truncation of the last C-terminal 49 amino-acids (F-BARΔ49) (Fig. S3A and Suppl. Material for details). Expression of F-BARΔ49-EGFP in COS7 cells results in significant membrane targeting (Fig. S4) but fails to induce filopodia in COS7 cells (Fig. S1M–O, S1P). We do not currently know the structural basis for the inability of this truncation to elicit filopodia but we can at least exclude two possibilities: (1) instability of the F-BARΔ49 protein since it expresses at level comparable to full-length srGAP2 or its F-BAR domain in cells (Fig. S5) and (2) this truncation does not disrupt its dimerization properties since F-BARΔ49 can dimerize with F-BAR or full-length srGAP2 (data not shown). Interestingly, these 49 amino acids reside in an extension specific to the srGAP subfamily (α6–8, Fig. S3A) that is C-terminal to the minimal, predicted F-BAR domain (amino acids 1–358; Itoh et al., 2005) (Fig. S3B). Indeed, we were unable to obtain stable protein expression of this minimal predicted F-BAR domain in mammalian cells or bacteria (data not shown). Furthermore, as shown for other F-BAR domains (Frost et al., 2008; Itoh et al., 2005; Kakimoto et al., 2006; Shimada et al., 2007), srGAP2 forms a stable dimer in solution as assessed by light scattering assays (Fig. S3C) and deletion of the FCH domain (green box Fig. S3A) which represents a significant portion of the dimerization interface, destroys the ability of srGAP2 to induce filopodia formation in COS7 cells (data not shown). Altogether, these data suggest that all 8 predicted α-helices are likely to be required for formation of the functional F-BAR domain of srGAP2.
The F-BAR domain of srGAP2 deforms membrane like an I-BAR domain
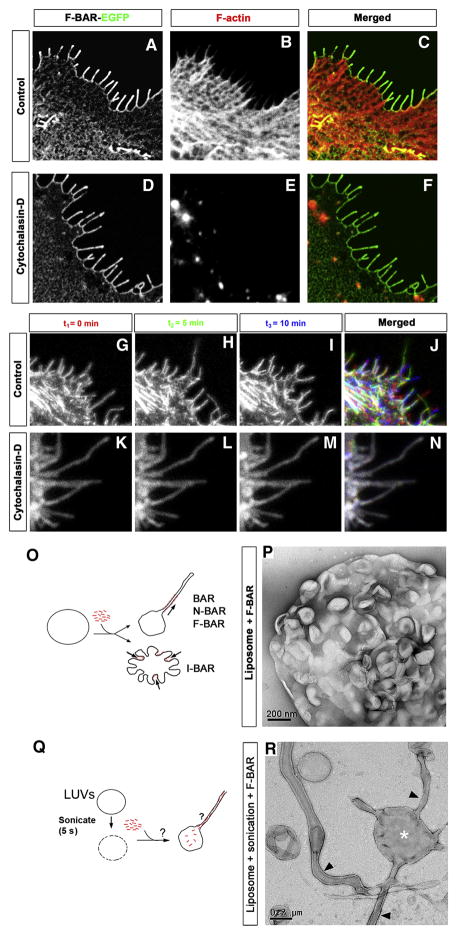
The ability of srGAP2 or its F-BAR domain to induce filopodia in COS7 cells is reminiscent of the activity of the structurally related, Inverse (I)-BAR domain-containing proteins such as IRSp53 and Missing-in-Metastasis (MIM) (Mattila et al., 2007; Millard et al., 2007; Saarikangas et al., 2009; Scita et al., 2008). The filopodia induced by I-BAR domains are dependent upon their ability to interact with and to deform the plasma membrane (Mattila et al., 2007; Saarikangas et al., 2009). Interestingly, F-actin depolymerization prevents the dynamics and formation of new filopodia but does not affect the maintenance of pre-existing filopodia induced by the I-BAR domains of IRSp53 or MIM (Mattila et al., 2007). Untreated COS7 cells expressing the F-BAR domain of srGAP2 developed F-actin rich filopodia (Fig. 2A–C), while cells treated with cytochalasin D were depleted of F-actin. Strikingly, this treatment had no effect on membrane localization of the F-BAR domain or on the maintenance of filopodia-like protrusions (Fig. 2D–F).
Figure 2. F-BAR induced filopodia required F-actin for their dynamic formation but not for their structural maintenance.
(A–C) COS7 cell expressing the F-BAR-EGFP fusion protein not treated with cytochalasin D (control). Note the cortical localization of the F-BAR domain and the numerous F-actin-rich filopodia (phalloidin in B and C).
(D–F) COS7 cell expressing the F-BAR-EGFP fusion protein incubated with 400μM cytochalasin D 30 minutes. Note that the complete loss of F-actin (phalloidin; E) had no effect on the localization of the F-BAR domain or on the structure of the F-BAR mediated protrusions.
(H–K) Time series showing the dynamics of F-BAR-EGFP-induced filopodia in COS7 cells. Time 0, 5, and 10 minutes are pseudo-colored in red, green, and blue respectively. Note there is little colocalization of filopodia at the cell periphery (K). This is in stark contrast to COS7 cells expressing F-BAR-EGFP treated with Cytochalasin D (30 minutes) (L–O) where the protrusions remain static and do not grow or retract for the same period of time shown in control cells.
(P) Schema depicting tubulation assay in Q.
(Q) F-BAR domain of srGAP2 added to preformed liposomes. Note the inward dimpling or “scalloping” of the liposome surface. (R) Schema depicting tubulation assay in (S) where F-BAR domain of srGAP2 was added to liposomes after extrusion. This results in a fraction of the F-BAR domain resident inside the liposome. Note the formation of tubule protrusion from the liposome.
(S) High magnification of liposome/F-BAR mixture after sonication. Note the absence of striations or an obvious protein coat on the lipid tubule, a hallmark of canonical F-BAR tubulation. These tubules are 83 nm +/−15 nm (average +/− SD, N=38) after being partially flattened by the negative staining procedure.
F-BAR-induced filopodia were highly dynamic in COS7 cells protruding and retracting within less than a minute (Fig. 2H–2K, and Movie S1). In contrast, F-BAR-induced filopodia formed prior to Cytochalasin-D treatment were totally resistant to F-actin depolymerization (Fig. 2L–2O, and Movie S2). These data suggest that the F-BAR domain of srGAP2 functions as an I-BAR domain in living cells, by inducing filopodia that require F-actin for their dynamics but is independent of F-actin for their structural maintenance.
In order to directly test the membrane deformation properties of the F-BAR domain of srGAP2, we incubated purified F-BAR domain with preformed liposomes. As visualized by negative stain transmission electron microscopy (TEM), this did not result in liposome outward tubulation as has been reported for other F-BAR domains (see Fig. S5B). Rather, the F-BAR domain of srGAP2 induced an inward dimpling or “scalloping” of the liposome surface (Fig. 2P–Q), which is reminiscent of the activity of I-BAR domains in the same conditions (Suetsugu et al., 2006), suggesting that the F-BAR domain of srGAP2 can induce “inverse” membrane tubulation.
This suggested the possibility that if the purified F-BAR domain of srGAP2 could be exposed to the inside surface of liposomes, then protrusive tubules would form (Fig. 2R). To test this hypothesis, mixtures of the F-BAR domain with intact, large unilamellar vesicles (LUVs) were briefly sonicated (5sec) which presumably resulted in transient pore formation in liposomes and introduction of the recombinant F-BAR inside LUVs. Following a wash, liposomes were fixed, negatively stained and imaged using TEM. As predicted by the I-BAR model, this resulted in numerous long tubular extensions emerging from LUVs (Fig. 2S) which is in stark constrast with control sonicated liposomes not incubated with recombinant protein (Fig. S6A). Consistent with the dimensions of tubules induced by other members of the F-BAR and I-BAR families (Frost et al., 2008; Mattila et al., 2007), the srGAP2 F-BAR-induced tubules were 83 nm +/− 15 nm (average +/− SD, n=38) in diameter. Importantly, at higher magnification, the tubules observed by negative staining electron microscopy after sonication do no have an obvious protein coat surrounding the liposomes (Fig. 2S). This is in contrast with tubules induced by other F-BAR and BAR domains that coat the outer surface of the tubule (Fig. S6B; Frost et al., 2008; Shimada et al., 2007). Together, these results suggest that unlike previously characterized F-BAR domains, the F-BAR domain of srGAP2 functions as an I-BAR domain (Mattila et al., 2007; Suetsugu et al., 2006).
