Abstract
Cytochrome P450 reductase (CPR) is a tethered membrane protein which transfers electrons from NADPH to microsomal P450s. We show that the lipid bilayer has a role in defining the redox potential of the CPR flavin domains. In order to quantitate the electrochemical behavior of this central redox protein, full length CPR was incorporated into soluble nanometer scale discoidal membrane bilayers (Nanodiscs) and potentials measured using spectro-potentiometry. The redox potentials of both FMN and FAD were found to shift to more positive values when in a membrane bilayer as compared to a solubilized version of the reductase. The potentials of the semiquinone/hydroquinone couple of both FMN and FAD are altered to a larger extent than the oxidized/semiquinone couple which is understood by a simple electrostatic model. When anionic lipids were used to change the membrane composition of the CPR-Nanodisc, the redox potential of both flavins became more negative, favoring electron transfer from CPR to cytochrome P450.
Cytochrome P450s (CYPs) are heme containing mono-oxygenases which activate atmospheric dioxygen and insert one oxygen atom into an organic substrate and reduce the other to water (1). For efficient catalysis, a tight coupling between electron transfer and P450 substrate oxidation is required to inhibit the formation of deleterious reactive oxygen species. For eukaryotic microsomal P450s, the membrane-bound form of cytochrome P450 reductase (CPR) orchestrates the stepwise electron transfer from NADPH to the cytochrome P450 heme center (2-4). CPR is an obligate electron donor to all microsomal P450s and is a critical partner in drug metabolism (5) and steroid synthesis (6).
CPR is a ~78 kDa, multidomain protein which belongs to the family of electron transfer flavoproteins (7) that includes methionine synthase reductase (MSR) (8), novel reductase (NR1) (9), nitric oxide synthases (NOS) (10-11), P450BM3 (12) and sulfite reductase (SR) (13) all of which utilize both FAD and FMN as tightly bound cofactors. CPR is the only known member of this family that is membrane associated.
In order to solubilize CPR, the N-terminal membrane spanning domain can be cleaved by trypsin. Subsequent experiments, however, demonstrated that membrane spanning domains are essential for efficient electron transfer to microsomal P450s (14). Interestingly, yeast CPR is able to function without requiring any membrane anchor. The structure of both yeast (15-16) and mammalian CPR (17) are similar but their redox potential and electron transfer kinetics differ (18). For microsomal CPR it is possible that, in addition to anchoring and orienting the protein, the membrane modulates the redox potential of the FMN and FAD prosthetic groups, facilitating electron transfer from NADPH to P450.
The structure of soluble CPR, devoid of the N-terminal membrane-anchoring sequence, is available in multiple conformations (17, 19-20). The protein has three distinct domains; FAD binding, FMN binding and a linker region that dictates the relative orientation of the FAD and FMN domains to facilitate interflavin and FMN-heme electron transfer. In the ‘closed’ conformation (17), the FAD and FMN domains are near each other, facilitating inter-flavin electron transfer. In the ‘open’ conformation (20), the structure of which was achieved by deleting the residues in the linker region, the FMN and heme domain are in proximity to each other, facilitating electron transfer from FMN to heme. In nitric oxide synthase (NOS), the redox potential of the ‘open’ and ‘close’ forms are very different, suggesting a conformationally gated redox potential (21).
It has been shown in reconstituted systems that the membrane charge is an important determinant for rates of substrate oxidation by CYP3A4, the major membrane bound drug metabolizing P450 in humans (22). Kinetic analysis demonstrated that a negative membrane charge facilitates electron flow and the interactions between CPR and P450 (22). Further, it has been shown that anionic lipids help in the insertion of CYP3A4 (23) and CYP1A2 (24) into the membrane. In CPR, there is a cluster of basic residues in the membrane anchor domain, and it is suggested that anionic lipids might aid in anchoring the protein to the membrane (17). Moreover, the FMN domain has clusters of positive and negative residues on opposite sides, forming a strong electric dipole which potentially could aid in orienting the reductase. A patch of positively charged residues on the FMN domain may increase membrane affinity. Anionic lipids may not only help in anchoring the CPR into the membrane but may also play a role in regulating the redox potential such that the electron transfer from CPR to P450 is more favorable.
The redox potential of human CPR has been determined in solution without its membrane spanning domain (25). The pH dependence of the redox potentials of the CPR redox couples has also been investigated (26). CPR regulates electron transfer from NADPH to P450 and is thought to operate between the 1-3-2-1 redox states (4). As the oxidized/semiquinone (ox/sq) redox potential is very high for FMN, this prosthetic group favors the formation of the neutral blue semiquinone, FAD/FMNH*, generating the resting state of the enzyme. During catalysis, NADPH donates a hydride to the FAD, forming a three electron reduced state, FADH2/FMNH*. Through fast inter-flavin electron transfer, the enzyme forms FADH*/FMNH2. The hydroquinone species of the FMN then donates one electron to the P450 heme, leaving the di-semiquinone species FADH*/FMNH* which equilibrates to form FAD/FMNH2. The fully reduced FMN donates the second electron to the P450 recycling to the original resting state, FAD/FMNH*. In CPR, it is the hydroquinone form of FMN which gives electrons to P450, and electron flow from NADPH to P450 is controlled by the redox potentials of the different states of the FMN and FAD.
In this work, we show that the membrane has a role in determining the redox potential of CPR. Further, anionic lipids change the redox potential of CPR to make several processes in the electron transfer from CPR to P450 more thermodynamically feasible.
