Abstract
Current methods for annotating biomedical data resources rely on simple mappings between data elements and the contents of a variety of biomedical ontologies and controlled vocabularies. Here we point out that such simple mappings are inadequate for large-scale multiscale, multidomain integrative “virtual human” projects. For such integrative challenges, we describe a “composite annotation” schema that is simple yet sufficiently extensible for mapping the biomedical content of a variety of data sources and biosimulation models to available biomedical ontologies.
I. Introduction
Akey strategy for integrating biomedical knowledge in service to solving biomedical research and clinical problems is the annotation of various knowledge resources — images, databases, electronic medical records (EMR), biosimulation models — against controlled vocabularies and reference ontologies. The usefulness and success of such a strategy depends on how well the annotations capture and disambiguate the biomedical meaning (the semantics) of the elements of the knowledge resources which the users can apply for searching, integrating and reusing data and models. Such an approach is central to integrative biology and, in particular, to efforts such as the Physiome [1] and the Virtual Physiological Human (see http://www.vph-noe.eu/home).
Such efforts at integrative biology depend on a number of different data and knowledge resources whose contents must be available for searching, reuse and computation. For example, images which are ubiquitous in biomedical enterprise include clinical scans (e.g., MRIs and CTs), gene expression maps, and electrophoresis gels. Quantitative data are derived from such images (e.g., volume of scanned regions, rates of gene expression) as well as from experimental and clinical measures (e.g., blood pressure, body weight). These data may be embedded within an original data source (image, EMR, etc.) or compiled in databases. Integrative biologists, particularly those who construct biosimulation models, depend on such data to parameterize and test their models against the biological “reality” they seek to represent, analyze, and explain.
In this paper we develop a representational schema for the systematic annotation of data from many diverse sources such as images, clinical and basic science databases and biosimulation models. With some use-case examples, we will show how simple annotations (e.g., a pointer to a single ontology class) are insufficient for annotating the variety data sources (and medical terminologies) that must be reused and integrated within the scope of current virtual human projects.
We propose a composite annotation schema designed to span and integrate multiple structural scales and apply to multiple biophysical domains. We demonstrate how composite annotations, founded on sound principles of biomedical ontology, apply to a wide range of biomedical clinical and research tasks and, we propose, may provide a central strategy for interrelating the various computational parts of multiscale virtual human projects.
II. Background — biomedical ontologies
Because the development of biomedical ontologies has occurred in parallel to, but largely independently of, the maturation of biosimulation technologies, a brief review of relevant ontologies is given below.
A. Biomedical ontologies
Biomedical ontology is a burgeoning field in which experts encode knowledge in terms of ontological classes for biological entities in various domains (e.g., anatomy, diseases, physics). Open Biomedical Ontologies (OBO; see http://www.obofoundry.org/) advocates that ontologies should be orthogonal with respect to each other in that one ontology ought not to include classes encompassed in other ontologies. Ontologies do, however, overlap having been independently developed for different sub-domains for different purposes based on different assumptions. This is illustrated in Fig. 1 where several structural ontologies span a broad range of structural granularity in which the CL cell types overlap with the FMA, and GO molecules with ChEBI.
Fig 1.
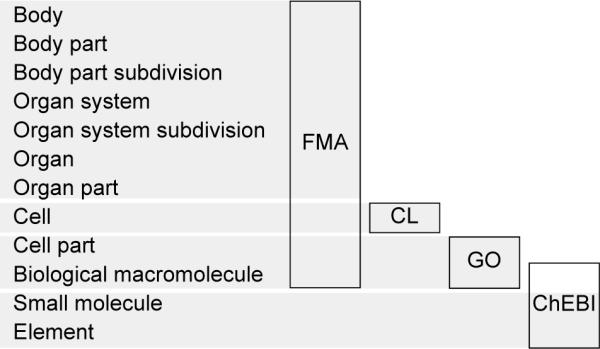
A spectrum of multiscale biomedical structures (arrayed vertically) is encoded by a set of overlapping biomedical structural ontologies (outlined boxes). To assure orthogonality across candidate ontologies, the spectrum may be partitioned, for example, by gray horizontal bars. FMA = Foundational Model of Anatomy Ontology [2]. CL = Cell Type Ontology [3]. GO = Gene Ontology [4]. ChEBI = Chemical Entities of Biological Interest [5].