srGAP2 regulates neurite formation and branching through the ability of its F-BAR domain to form filopodia
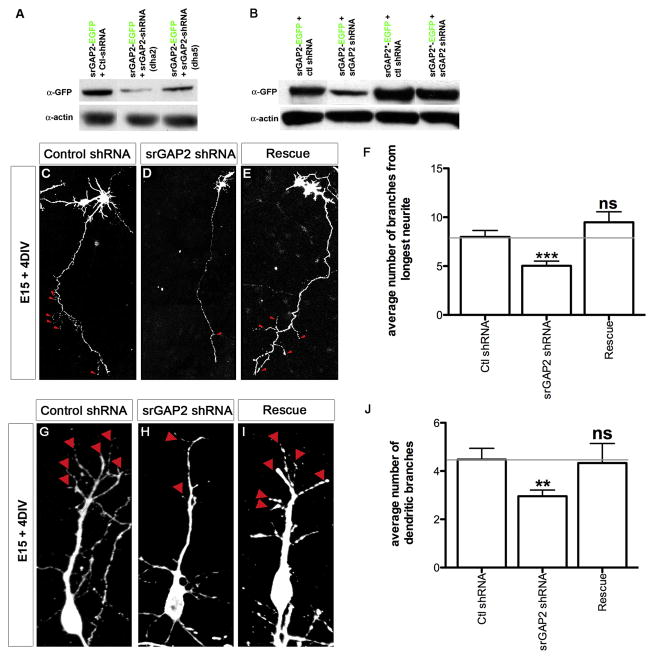
We next tested the function of srGAP2 in neuronal morphogenesis by designing short-hairpin interfering RNA (shRNA) in order to acutely knockdown srGAP2 expression (Fig. 3A). We found that srGAP2 knockdown in E15 cortical neurons led to a significant decrease in both axonal (Fig. 3C–D and 3F) and dendritic branching after 5 div (Fig. 3G–H and 3J). Both of these effects were rescued by co-transfection of a shRNA-resistant form of srGAP2 (srGAP2*; Fig. 3E and 3I; 3F and 3J) demonstrating that this is not an off-target effect. The fact that srGAP2 knockdown reduced branching in cortical neurons, a process previously shown to require filopodia formation (Dent et al., 2004; Gallo and Letourneau, 1998), suggest that srGAP2 may promote neurite branching through its ability to induce filopodia in neurons.
Figure 3. Knockdown of srGAP2 in cortical neurons reduces axonal and dendritic branching.
(A) Western blot probed with ant-GFP and anti-actin antibodies from COS7 cells co-transfected with either control shRNA plus srGAP2-EGFP (lane 1), srGAP2 shRNA plus srGAP2-EGFP (Dha2, lane 2) or (Dha5, lane3)
(B) Western blot probed with anti-GFP and anti-actin antibodies from COS7 cells co-transfected with either control shRNA plus srGAP2-EGFP (lane 1), srGAP2 shRNA plus srGAP2-EGFP (lane 2), a mutated form of srGAP2*-EGFP (resistant to srGAP2 shRNA) plus control shRNA (lane 3), or srGAP2*-EGFP plus srGAP2 shRNA (lane 4). srGAP2 shRNA significantly knocks down srGAP2 expression compared to control shRNA which can be rescued by expression of srGAP2*-EGFP (compare lanes 3 and 4).
(C–E, G–I) E15 dissociated cortical neurons were cultured for 4 days after ex vivo electroporation with control shRNA, srGAP2 shRNA, or srGAP2 shRNA + srGAP2*-EGFP. Control shRNA transfected neurons display frequent primary branches from the axon (arrowheads in B) and the primary dendrite (arrowheads in F). Both effects were markedly reduced in srGAP2 shRNA transfected neurons (D and H) and rescued by co-transfection of srGAP2 shRNA with srGAP2*-EGFP (E and I).
(E) Quantification of the number of branches from the longest neurite (axon) as shown in C–E.
(I) Quantification of the number of primary dendritic branches as shown in G–I. (Control shRNA, n=42 cells; srGAP2 shRNA, n=95; srGAP2*-EGFP + srGAP2 shRNA, n=39. Cells were taken from 3 independent experiments and analyzed blind to the treatment.
Mann-Whitney Test * p<0.05; ** p<0.01; *** p<0.001.
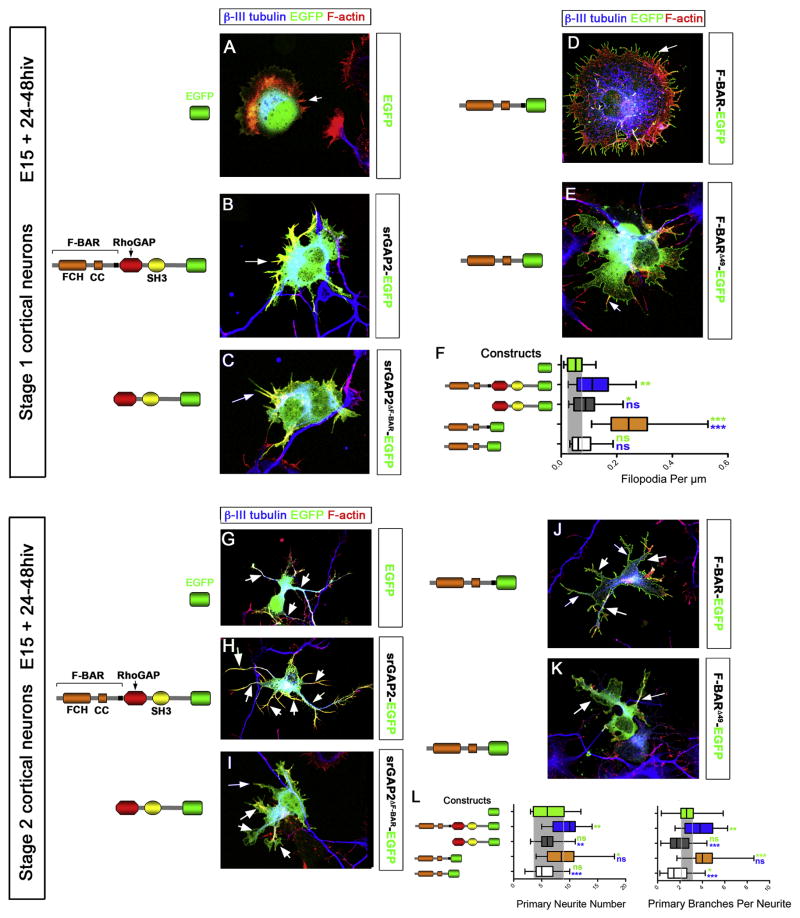
To test this hypothesis, we performed structure/function analysis using electroporation of E15 cortical progenitors with various srGAP2 constructs followed by dissociation and culture, which induces rapid differentiation. First, we restricted our analysis to Stage 1 neurons (Dotti et al., 1988) when immature neurons produce a significant number of filopodia-like protrusions (Dent et al., 2007; Kwiatkowski et al., 2007). Our analysis show that expression of full-length srGAP2 induced a significant increase in filopodia-like protrusions in Stage 1 cortical neurons compared to control EGFP (Fig. 4A–C and 4F). This effect requires the F-BAR domain since deletion of the F-BAR domain (srGAP2ΔF-BAR) significantly reduced the ability of srGAP2 to induce filopodia in Stage 1 neurons (Fig. 4C and 4F). As in COS7 cells, expression of the F-BAR domain alone potently induces formation of F-actin-rich filopodia (Fig. 4D and 4F). Again, the effect of the F-BAR domain requires its membrane deformation properties and not simply its membrane targeting property since expression of F-BARΔ49 does not induce filopodia in Stage 1 cortical neurons (Fig. 4E and 4F) and instead induces large lamellipodia (arrowhead in Fig. 4E). These data suggest that srGAP2, through its F-BAR domain, induces filopodia in cortical neurons as shown in COS7 cells.
Figure 4. srGAP2 promotes filopodia formation and neurite outgrowth in an F-BAR dependent manner.
(A–E) Stage 1 cortical neurons expressing various srGAP2 constructs. All cells are stained with neuron-specific β-III tubulin (blue) to reveal presence of microtubules (see also Fig. S7) and phalloidin (red) to visualize F-actin.
(F) Quantification of filopodia normalized per cell perimeter in all conditions. EGFP n= 20 cells; srGAP2-EGFP n= 21; srGAP2ΔF-BAR-EGFP n= 20; F-BAR-EGFP n= 20; F-BARΔ49-EGFP n= 20. Cells were taken from 3 independent experiments and analyzed blind to the treatment.
(G–K) Stage 2 cortical neurons expressing various srGAP2 constructs. All cells are stained with β-III tubulin (blue) and phalloidin (red) as in panels A–F. Arrows point to primary neurites and arrowheads point to neurite branches.
(L) Quantification of neurite number normalized per cell perimeter in all conditions and primary branch number per neurite. Note srGAP2 and F-BAR are potent inducers of neurite outgrowth while srGAP2 ΔF-BAR and F-BARΔ49 are not.
Mann-Whitney Test * p<0.05; ** p<.001; *** p<0.001. Green stars indicates comparison to EGFP and blue stars indicates comparison to srGAP2-EGFP.
We then analyzed Stage 2 neurons i.e. prior the emergence of a single axon (Dotti et al., 1988) in order to test if srGAP2 and its F-BAR domain were sufficient to promote the transition between filopodia and elongating neurites defined by the presence of bundled microtubules (see also Fig. S7 for isolated βIII-tubulin staining). As shown in Figure 4G–K, both full-length srGAP2 and the F-BAR domain significantly increased the total number of primary neurites emerging from the cell body as well as the number of primary neurite branches (Fig. 4L). Expression of srGAP2ΔF-BAR as well as F-BARΔ49 fail to increase primary neurite number and neurite branching compared to control EGFP (Fig. 4L–4J and 4L).
Reduction of srGAP2 expression promotes neuronal migration
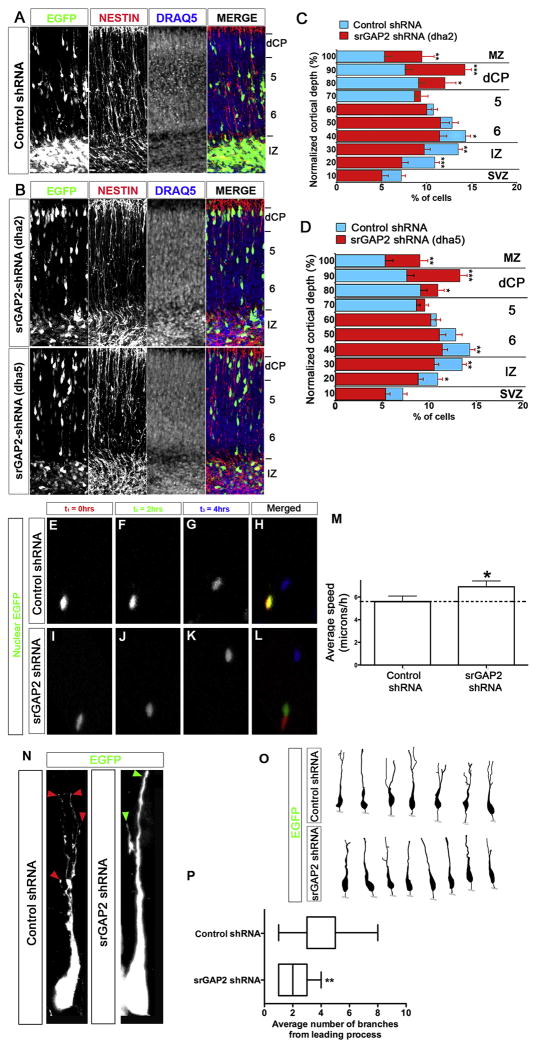
To determine more generally the function of srGAP2 during cortical development, we introduced our shRNA constructs directed against srGAP2 (Dha2 and Dha5; Fig. 3A) into radial glial progenitors at E15 using ex vivo electroporation coupled with organotypic slice culture (Hand et al., 2005). Interestingly, after 3 days in culture, at a time point when few control-shRNA electroporated neurons have migrated yet (Fig. 5A and 5C–D), slices expressing srGAP2 shRNA showed a significant increase in the percentage of neurons that have reached the dense cortical plate (dCP) and a corresponding decreased percentage of neurons in the intermediate zone (IZ) (Fig. 5B and 5C–D) suggesting that reduction of srGAP2 expression accelerated radial migration. To test this directly, we used time-lapse confocal microscopy to visualize neurons coexpressing nuclear-(n) EGFP (to ease cell tracking) and control shRNA (Fig. 5E–-H and Movie S3) or srGAP2 shRNA (Fig. 5E–L and Movie S4) in slice culture. We found that srGAP2 shRNA expressing neurons migrated approximately 25% faster than those expressing control shRNA (Fig. 5M) suggesting that reduction of srGAP2 increased the actual rate of cell translocation.
Figure 5. Knockdown of srGAP2 promotes neuronal migration and reduces leading process branching.
(A) E15 cortical slices cultured for 3 days after electroporation with EGFP + control shRNA. Slices were stained with anti-Nestin antibody revealing radial glial scaffold and Draq5 to illustrate cytoarchitecture.
(B) E15 cortical slices cultured for 3 days after electroporation with EGFP + Dha2 (B, top panel) or Dha5 (B lower panel). Slices were stained with anti-Nestin antibody revealing radial glial scaffold and Draq5 to illustrate cytoarchitecture.
(E–L) E15 cortical slices cultured for 2 days ex vivo after electroporation with Nuclear EGFP (3NLS) along with control shRNA (E–H) or srGAP2 shRNA (I–L) were imaged using time-lapse confocal microscopy. Neurons transfected with srGAP2 shRNA undergo faster translocation within 4 hrs (I–L and no colocalization in L) than control shRNA-transfected neurons.
(M) Quantification of effects of srGAP2 knockdown on cell speed. Neurons with reduced level of srGAP2 (shRNA) migrated approximately 23% faster (6.91 μicrons/h compared to 5.59 μicrons/h) compared to control shRNA-transfected neurons. Control shRNA, n=95 cells; srGAP2 shRNA n=84. Cells were taken from 3 independent experiments. Mann-Whitney Test * p<0.05; ** p<.001; *** p<0.001.
(N–O) High magnification images (N) and reconstructions (O) of control shRNA (left panel) or srGAP2 shRNA (right panel) expressing neurons in layers 5/6. Note the branched morphology of the leading process of control shRNA expressing neurons (red arrowheads pointing to leading process tips in N) whereas srGAP2 shRNA expressing neurons displayed a simpler, less branched morphology (green arrowhead in N pointing to single leading process tip).
(P) Quantification of the leading process branch number in control shRNA or srGAP2 shRNA expressing neurons. Control shRNA, n=19 cells; srGAP2 shRNA n=17 cells. Cells were taken from 3 independent experiments. Mann-Whitney Test * p<0.05; ** p<.001; *** p<0.001.
Excessive LP branching in migrating cortical neurons can have strong inhibitory effects on neuronal migration (Gupta et al., 2003; Ohshima et al., 2007). Indeed, the leading process of srGAP2 knockdown neurons in layers 5/6 was significantly less branched compared to control shRNA neurons (Fig. 5N–P). These data suggest that srGAP2 may negatively regulate the rate of radial migration by regulating LP branching and dynamics.
The F-BAR domain is necessary and sufficient for srGAP2-mediated inhibition of radial migration
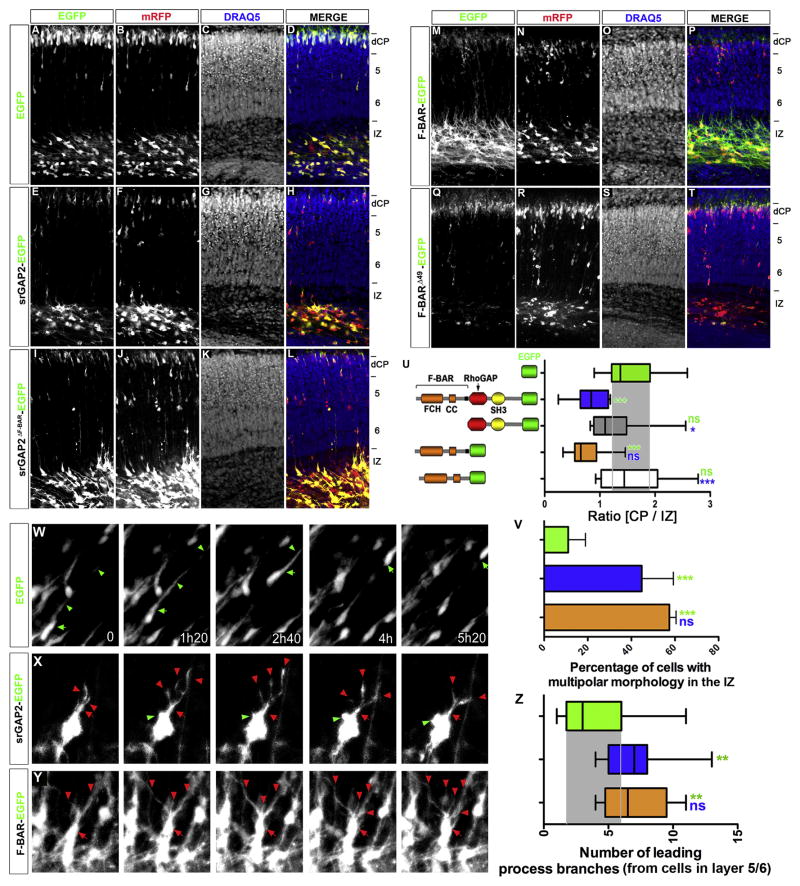
We hypothesized that over-expression of srGAP2 or its F-BAR domain should be sufficient to block migration by increasing filopodia formation and LP dynamics. Indeed, overexpression of srGAP2 severely inhibited radial migration compared to control EGFP-expressing slices electroporated at E15 and cultured for 5 div (Fig. 6E–H). We quantified radial migration by determining the ratio of neurons in the dense cortical plate (dCP; where pyramidal neurons complete migration) and in the IZ, where they initiate radial migration (see Fig. S8 for definition of cytoarchitecture). This CP/IZ ratio (Fig. 6U) is significantly decreased by srGAP2 overexpression (Fig. 6E–H) when compared to control EGFP expressing neurons (Fig. 6A–D) demonstrating that srGAP2 overexpression inhibits neuronal migration. Expression of srGAP2ΔF-BAR did not significantly reduce the CP/IZ ratio compared to EGFP control (Fig. 6I–L, 6U) and is significantly different from the ratio measured by srGAP2 overexpression (Fig. 6U) suggesting that the F-BAR domain is partially required for srGAP2’s ability to inhibit migration. Moreover, expression of the F-BAR domain alone was sufficient to reduce neuronal migration to the same extent than srGAP2 while expression of F-BARΔ49 had no effect on the ability of neurons to migrate (Fig. 6M–P and Fig. 6Q–T and 6U) suggesting that the ability of the F-BAR domain to induce filopodia is required for the ability of srGAP2 to inhibit neuronal migration.