Whereas the structural ontologies encode and classify the “stuff” of biological systems, databases and biosimulation variables encode the values of measured (or calculated) physical properties (e.g., volume, flow rate) of such entities. Thus model variables must be mapped to their inherent physical properties as well as to the physical entities that are bearers of the properties. Toward this, we have introduced the Ontology of Physics for Biology (OPB [6]) that is a formal ontology encoding both the properties and the laws of systems dynamics (Fig. 2). The OPB includes a comprehensive taxonomy of Physical property classes that represent both system dynamic variables (e.g., flows, displacements) as well as constitutive properties (e.g., flow resistances, vessel compliances) of biophysical systems. The OPB spans the multiple physical domains (Fluid domain, Chemical kinetic domain, etc.) that occur in biosimulation modeling.
Fig 2.
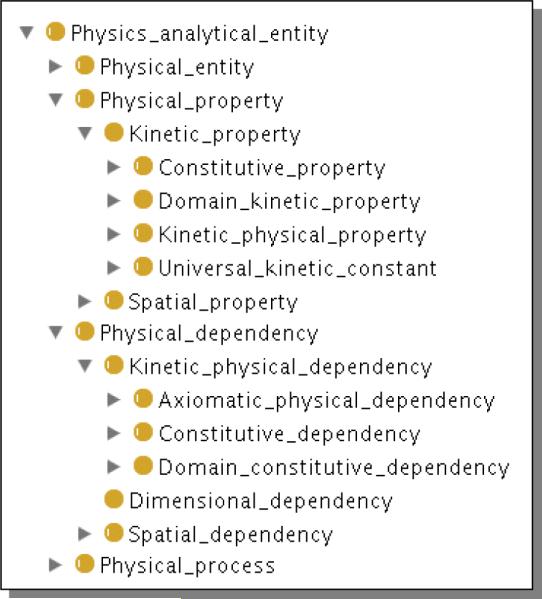
Protégé screenshot a part of the class hierarchy of the Ontology of Physics for Biology (OPB [6]), an ontology that encodes the physical properties and laws required for modeling dynamical biological systems.
B. Nomenclature and typography
Although the relative merits of various controlled vocabularies, terminologies and formal ontologies continue to be debated, in this paper we simplify our nomenclature by using the term “ontology” in its most general and inclusive sense to refer to both controlled vocabularies and to formal ontologies. Accordingly, we use “classes” to refer to both vocabulary terms and ontology classes (types or universals) and are in Courier font. Furthermore, we describe composite annotations in schematic terms that could be implemented in a variety of formalisms (e.g., OWL, obo.edit).
III. Annotating anatomical structures
Whereas a region of a scanned image may be annotated with a single FMA class (e.g., FMA:Blood in aorta), not all structural entities have been entered in the FMA (despite its ≈80K classes). For example, there is a class Blood in aorta yet there is no class Urine in ureter even though such a class would fall within the FMA's representational guidelines. Blood in aorta exists as a single class because the FMA curators pre-coordinated two classes FMA:Lumen of aorta and FMA:Portion of blood logically with the structural relation contains. Although the same class construct can be pre-coordinated for urine in the ureter or in the loop of Henle, it would be impractical and impossible to anticipate and include all such combinations as may be required by users. Furthermore, other bone fide structural classes cannot be pre-coordinated from a single ontology. For example, the oxygen content of erythrocytes in the blood in the aorta requires annotation classes from 3 ontologies: ChEBI, CL, and FMA. For example, a post-coordinated annotation for the oxygen in aortic blood could be:
| (1) |
To build an ontology that precoordinates classes for all molecular components in all cell parts of all cells in all organs…and so on, leads to an impossible combinatorial explosion. Yet annotating images, databases and biosimulation models requires such specific combinations. One solution to the combinatorial problem is to post-coordinate (or, post-compose) annotations as required for solving specific problems. A post-coordinated annotation references specific classes and logically relates them using relations as, say, defined in the OBO Relations Ontology (see http://www.obofoundry.org/ro/).
Furthermore, a biosimulation models often define structural participants using functional, not structural, criteria. For example a modeler may bifurcate the aortic blood into a turbulent boundary flow region and a laminar-flow central region. Because neither region is represented in the FMA, annotating such a biosimulation entity in a machine-readable fashion requires creating two new classes (Boundary region of aortic blood and Central region of aortic blood) and relating them to Blood in aorta via structural relation part of as in:
| (2) |
As we will show below, the composite annotation schema we have developed has the capacity for creating post-coordinated and composed annotations as exemplified in (1) and (2). Next we show how post-coordinated composite annotations are required for annotating physically-measured (or calculated) properties of physical entities.