Figure 6. srGAP2 mediated inhibition of migration requires F-BAR-mediated membrane deformation.
(A–T) E15 cortical slices cultured for 5 days after co-electroporation of monomeric Red Fluorescence Protein (mRFP for cytoplasmic filing) together with EGFP (A–D), full-length srGAP2-EGFP (E–H), srGAP2ΔF-BAR-EGFP (I–L), F-BAR-EGFP (M–P) and F-BARΔ49-EGFP fusion proteins (Q–T). Slices were stained with nuclear marker Draq5 in order to reveal the cytoarchitecture.
(U) Quantification of CP/IZ ratio. EGFP n= 13 slices; srGAP2-EGFP n= 14 slices; srGAP2ΔF-BAR-EGFP n= 8 slices; F-BAR-EGFP n= 10 slices; F-BARΔ49-EGFP n= 6 slice. Slices were taken from 4 different experiments and CP/IZ ratio analyzed using Mann-Whitney Test * p<0.05; ** p<.001; *** p<0.001. Green stars indicates comparison to EGFP and blue stars indicates comparison to srGAP2-EGFP.
(V) Quantification of percentage of cells with multipolar morphology in EGFP, srGAP2-EGFP, or F-BAR-EGFP transfected slices. Multipolar cells were defined as cells possessing ≥ 3 processes. EGFP n= 66 cells; srGAP2-EGFP n= 42; F-BAR-EGFP n= 57. Cells were taken from 3 different experiments and analyzed using Fisher’s exact test * p<0.05; ** p<.001; *** p<0.001.
(W–Y) Individual frames using time-lapse confocal microscopy of E15 cortical slices cultured for 3 days after electroporation with EGFP, srGAP2-EGFP, or F-BAR-EGFP (co-transfected with Venus plasmid). Arrows indicate leading processes and arrowheads the cell body.
(Z) Quantification of leading process branch number from cells expressing EGFP, srGAP2-EGFP, or F-BAR-EGFP in layer 5/6. EGFP n= 17 cells; srGAP2-EGFP n= 21 cells; F-BAR-EGFP n= 9 cells. Cells were taken from 3 independent slices. Mann-Whitney Test * p<0.05; ** p<.001; *** p<0.001. Green stars indicates comparison to EGFP and blue stars indicates comparison to srGAP2-EGFP.
srGAP2 inhibits migration by increasing leading process dynamics and branching
The accumulation of neurons expressing srGAP2 or its F-BAR domain in the IZ suggested that the neurons might be partially blocked in the multipolar to unipolar transition (Noctor et al., 2004). Indeed quantification of the percentage of multipolar cells (cells displaying ≥ 3 neurites) in the IZ in slices electroporated srGAP2 or the F-BAR domain revealed a significant increase in the percentage of neurons with multiple processes emerging from the cell body compared to control (Fig. 6V). This is consistent with the ability of srGAP2 to induce filopodia and neurite initiation/branching in dissociated neurons (see Fig. 4).
Our time-lapse confocal microscopy analysis show that control neurons engaging radial migration in the IZ, form a stable LP upon initiating radial migration (green arrowheads in Fig. 6W and Movie S5) and undergo efficient cell body translocation (green arrows in Fig. 6W and Movie S5). In contrast, neurons overexpressing srGAP2 or the F-BAR domain alone, do not undergo cell body translocation (red arrows in Fig. 6X and 6Y and Movies S6 and S7), and instead form multiple processes that are highly dynamic and unstable (red arrowheads in Fig. 6X and 6Y and Movies S6 and S7). The plasma membrane of these cells appears highly dynamic showing large, transient protrusions (green arrowheads in Fig. 6X). While many neurons accumulate in the IZ, some neurons did manage to translocate into layers 5/6 (Fig 6E–H and 6M–P), where expression of srGAP2 or its F-BAR domain significantly increases LP branching compared to EGFP control (Fig 6Z and Fig. 7A–C). Taken together these data suggest that srGAP2 increases neurite initiation and branching through the ability of its F-BAR domain to induce filopodia, which in turn negatively regulates neuronal migration.
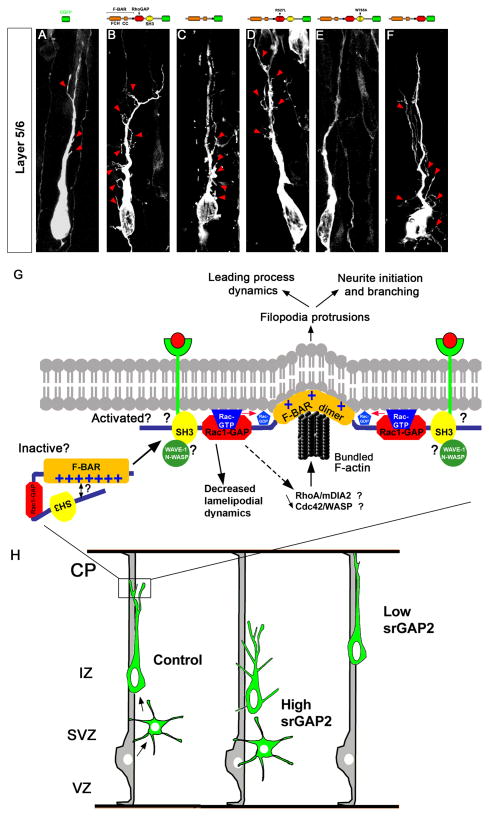
Figure 7. Model for srGAP2 regulated membrane protrusion in neuronal migration.
(A–F) Representative images of optically isolated neurons translocating radially through layer 5/6 following electroporation at E15 (+5div) with indicated srGAP2 constructs containing a F-BAR domain.
(G–H) Hypothetical model of the molecular mechanisms underlying srGAP2 function in membrane protrusion during neuronal migration and morphogenesis (G). Summary of srGAP2 effects on neuronal migration and morphogenesis during cortical development (H). See text for details.
Finally, to ensure that the reduction in the number of migrating neurons was not due to an indirect effect of srGAP2 expression on progenitor proliferation and/or cell cycle exit, we designed a vector allowing us to express srGAP2 in early post-mitotic neurons using the 2.2kB NeuroD promoter (Fig. S9A). NeuroD is a bHLH transcription factor and a direct transcriptional target of Ngn2 (Hand et al., 2005; Heng et al., 2008) thereby inducing cDNA expression in intermediate progenitors and early post-mitotic neurons in the SVZ and IZ (Fig. S10E–H) but not by Nestin+ radial glial progenitors in the VZ as obtained by the chicken β-actin promoter (Fig. S10A–D). Furthermore, the level of protein expression in neurons obtained with this promoter is significantly lower than using the chicken β-actin promoter (data not shown; (Heng et al., 2008)). Expression of srGAP2 using this NeuroD promoter significantly reduced the number of cells reaching the CP compared to control (Fig. S9B–I and S9J).
srGAP2 partially requires its RhoGAP and SH3 domains to inhibit migration
We next wanted to determine the contributions of the RhoGAP and SH3 domains to srGAP2 function in neuronal migration and morphogenesis. In order to determine the substrate specificity of the GAP domain of srGAP2, we purified its GAP domain as a GST-fusion (Fig. S11A). We then performed fluorescence-based GTP hydrolysis assays (Fig. S11B; (Shutes and Der, 2006)). The GAP domain of srGAP2 increased the rate of GTP hydrolysis on Rac1, but had no effect on RhoA or Cdc42 (Fig. S11B) or RhoG (data not shown). In addition, full-length srGAP2 strongly interacted with activated Rac1 (Rac1Q61L) but only weakly interacted with activated Cdc42Q61L (Fig. S11C) and activated RhoAQ63L (data not shown). These two independent approaches demonstrate that the GAP domain of srGAP2 is specific for Rac1.
To determine the contribution of the Rac1-GAP domain on srGAP2’s ability to regulate neuronal morphogenesis and migration, we engineered a catalytically-inactive form of srGAP2 (srGAP2R527L). Indeed this mutant was unable to accelerate GTP hydrolysis of Rac1 (Fig. S11D). Expression of the ‘GAP-dead’, srGAP2R527L was as potent as srGAP2 at inducing filopodia-like membrane protrusions in Stage 1 cortical neurons (compare Fig. S12B and S12C; quantified in S12F) and at promoting primary neurite initiation (Fig. S12H and S12I; quantified in S12L). While, this mutant was competent to increase neurite formation, there were significantly fewer (2-fold) srGAP2R527L expressing neurons at stage 2 when compared to srGAP2 (Fig. S13). In addition, srGAP2R527L displays a reduced ability to induce neurite branching when compared to srGAP2 (Fig. S12L), suggesting that the Rac1-GAP activity of srGAP2 might be required for some aspects of neurite dynamics such as neurite branching.
We next wanted to determine the contribution of the Rac1-GAP activity of srGAP2 in its ability to inhibit neuronal migration. Expression of srGAP2R527L in E15 cortical progenitors significantly inhibits migration compared to control EGFP (Fig. S14A–D and S14I–L and S14U) although not as potently as full-length srGAP2 over-expression (Fig. S14E–H and SU) suggesting that the Rac1-GAP activity of srGAP2 contributes to its ability to inhibit migration. In addition, similarly to srGAP2, expression of srGAP2R527L increased the percentage of multipolar cells in the IZ (Fig. S14V) and increased LP branching of radially migrating neurons in layer 5/6 (Fig. S14X and Fig. 6Z). These data suggest that the Rac1-GAP activity may act to modulate protrusion formation induced by the F-BAR domain of srGAP2, but is not absolutely required to induce filopodia-like membrane protrusions and inhibit neuronal migration.
The SH3 domain of the closely related proteins, srGAP1 and srGAP3, have been shown to interact with the Robo1 receptor, a known axon guidance receptor (Li et al., 2006; Wong et al., 2001), and the WAVE-1 complex, an actin-polymerizing complex (Soderling et al., 2002). To test the contribution of the SH3 domain of srGAP2, we engineered a mutant to a conserved tryptophan residue (srGAP2W765A), which was shown to be required for the ability of the SH3 domain of srGAP1 to bind to Robo1 and for the SH3 domain of srGAP3 to bind to WAVE-1 (Li et al., 2006; Soderling et al., 2002). Expression of srGAP2W765A, unlike the expression of full-length srGAP2 or its F-BAR domain, did not efficiently induce filopodia-like membrane protrusions in Stage 1 cortical neurons (Fig. S12D and S12F), and had a significantly decreased ability to induce primary neurite branching compared to full-length srGAP2 (Fig. S12J and S12L). Expression of srGAP2W765A increased primary neurite initiation, however showed a significantly reduced percentage (2 fold) of neurons transitioning from Stage 1 to Stage 2 compared to srGAP2 (Fig. S13) suggesting that full-length (i.e. all domains functionally intact) srGAP2 is required for optimal transition between a filopodia-like protrusion to an elongating neurite.
Interestingly, using our slice migration assay, expression of srGAP2W765A had no effect on cortical neuron migration (Fig. S14M–P and S14U), although there was a slight increase in cells with multipolar morphology in the IZ compared to EGFP (Fig. S14V). The lack of effect of srGAP2W765A over-expression on the CP/IZ ration prompted us to use time-lapse microscopy to observe LP dynamics in radially migrating neurons. This analysis revealed that migrating neurons expressing srGAP2W765A did not display increased leading process branching and dynamics but instead had a single, stable leading processes (red arrowheads Fig. S14W and Movie S8) and translocated efficiently (green arrowheads Fig. S14W and Movie S8) which is strikingly different from neurons overexpressing full-length srGAP2 (Fig. 6X). Moreover analysis of neurons in layer 5/6 showed no significant increase in LP branching as demonstrated with other constructs containing an F-BAR domain (Fig. 6 and S14X and Fig. 7E).
The fact that srGAP2W765A showed weak filopodia formation compared to full-length srGAP2 and no increase in neurite branching suggested that the F-BAR domain function might be inhibited in srGAP2W765A. By analogy to the molecular mechanisms controlling the activation of other RhoGAP and RhoGEF proteins (Eberth et al., 2009; Mitin et al., 2007; Yohe et al., 2007), we hypothesized that srGAP2 might normally be in an auto-inhibited conformation through structural interaction between the N-terminal F-BAR domain and of the C-terminal region (including the SH3 domain) that is released upon binding of protein interactors to its SH3 domain (see model in Fig. 7G).
To test this model, we performed a C-terminal deletion of srGAP2 (srGAP2ΔC-term), which deletes the entire C-terminal portion starting from the SH3 domain to the C-terminal end. Expression of srGAP2ΔC-term potently induced filopodia formation in Stage 1 neurons (Fig. S12E and S12F) and neurite outgrowth and branching in Stage 2 neurons (Fig. S12K–L). In sharp contrast to srGAP2W765A, expression of srGAP2ΔC-term potently inhibited migration (Fig. S14Q–T and S14U) resulting in increased multipolar cells in the IZ (Fig. S14V) as well as increased LP branching of migrating neurons in layer 5/6 similarly to other F-BAR containing constructs but unlike srGAP2W765A (Fig. S14X and Fig. 7F).
DISCUSSION
In this study, we provide loss-of-function, gain-of-function as well as structure-function analysis demonstrating that srGAP2 is a novel negative regulator of neuronal migration and a positive regulator of neurite initiation and branching through the ability of its F-BAR domain to induce filopodia-like membrane protrusions. Our findings demonstrate for the first time the function of a F-BAR domain-containing protein in cell migration and morphogenesis in vivo and more generally highlight the functional importance of proteins directly regulating membrane deformation.
srGAP2 is a novel F-BAR domain-containing protein
It is well-established that cytoskeletal dynamics produce forces to generate plasma membrane protrusions and invaginations, however recent evidence suggest that many membrane-associated proteins directly sculpt and deform biological membranes (Doherty and McMahon, 2008). Here we report that srGAP2 regulates neuronal migration as well as neurite initiation and branching through the ability of its F-BAR domain to deform membranes and form filopodia-like membrane protrusions. This is a surprising finding since F-BAR domains have been mostly characterized for their ability to induce membrane tabulation and invaginations (Frost et al., 2008; Habermann, 2004; Henne et al., 2007; Itoh and De Camilli, 2006; Peter et al., 2004; Shimada et al., 2007). F-BAR domains are composed of a series of alpha-helices forming a strong dimerization motif, which allow the homodimers to adopt a quaternary ‘banana-like’ structure (Frost et al., 2008; Henne et al., 2007; Peter et al., 2004; Shimada et al., 2007). One possibility for how srGAP2’s F-BAR domain may induce filopodia-like protrusions is by having a different curvature leading to a different surface distribution of positively charged residues than ‘canonical’ F-BAR domains. Interestingly, I-BAR domains present in proteins such as IRSp53 or MIM induce filopodia, a property linked to the inherent curvature of the I-BAR homodimer and the presence of phospholipid-binding residues on the convex side of the homodimers (Lim et al., 2008; Mattila et al., 2007; Millard et al., 2007; Saarikangas et al., 2009).
We hypothesize that the homodimer formed by the F-BAR domain of srGAP2 displays a general quaternary structure and charge distribution comparable to I-BAR domains. While this can only be proven by structural information, we provide several lines of evidence supporting an I-BAR like behavior: (1) the structural maintenance of filopodia induced by the F-BAR domain of srGAP2 is resistant to F-actin depolymerization, (2) over-expression of the F-BAR domain of srGAP2 does not inhibit endocytosis, (3) the F-BAR domain of srGAP2 induces similar liposome deformations compared to IRSp53 (Suetsugu et al., 2006).
Interestingly, srGAP2 is not the only predicted F-BAR domain containing protein inducing filopodia formation: Gas7 and PSTPIP2 (MAYP) have also been shown to induce filopodia in cell lines (Chitu et al., 2005; She et al., 2002). However, these proteins and more importantly their predicted F-BAR domains have not been directly tested for their ability to deform membranes. Our results suggest that the F-BAR domain subfamily could be functionally diverse and that this diversity might be due to subtle structural differences.
A role for srGAP2 during neuronal development
It is well established that filopodia promote neurite outgrowth and it was recently shown that filopodia were required for neurite initiation in cortical neurons (Dent et al., 2007). The absence of effect of srGAP2 knockdown on neurite initiation is likely due to the presence of many other proteins involved in filopodia formation such as I-BAR-containing proteins such as IRSp53 or ABBA (Mattila and Lappalainen, 2008; Saarikangas et al., 2008) or other classes of proteins previously shown to promote filopodia formation and neurite initiation through distinct mechanisms such as the ENA/VASP proteins (Dent et al., 2007; Kwiatkowski et al., 2007).
The ability of srGAP2 to promote neurite initiation and branching appears to also be important for its regulation of migration (Fig. 7H). Knockdown of srGAP2 increased the rate of migration and significantly reduced leading process complexity and branching (Fig. 7H). This could potentially explain the increase in the rate of cell migration since in fibroblast reduction of ENA/VASP proteins activity (i.e. proteins that promote filopodia formation) increased lamellipodia persistence and increased cell speed (Bear et al., 2000; Bear et al., 2002). In addition, it was recently shown that loss of ENA/VASP proteins in cortical neurons lead to more superficial laminar position, which could be the result of increased migration (Goh et al., 2002; Kwiatkowski et al., 2007). Therefore decreased branching in srGAP2 knockdown neurons may increase LP persistence and increase translocation efficiency. Finally, recent siRNA screens in cancer cell lines revealed that downregulation of the srGAP2 homolog, srGAP3 also increased the rate of cell migration suggesting that negative regulation of cell migration may be a conserved function of the srGAP family (Simpson et al., 2008).
Regulation of srGAP2: GAP and SH3 domains
The BAR superfamily of proteins are involved in a wide range of functions and this diversity arises from the different functional domains associated with BAR-like domain (Itoh and De Camilli, 2006). We demonstrate that srGAP2 is a Rac1-specific GAP and recent work has highlighted the importance of Rac1 regulation in neuronal development (Govek et al., 2005). Mutation of the Rac1/Cdc42 GEF, ARHGEF6 (also called Cool-2, α̃PIX) results in X-linked mental retardation suggesting the importance of properly regulating Rac1 activity during neuronal development (Kutsche et al., 2000). Interestingly, the BAR domain containing protein Oligophrenin-1 as well as the F-BAR containing protein srGAP3 (also called MEntal retardation GAP or MEGAP), are both Rac1-GAPs that have been involved in severe forms of mental retardation (Billuart et al., 1998; Endris et al., 2002; Govek et al., 2004).