IV. Annotating physical properties
In addition to annotating the kinds of biomedical entities, the properties of such entities must be identified and annotated as well. Clinical data embedded in EMRs, gene expression rates in genomic databases, and simulation variables are different kinds of properties whose physical meaning must be annotated in order to interrelate the biological content of the various data sources. To do so in a pre-coordinated fashion only exacerbates the combinatorial problem because each physical entity has several physical properties. For example, aortic blood can have four dynamic variables (volume, flow rate, pressure, and fluid momentum) as well as several so-called “constitutive” parameters (e.g., viscosity) that characterize the blood itself. The biophysical meaning of such variables in a biosimulation model may be suggested by mnemonic names (e.g., “Paorta”, “AoP”), or revealed to human readers by cryptic code comments (e.g., “// aortic pressure”). Consequently, it is increasingly being recognized that informal annotations are insufficient for model reuse and integration for large-scale efforts such as the IUPS Physiome and EU Virtual Physiological Human.
The solution to this annotation problem is post-coordination; in this case between an annotated physical entity (as described above) and a class that represents a measurable physical property. Thus, a variable or datum encoding aortic blood pressure can be post-coordinated as follows using the relation is_property_of:
| (3) |
Similarly, the flow rate of blood in the central region of blood in the aorta (as in (2)) would be:
| (4) |
In this fashion, the physical properties (or any kind of property, given a suitable ontology) can be attributed to any kind of physical entity as required.
V. Annotating physical laws
The values of physical properties (of physical entities) are not, of course, arbitrarily independent of each other but are constrained by the laws of physics; laws which apply at all levels of structural granularity. It is precisely these constraints that are encoded in biosimulation models to simulate the behavior of multiscale biological systems. Many such models, for example, simulate cardiovascular dynamics that relate systemic properties like the blood pressure in the aorta to neuroregulatory properties to contractile properties of arteriolar smooth muscle, and so on. Models can differ, of course, in the modelers choice of which physical properties will be calculated as well as the kinds of physical laws that are encoded in the model computations. One model may use the linear form of the fluid Ohm's Law whereas another model may employ one of a number of non-linear versions. Such distinctions are important to make but are, typically, only knowable by human-readable documentation.
To make these model-model distinctions available in machine-readable form requires annotating the kinds of physical dependencies that are encoded in a model. For example:
| (5) |
The representation of specific physical laws in terms of computational dependencies is a key step in modularizing, reusing and re-encoding biosimulation models [].
VI. composite annotation schema
We propose that the annotations as exemplified above may be implemented in a machine-readable fashion using the composite annotation schema (Fig. 3). A composite annotation is a nested representational structure that is a generalization of the SemSim (semantic simulation) model architecture that we have developed and tested in the domain of biosimulation modeling [7]. Each composite annotation model is a light-weight ontology (SemSim models are encoded in OWL) consisting the same three main classes as in the OPB (Fig. 2): Physical entity, Physical property, and Physical dependency. A composite annotation therefore consists of instances of these classes that are related by various structural relations as found in the OBO Relation Ontology (RO [8]). This ontology provides the formal relations needed to describe how the structural entities in a composite annotation relate to each other as in the prior examples. However, RO does not yet include relations appropriate for connecting non-structural ontologies used in OPB and the composite annotation schema. Thus, we currently use the has_property and has_player relations for such links.
Fig 3.
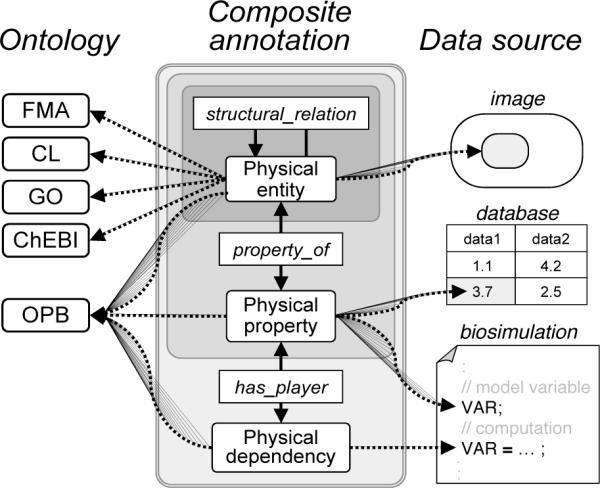
A nested schema for composite annotations for instances of biomedical data from a variety of data sources (images, databases, biosimulation models) to relevant domain entities in biomedical ontologies. Solid lines map logical relations between class instances; dotted lines are pointers to ontology classes (left) or to specific data sources (right).