Rac1 has also been implicated in regulating radial migration and neurite outgrowth (Causeret et al., 2008; Govek et al., 2005; Kawauchi et al., 2003; Konno et al., 2005; Yoshizawa et al., 2005). Although not required, the GAP activity of srGAP2 might play a role in neurite formation in two ways: (1) local inactivation of Rac1 could result in increased Cdc42 activity which could in turn activate pathways that promote bundled F-actin which are required for filopodia formation (Raftopoulou and Hall, 2004) and/or (2) alternatively, Rac1 inactivation could lead to increased activation of RhoA (since Rac1 activation has been shown to inactivate RhoA (Nimnual et al., 2003), which in turn could lead to the activation of the formin, mDia2 and increased actin nucleation (Fig. 7G).
A high percentage of F-BAR domain-containing proteins possess SH3 domains (Itoh and De Camilli, 2006) which bind to a myriad of effectors ranging from regulators of endocytosis such as dynamin (Itoh and De Camilli, 2006), to regulators of the actin cytoskeleton (Aspenstrom et al., 2006; Chitu et al., 2005) such as WAVE1 (Soderling et al., 2002). The SH3 domain of srGAP2 has been shown to bind the Robo1 receptor (Wong et al., 2001) and has also been shown to bind N-WASP (Linkermann et al., 2009), but the functional relevance of these interactions has yet to be determined. Our results strongly suggest that srGAP2 is auto-inhibited at resting state which is a commonly accepted model of regulation of many RhoGEF and RhoGAP proteins (Rossman et al., 2005), the BAR domain-containing proteins GRAF and Oligophrenin-1 (Eberth et al., 2009). Future experiments will test if this auto-inhibition can be released by effector-binding to the SH3 domain exposing the F-BAR domain to facilitate membrane protrusion (Fig. 7G).
MATERIAL AND METHDOS
Animals
Mice were used according to a protocol approved by the Institutional Animal Care and Use Committee (IA-CUC) at the University of North Carolina-Chapel Hill and in accordance with NIH guidelines. Time-pregnant females were maintained in a 12 hr light/dark cycle and obtained by overnight breeding with males of the same strain. Noon following breeding is considered as E0.5.
Protein purification
srGAP2 (amino acids 1–785) and the F-BAR (amino acids 22–501) were cloned into pLIC vectors and expressed in Escherichia coli BL21 (DE3) cells. Proteins were then purified on a Ni2+ affinity column. Proteins were further purified by cation exchange chromatography, using a Source S column, and concentrated in 20 mM Tris buffer, pH 8, 150 mM NaCl, 1 mM DTT and 5% glycerol. GAP (amino acids 502–676) and GAPR527L domain of srGAP2 was cloned into pGex-4T3 (Amersham). Recombinant GST-fusion proteins were then purified using glutathione sepharose and resuspended in 20 mM Tris buffer, pH 8, 150 mM NaCl, 1 mM DTT and 5% glycerol.
In vitro GAP assay
In vitro fluorescent-based GAP assay was performed as described previously (Shutes and Der, 2006).
Liposome Preparation
Folch Fraction I Brain Lipid Extract from bovine brain (B1502) in chloroform was obtained from Sigma-Aldrich (St. Louis, MO) and used without further purification. 10 mg of total lipid were added to a glass vial and dried at room temperature under streaming argon while vortexing in order to form a thin lipid film around the tube surface. The lipids were re-dissolved in absolute hexane, dried under argon again while vortexing, and then dessicated in vacuo for >2 hours to remove the last traces of chloroform. The dried lipid film was then pre-hydrated at RT with water-saturated N2 for 2 minutes until the film became transparent. Buffer (50mM KCl/10mM/HEPES/1mM DTT, pH 7.4) was added to the hydrated lipid film to a final lipid concentration of 2 mg/ml. The vial was sealed under argon and incubated at RT for 2 h, and then gently rocked overnight to disperse the lipids into solution.
Liposome tubulation assays
The liposomes described above were first subjected to 10 cycles of freeze-thaw, and then used immediately or stored in aliquots at −80° C. The liposomes were then equilibrated at RT for 1 hour before adding protein (either FBP17 F-BAR domain or srGAP2 F-BAR) at a lipid-to-protein ratio of 2:1 mass/mass and final concentrations of 0.2 mg/ml (lipid) and 0.1 mg/ml (protein). The tubulation reaction incubated for 30 minutes at RT before negative staining, as described below. In order to introduce the recombinant purified F-BAR into the liposomes, 250 μL of the tubulation reaction subjected to 5 seconds of bath sonication at RT, immediately after adding protein. Following sonication, the sample allowed to incubate for another 30 minutes before negative staining, as described below.
Electron Microscopy
Continuous carbon-coated Cu-grids were glow discharged in room air according to standard protocols. 4 μL of sample were added and allowed to sit for ~10 seconds before being blotted onto filter paper. The grid surface was then immediately stained with freshly prepared (<15 minutes) 0.8% uranyl formate. Images were acquired using a Philips Tecnai F12 microscope operating at 120 kV using nominal magnifications of 29–50,000x, and defocus values of −15,000 to −22,000 Å. Images were recorded on a Gatan 1K CCD. Image analysis, including tubule diameter measurements, were performed with NIH ImageJ.
Ex Vivo Cortical Electroporation and Primary Cortical Neuron Cultures
Mouse cortical progenitors were electroporated ex vivo at embryonic day (E) E15 as described previously (Hand et al., 2005). Following electroporation, cerebral hemispheres were either (1) dissected, enzymatically dissociated with papain and plated on poly-L-lysine and Laminin-coated glass coverslips as described previously ((Polleux and Ghosh, 2002)); (2) or sliced using a LEICA VT1000S vibratome and cultured organotypically as described previously ((Hand et al., 2005); see Supplementary Material for details).
Time Lapse confocal microscopy of cortical sections
Using a Leica TCS-SL confocal microscope (mounted on a DM-IRE2 inverted microscope stand) and equipped with a X-Y motorized Märzhäuser stage, time-lapse confocal microscopy was performed by imaging multiple Z-stacks at pre-selected positions on a given set of electroporated slices as described previously (Hand et al, 2005). Slices were cultured on confocal inserts (Millipore, 5mm height) and imaged using a long distance 20x/0.4 NA objective. For shRNA expressing sections, pictures were taken at a frequency of 1 picture every 12 minutes for 4hrs. In the case of srGAP2 over-expression experiments, sections were imaged every 16 minutes for a maximum of 10hrs 24 minutes.
Sequence alignments, shRNA and cDNA constructs and neuronal cultures and confocal microscopy are detailed in the supplementary methods.
Supplementary Material
Acknowledgments
We thank the members of the Polleux and Jim Bear labs for helpful comments. We thank Carol Schuurmans and Pierre Mattar for in situ hybridization and the original srGAP2 cDNA. We thank Gong Ju and Wei-Lin Jin for the srGAP2 antibody. We thank the Sondek (Rafael Rojas) and Der (Adam Shutes) labs for technical assistance with protein purification and fluorescent GTP hydrolysis assay. We thank Brenda Temple for technical assistance with protein modeling and sequence analysis. We thank Vinzenz Unger, Carsten Mim and Pietro De Camilli for helpful discussions. This work was supported by a Pre-doctoral Research Training Fellowship from the Epilepsy Foundation (AF) and a NIH MSTP award TG 5T32GM07205 (AF), a NIH Predoctoral Fellowship 5F31NS052969-03 (SG), UNC Developmental Biology Training Grant (T32 HD046369) (JCB) and a Pew Scholar Award in Biomedical Sciences (FP) and the NINDS Institutional Center Core Grant to Support Neuroscience Research (P30 NS45892-01).
References
- Aspenstrom P, Fransson A, Richnau N. Pombe Cdc15 homology proteins: regulators of membrane dynamics and the actin cytoskeleton. Trends Biochem Sci. 2006;31:670–679. doi: 10.1016/j.tibs.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Ayala R, Shu T, Tsai LH. Trekking across the brain: the journey of neuronal migration. Cell. 2007;128:29–43. doi: 10.1016/j.cell.2006.12.021. [DOI] [PubMed] [Google Scholar]
- Bacon C, Endris V, Rappold G. Dynamic expression of the Slit-Robo GTPase activating protein genes during development of the murine nervous system. J Comp Neurol. 2009;513:224–236. doi: 10.1002/cne.21955. [DOI] [PubMed] [Google Scholar]
- Bear JE, Loureiro JJ, Libova I, Fassler R, Wehland J, Gertler FB. Negative regulation of fibroblast motility by Ena/VASP proteins. Cell. 2000;101:717–728. doi: 10.1016/s0092-8674(00)80884-3. [DOI] [PubMed] [Google Scholar]
- Bear JE, Svitkina TM, Krause M, Schafer DA, Loureiro JJ, Strasser GA, Maly IV, Chaga OY, Cooper JA, Borisy GG, et al. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell. 2002;109:509–521. doi: 10.1016/s0092-8674(02)00731-6. [DOI] [PubMed] [Google Scholar]
- Billuart P, Bienvenu T, Ronce N, des Portes V, Vinet MC, Zemni R, Roest Crollius H, Carrie A, Fauchereau F, Cherry M, et al. Oligophrenin-1 encodes a rhoGAP protein involved in X-linked mental retardation. Nature. 1998;392:923–926. doi: 10.1038/31940. [DOI] [PubMed] [Google Scholar]
- Burnette DT, Schaefer AW, Ji L, Danuser G, Forscher P. Filopodial actin bundles are not necessary for microtubule advance into the peripheral domain of Aplysia neuronal growth cones. Nat Cell Biol. 2007;9:1360–1369. doi: 10.1038/ncb1655. [DOI] [PubMed] [Google Scholar]
- Causeret F, Terao M, Jacobs T, Nishimura YV, Yanagawa Y, Obata K, Hoshino M, Nikolic M. The p21-Activated Kinase Is Required for Neuronal Migration in the Cerebral Cortex. Cereb Cortex. 2008 doi: 10.1093/cercor/bhn133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitu V, Pixley FJ, Macaluso F, Larson DR, Condeelis J, Yeung YG, Stanley ER. The PCH family member MAYP/PSTPIP2 directly regulates F-actin bundling and enhances filopodia formation and motility in macrophages. Mol Biol Cell. 2005;16:2947–2959. doi: 10.1091/mbc.E04-10-0914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent EW, Barnes AM, Tang F, Kalil K. Netrin-1 and semaphorin 3A promote or inhibit cortical axon branching, respectively, by reorganization of the cytoskeleton. J Neurosci. 2004;24:3002–3012. doi: 10.1523/JNEUROSCI.4963-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent EW, Kwiatkowski AV, Mebane LM, Philippar U, Barzik M, Rubinson DA, Gupton S, Van Veen JE, Furman C, Zhang J, et al. Filopodia are required for cortical neurite initiation. Nat Cell Biol. 2007;9:1347–1359. doi: 10.1038/ncb1654. [DOI] [PubMed] [Google Scholar]
- Doherty GJ, McMahon HT. Mediation, modulation, and consequences of membrane-cytoskeleton interactions. Annu Rev Biophys. 2008;37:65–95. doi: 10.1146/annurev.biophys.37.032807.125912. [DOI] [PubMed] [Google Scholar]
- Dotti CG, Sullivan CA, Banker GA. The establishment of polarity by hippocampal neurons in culture. J Neurosci. 1988;8:1454–1468. doi: 10.1523/JNEUROSCI.08-04-01454.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberth A, Lundmark R, Gremer L, Dvorsky R, Koessmeier KT, McMahon HT, Ahmadian MR. A BAR domain-mediated autoinhibitory mechanism for RhoGAPs of the GRAF family. Biochem J. 2009;417:371–377. doi: 10.1042/BJ20081535. [DOI] [PubMed] [Google Scholar]
- Endris V, Wogatzky B, Leimer U, Bartsch D, Zatyka M, Latif F, Maher ER, Tariverdian G, Kirsch S, Karch D, et al. The novel Rho-GTPase activating gene MEGAP/srGAP3 has a putative role in severe mental retardation. Proc Natl Acad Sci U S A. 2002;99:11754–11759. doi: 10.1073/pnas.162241099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost A, Perera R, Roux A, Spasov K, Destaing O, Egelman EH, De Camilli P, Unger VM. Structural basis of membrane invagination by F-BAR domains. Cell. 2008;132:807–817. doi: 10.1016/j.cell.2007.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost A, Unger VM, De Camilli P. The BAR domain superfamily: membrane-molding macromolecules. Cell. 2009;137:191–196. doi: 10.1016/j.cell.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo G, Letourneau PC. Localized sources of neurotrophins initiate axon collateral sprouting. J Neurosci. 1998;18:5403–5414. doi: 10.1523/JNEUROSCI.18-14-05403.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo G, Letourneau PC. Regulation of growth cone actin filaments by guidance cues. J Neurobiol. 2004;58:92–102. doi: 10.1002/neu.10282. [DOI] [PubMed] [Google Scholar]
- Goh KL, Cai L, Cepko CL, Gertler FB. Ena/VASP proteins regulate cortical neuronal positioning. Curr Biol. 2002;12:565–569. doi: 10.1016/s0960-9822(02)00725-x. [DOI] [PubMed] [Google Scholar]
- Govek EE, Newey SE, Akerman CJ, Cross JR, Van der Veken L, Van Aelst L. The X-linked mental retardation protein oligophrenin-1 is required for dendritic spine morphogenesis. Nat Neurosci. 2004;7:364–372. doi: 10.1038/nn1210. [DOI] [PubMed] [Google Scholar]
- Govek EE, Newey SE, Van Aelst L. The role of the Rho GTPases in neuronal development. Genes Dev. 2005;19:1–49. doi: 10.1101/gad.1256405. [DOI] [PubMed] [Google Scholar]
- Gupta A, Sanada K, Miyamoto DT, Rovelstad S, Nadarajah B, Pearlman AL, Brunstrom J, Tsai LH. Layering defect in p35 deficiency is linked to improper neuronal-glial interaction in radial migration. Nat Neurosci. 2003;6:1284–1291. doi: 10.1038/nn1151. [DOI] [PubMed] [Google Scholar]
- Gupton SL, Gertler FB. Filopodia: the fingers that do the walking. Sci STKE 2007. 2007:re5. doi: 10.1126/stke.4002007re5. [DOI] [PubMed] [Google Scholar]
- Habermann B. The BAR-domain family of proteins: a case of bending and binding? EMBO Rep. 2004;5:250–255. doi: 10.1038/sj.embor.7400105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand R, Bortone D, Mattar P, Nguyen L, Heng JI, Guerrier S, Boutt E, Peters E, Barnes AP, Parras C, et al. Phosphorylation of Neurogenin2 specifies the migration properties and the dendritic morphology of pyramidal neurons in the neocortex. Neuron. 2005;48:45–62. doi: 10.1016/j.neuron.2005.08.032. [DOI] [PubMed] [Google Scholar]
- Heng JI, Nguyen L, Castro DS, Zimmer C, Wildner H, Armant O, Skowronska-Krawczyk D, Bedogni F, Matter JM, Hevner R, et al. Neurogenin 2 controls cortical neuron migration through regulation of Rnd2. Nature. 2008 doi: 10.1038/nature07198. [DOI] [PubMed] [Google Scholar]
- Henne WM, Kent HM, Ford MG, Hegde BG, Daumke O, Butler PJ, Mittal R, Langen R, Evans PR, McMahon HT. Structure and analysis of FCHo2 F-BAR domain: a dimerizing and membrane recruitment module that effects membrane curvature. Structure. 2007;15:839–852. doi: 10.1016/j.str.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Higginbotham HR, Gleeson JG. The centrosome in neuronal development. Trends Neurosci. 2007;30:276–283. doi: 10.1016/j.tins.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Itoh T, De Camilli P. BAR, F-BAR (EFC) and ENTH/ANTH domains in the regulation of membrane-cytosol interfaces and membrane curvature. Biochim Biophys Acta. 2006;1761:897–912. doi: 10.1016/j.bbalip.2006.06.015. [DOI] [PubMed] [Google Scholar]
- Itoh T, Erdmann KS, Roux A, Habermann B, Werner H, De Camilli P. Dynamin and the actin cytoskeleton cooperatively regulate plasma membrane invagination by BAR and F-BAR proteins. Dev Cell. 2005;9:791–804. doi: 10.1016/j.devcel.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Kakimoto T, Katoh H, Negishi M. Regulation of neuronal morphology by Toca-1, an F-BAR/EFC protein that induces plasma membrane invagination. J Biol Chem. 2006;281:29042–29053. doi: 10.1074/jbc.M604025200. [DOI] [PubMed] [Google Scholar]
- Kawauchi T, Chihama K, Nabeshima Y, Hoshino M. The in vivo roles of STEF/Tiam1, Rac1 and JNK in cortical neuronal migration. EMBO J. 2003;22:4190–4201. doi: 10.1093/emboj/cdg413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno D, Yoshimura S, Hori K, Maruoka H, Sobue K. Involvement of the phosphatidylinositol 3-kinase/rac1 and cdc42 pathways in radial migration of cortical neurons. J Biol Chem. 2005;280:5082–5088. doi: 10.1074/jbc.M408251200. [DOI] [PubMed] [Google Scholar]