The flexibility and, hence, the generalizability of composite annotations lies, in part, in the ability to create post-coordinated composites of Physical entity from existing ontology classes. Furthermore, the schema allows the fabrication of novel Physical entity instances composed according to the same structural relations as used in domain structural ontologies.
The nested structure in the center of Fig 3 shows that composite annotations can increase in complexity. Indeed, the presentation order of this paper follows exactly this structure: equations (1) & (2) show examples of physical entities and their structural relationships; equations (4) & (5) show the next level, where we use “property_of”, and annotations of computations require the most complex sort, encompassing all elements in the figure. This nested structure reflects the principle of ontological dependence: an instance of Physical dependency can exist only if instances Physical property exist (its players), and an instance of Physical property can exist only if an instance of Physical entity exists (the bearer of the property).
VII. Discussion
We demonstrate here a flexible and generalizable schema for the machine-readable annotation of images, datasets and biosimulation variables in terms of available biomedical ontologies. We are motivated by the computational needs of large-scale efforts at data and model integration that require flexible and efficient methodology and tools for deriving and accessing data and the means to reusing mathematical models. The composite annotation schema, adapted from the OPB [6], is a component of the SemSim (semantic simulation) modeling approach that we use to annotate, merge, and re-encode biosimulation models [9,7].
We offer composite annotation as a solution to the problem of integrating biomedical data and knowledge across large scale integrative projects such as Physiome and the Virtual Physiological Human. We argue that, given a set of orthogonal reference ontologies and a generalizable annotation schema as described here could significantly assist such work. A key first step for data and model integration is a solid, machine-encodable semantics of model variables and equations. We propose that a repository of composite annotations could allow researchers to find variables that share common semantics across biosimulation models and datasets.
Acknowledgment
This work was partially funded by NIH grants #R01 HL087706-01 and #T15 LM007442-06. We also thank Michal Galdzicki for contributions to these research ideas.
This work was supported in part by the National Institutes of Health under Grant R01HL087706-01.
References
- [1].Hunter PJ, Borg TK. Integration from proteins to organs: the Physiome Project. Nat Rev Mol Cell Biol. 2003;4:237–43. doi: 10.1038/nrm1054. [DOI] [PubMed] [Google Scholar]
- [2].Rosse C, Mejino JLV., Jr. The Foundational Model of Anatomy Ontology. In: Burger A, Davidson D, Baldock R, editors. Anatomy Ontologies for Bioinformatics: Principles and Practice. Springer; New York: 2007. [Google Scholar]
- [3].Bard J, Rhee SY, Ashburner M. An ontology for cell types. Genome Biology. 2005;6:R21.1–R21.5. doi: 10.1186/gb-2005-6-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–61. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Degtyarenko K, de Matos P, Ennis M, Hastings J, Zbinden M, McNaught A, Alcantara R, Darsow M, Guedj M, Ashburner M. ChEBI: a database and ontology for chemical entities of biological interest. Nucleic Acids Res. 2008;36:D344–50. doi: 10.1093/nar/gkm791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Cook DL, Mejino JL, Neal ML, Gennari JH. Bridging biological ontologies and biosimulation: the Ontology of Physics for Biology. AMIA Annu Symp Proc. 2008:136–40. [PMC free article] [PubMed] [Google Scholar]
- [7].Neal ML, Gennari JH, Arts T, Cook DL. Advances in semantic representation for multiscale biosimulation: a case study in merging models. Pac Symp Biocomput. 2009:304–15. [PMC free article] [PubMed] [Google Scholar]
- [8].Smith B, Ceusters W, Klagges B, Kohler J, Kumar A, Lomax J, Mungall C, Neuhaus F, Rector AL, Rosse C. Relations in biomedical ontologies. Genome Biol. 2005;6:R46. doi: 10.1186/gb-2005-6-5-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gennari JH, Neal ML, Carlson BE, Cook DL. Integration of multi-scale biosimulation models via light-weight semantics. Pac Symp Biocomput. 2008:414–25. [PMC free article] [PubMed] [Google Scholar]