- Kutsche K, Yntema H, Brandt A, Jantke I, Nothwang HG, Orth U, Boavida MG, David D, Chelly J, Fryns JP, et al. Mutations in ARHGEF6, encoding a guanine nucleotide exchange factor for Rho GTPases, in patients with X-linked mental retardation. Nat Genet. 2000;26:247–250. doi: 10.1038/80002. [DOI] [PubMed] [Google Scholar]
- Kwiatkowski AV, Rubinson DA, Dent EW, Edward van Veen J, Leslie JD, Zhang J, Mebane LM, Philippar U, Pinheiro EM, Burds AA, et al. Ena/VASP Is Required for neuritogenesis in the developing cortex. Neuron. 2007;56:441–455. doi: 10.1016/j.neuron.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Lambert de Rouvroit C, Goffinet AM. Neuronal migration. Mech Dev. 2001;105:47–56. doi: 10.1016/s0925-4773(01)00396-3. [DOI] [PubMed] [Google Scholar]
- Li X, Chen Y, Liu Y, Gao J, Gao F, Bartlam M, Wu JY, Rao Z. Structural basis of Robo proline-rich motif recognition by the srGAP1 Src homology 3 domain in the Slit-Robo signaling pathway. J Biol Chem. 2006;281:28430–28437. doi: 10.1074/jbc.M604135200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KB, Bu W, Goh WI, Koh E, Ong SH, Pawson T, Sudhaharan T, Ahmed S. The Cdc42 effector IRSp53 generates filopodia by coupling membrane protrusion with actin dynamics. J Biol Chem. 2008;283:20454–20472. doi: 10.1074/jbc.M710185200. [DOI] [PubMed] [Google Scholar]
- Linkermann A, Gelhaus C, Lettau M, Qian J, Kabelitz D, Janssen O. Identification of interaction partners for individual SH3 domains of Fas ligand associated members of the PCH protein family in T lymphocytes. Biochim Biophys Acta. 2009;1794:168–176. doi: 10.1016/j.bbapap.2008.10.013. [DOI] [PubMed] [Google Scholar]
- Luo L. Actin cytoskeleton regulation in neuronal morphogenesis and structural plasticity. Annu Rev Cell Dev Biol. 2002;18:601–635. doi: 10.1146/annurev.cellbio.18.031802.150501. [DOI] [PubMed] [Google Scholar]
- Mattar P, Britz O, Johannes C, Nieto M, Ma L, Rebeyka A, Klenin N, Polleux F, Guillemot F, Schuurmans C. A screen for downstream effectors of Neurogenin2 in the embryonic neocortex. Dev Biol. 2004;273:373–389. doi: 10.1016/j.ydbio.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Mattila PK, Lappalainen P. Filopodia: molecular architecture and cellular functions. Nat Rev Mol Cell Biol. 2008;9:446–454. doi: 10.1038/nrm2406. [DOI] [PubMed] [Google Scholar]
- Mattila PK, Pykalainen A, Saarikangas J, Paavilainen VO, Vihinen H, Jokitalo E, Lappalainen P. Missing-in-metastasis and IRSp53 deform PI(4,5)P2-rich membranes by an inverse BAR domain-like mechanism. J Cell Biol. 2007;176:953–964. doi: 10.1083/jcb.200609176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard TH, Dawson J, Machesky LM. Characterisation of IRTKS, a novel IRSp53/MIM family actin regulator with distinct filament bundling properties. J Cell Sci. 2007;120:1663–1672. doi: 10.1242/jcs.001776. [DOI] [PubMed] [Google Scholar]
- Mitin N, Betts L, Yohe ME, Der CJ, Sondek J, Rossman KL. Release of autoinhibition of ASEF by APC leads to CDC42 activation and tumor suppression. Nat Struct Mol Biol. 2007;14:814–823. doi: 10.1038/nsmb1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimnual AS, Taylor LJ, Bar-Sagi D. Redox-dependent downregulation of Rho by Rac. Nat Cell Biol. 2003;5:236–241. doi: 10.1038/ncb938. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Martinez-Cerdeno V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- Ohshima T, Hirasawa M, Tabata H, Mutoh T, Adachi T, Suzuki H, Saruta K, Iwasato T, Itohara S, Hashimoto M, et al. Cdk5 is required for multipolar-to-bipolar transition during radial neuronal migration and proper dendrite development of pyramidal neurons in the cerebral cortex. Development. 2007;134:2273–2282. doi: 10.1242/dev.02854. [DOI] [PubMed] [Google Scholar]
- Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- Polleux F, Ghosh A. The slice overlay assay: a versatile tool to study the influence of extracellular signals on neuronal development. Sci STKE 2002. 2002:PL9. doi: 10.1126/stke.2002.136.pl9. [DOI] [PubMed] [Google Scholar]
- Raftopoulou M, Hall A. Cell migration: Rho GTPases lead the way. Dev Biol. 2004;265:23–32. doi: 10.1016/j.ydbio.2003.06.003. [DOI] [PubMed] [Google Scholar]
- Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6:167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- Saarikangas J, Hakanen J, Mattila PK, Grumet M, Salminen M, Lappalainen P. ABBA regulates plasma-membrane and actin dynamics to promote radial glia extension. J Cell Sci. 2008;121:1444–1454. doi: 10.1242/jcs.027466. [DOI] [PubMed] [Google Scholar]
- Saarikangas J, Zhao H, Pykalainen A, Laurinmaki P, Mattila PK, Kinnunen PK, Butcher SJ, Lappalainen P. Molecular mechanisms of membrane deformation by I-BAR domain proteins. Curr Biol. 2009;19:95–107. doi: 10.1016/j.cub.2008.12.029. [DOI] [PubMed] [Google Scholar]
- Scita G, Confalonieri S, Lappalainen P, Suetsugu S. IRSp53: crossing the road of membrane and actin dynamics in the formation of membrane protrusions. Trends Cell Biol. 2008;18:52–60. doi: 10.1016/j.tcb.2007.12.002. [DOI] [PubMed] [Google Scholar]
- She BR, Liou GG, Lin-Chao S. Association of the growth-arrest-specific protein Gas7 with F-actin induces reorganization of microfilaments and promotes membrane outgrowth. Exp Cell Res. 2002;273:34–44. doi: 10.1006/excr.2001.5435. [DOI] [PubMed] [Google Scholar]
- Shimada A, Niwa H, Tsujita K, Suetsugu S, Nitta K, Hanawa-Suetsugu K, Akasaka R, Nishino Y, Toyama M, Chen L, et al. Curved EFC/F-BAR-domain dimers are joined end to end into a filament for membrane invagination in endocytosis. Cell. 2007;129:761–772. doi: 10.1016/j.cell.2007.03.040. [DOI] [PubMed] [Google Scholar]
- Shutes A, Der CJ. Real-time in vitro measurement of intrinsic and Ras GAP-mediated GTP hydrolysis. Methods Enzymol. 2006;407:9–22. doi: 10.1016/S0076-6879(05)07002-3. [DOI] [PubMed] [Google Scholar]
- Simpson KJ, Selfors LM, Bui J, Reynolds A, Leake D, Khvorova A, Brugge JS. Identification of genes that regulate epithelial cell migration using an siRNA screening approach. Nat Cell Biol. 2008 doi: 10.1038/ncb1762. [DOI] [PubMed] [Google Scholar]
- Soderling SH, Binns KL, Wayman GA, Davee SM, Ong SH, Pawson T, Scott JD. The WRP component of the WAVE-1 complex attenuates Rac-mediated signalling. Nat Cell Biol. 2002;4:970–975. doi: 10.1038/ncb886. [DOI] [PubMed] [Google Scholar]
- Suetsugu S, Murayama K, Sakamoto A, Hanawa-Suetsugu K, Seto A, Oikawa T, Mishima C, Shirouzu M, Takenawa T, Yokoyama S. The RAC binding domain/IRSp53-MIM homology domain of IRSp53 induces RAC-dependent membrane deformation. J Biol Chem. 2006;281:35347–35358. doi: 10.1074/jbc.M606814200. [DOI] [PubMed] [Google Scholar]
- Tsujita K, Suetsugu S, Sasaki N, Furutani M, Oikawa T, Takenawa T. Coordination between the actin cytoskeleton and membrane deformation by a novel membrane tubulation domain of PCH proteins is involved in endocytosis. J Cell Biol. 2006;172:269–279. doi: 10.1083/jcb.200508091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K, Ren XR, Huang YZ, Xie Y, Liu G, Saito H, Tang H, Wen L, Brady-Kalnay SM, Mei L, et al. Signal transduction in neuronal migration: roles of GTPase activating proteins and the small GTPase Cdc42 in the Slit-Robo pathway. Cell. 2001;107:209–221. doi: 10.1016/s0092-8674(01)00530-x. [DOI] [PubMed] [Google Scholar]
- Yao Q, Jin WL, Wang Y, Ju G. Regulated shuttling of Slit-Robo-GTPase activating proteins between nucleus and cytoplasm during brain development. Cell Mol Neurobiol. 2008;28:205–221. doi: 10.1007/s10571-007-9187-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yohe ME, Rossman KL, Gardner OS, Karnoub AE, Snyder JT, Gershburg S, Graves LM, Der CJ, Sondek J. Auto-inhibition of the Dbl family protein Tim by an N-terminal helical motif. J Biol Chem. 2007;282:13813–13823. doi: 10.1074/jbc.M700185200. [DOI] [PubMed] [Google Scholar]
- Yoshizawa M, Kawauchi T, Sone M, Nishimura YV, Terao M, Chihama K, Nabeshima Y, Hoshino M. Involvement of a Rac activator, P-Rex1, in neurotrophin-derived signaling and neuronal migration. J Neurosci. 2005;25:4406–4419. doi: 10.1523/JNEUROSCI.4955-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.