Materials and Methods
Materials
Imidazole, sodium cholate, Triton X-100 and Amberlite (XAD-2) were purchased from Sigma. POPC (1-Palmitoyl-2-Oleoyl-sn-Glycero-3-Phosphocholine) and POPS ((1-Palmitoyl-2-Oleoyl-sn-Glycero-3-Phosphoserine) were from Avanti Polar Lipids, Inc. (Alabaster, AL). All other chemicals, purchased from Fisher, were at least ACS grade and used without further purification.
Expression, purification and characterization of cytochrome P450 reductase
CPR from rat was expressed in the pOR262 vector and purified as described previously (27). The rat CPR/pOR262 plasmid was a generous gift from Dr. Todd D. Porter (University of Kentucky, Lexington, KY). CPR purity was ascertained by a single band when analyzed by SDS-PAGE with an Rz ratio (A276 nm/A456 nm) of 8.7 (28). The specific activity of CPR was measured using the cytochrome c activity assay (14), using an extinction coefficient of 24.1 mM–1 cm–1 at 456 nm for CPR (7).
Self-Assembly and Purification of CPR Nanodiscs
To self-assemble Nanodiscs containing CPR, the protein at ~ 10μM concentration was added to cholate-solubilized lipid and the membrane scaffold protein MSP1D2 (29)(30). Phospholipids were quantitated by inorganic phosphate analysis, dried under a stream of nitrogen, and stored in a vacuum desiccator overnight. The final molar ratio during reconstitution was 0.1:130:65:1 (CPR:cholate:POPC:MSP1D2). One Nanodisc is formed by two MSP1D2 molecules and 130 POPC molecules. For a 50% POPC-50% POPS Nanodisc, there are about 65 molecules of POPC and 65 molecules of POPS. Thus, on average, the assembly mixture produced an excess of empty Nanodiscs and favors the formation of a single CPR per disc. Once the total reconstitution mixture was added at the appropriate ratios, it was incubated on ice for 1 h. To remove the detergents, and concomitantly initiate the self-assembly process, an excess of Amberlite was added (1g of amberlite per mL of reconstitution solution) and incubated on ice with shaking for an additional 4 h. The formed Nanodisc assemblies were separated from the Amberlite by filtering through a 33-mm 0.22 μM syringe filter (Millipore Corp.). To purify homogenous CPR-Nanodiscs the mixture was applied to a nickel-nitrilotriacetic acid (Ni-NTA) column (0.6 × 3.5 cm; Qiagen Inc.) equilibrated with 10 mM Tris-HCl (pH 7.4) and 100 mM NaCl. The nickel affinity column selectively binds to the his-tag on MSP1D2. The column was washed with 5 column volumes of 10 mM Tris-HCl (pH 7.4), 100 mM NaCl, and 15 mM imidazole. The nickel affinity column was then eluted with 5 column volumes of 10 mM Tris-HCl (pH 7.4), 100 mM NaCl, and 250 mM imidazole, and the colored fractions pooled, concentrated and injected onto a calibrated Superdex 200 HR 10/30 size exclusion column (GE Healthcare) equilibrated with 10 mM Tris-HCl (pH 7.4) and 100 mM NaCl and connected to a Waters HPLC system with a photodiode array detector at a flow rate of 0.5 ml/min. The Nanodisc fraction was pooled, concentrated and buffer exchanged into 100 mM phosphate buffer, frozen and stored at −80°C. The CPR-Nanodiscs concentration was measured by UV–Vis spectroscopy using a CARY BIO300 spectrophotometer. CPR-Nanodiscs were in 100 mM potassium phosphate buffer (pH 7.4) for the spectroelectrochemical titrations. All titrations were performed at 25° C using CPR-Nanodiscs at a concentration of ~30-35 μM.
In order to demonstrate the stability of CPR-Nanodiscs to proteolysis, SDS-PAGE gels were run before and after each redox titration. No evidence of proteolytic degradation of MSP or CPR was observed.
Determination of FMN and FAD content of soluble CPR and CPR-Nanodiscs
Quantification of flavin cofactors was performed by fluorescence titration. This method is based on the markedly different behavior of FAD and FMN when the fluorescence of each component is examined as a function of pH (31-33). All solutions were prepared in a standard buffer, 0.1 M phosphate buffer pH 7.7, 0.1 mM EDTA. The FAD and FMN stock solutions were prepared at 10 μM and their concentrations were measured using the extinction coefficients 1.22 × 10−4 M−1 cm−1 for FMN and 1.13 × 10−4 M−1 cm−1 for FAD. The solutions were diluted to 100 nM concentration before assay. The protein samples (CPR and CPR-Nanodiscs) were diluted with standard buffer so that the final flavin concentration was ~100 nM. Aliquots of the protein sample were placed in aluminum foiled covered centrifuge tubes and immersed in a boiling water bath for 10 minutes and then rapidly cooled, followed by centrifugation at 20,000 g for 10 minutes to remove the denatured proteins. The fluorescence of all solutions were measured at 22° C using an excitation wavelength of 450 nm and the emission at 524 nm was used to calculate the FMN and FAD ratio. After fluorescence measurement in standard buffer, the pH of the solution was changed to 2.6 by addition of 1.0 N HCL and the experiment repeated. The concentrations and ratios were determined from the fluorimetric data (31). The ratio of FMN to FAD was found to be 1.00 in CPR and 0.95 in CPR-Nanodiscs, demonstrating that there is no significant loss of FMN during Nanodisc assembly.
Spectroelectrochemical Titrations
Redox titrations were carried out under a continuous flow of nitrogen in a cuvette that was assembled in the anaerobic glove box (COY Laboratory Products, Inc. Grass Lake, MI). The cuvette has an open top through which the Ag/AgCl reference electrode, the gold working electrode, an argon purging tube and gas-tight Hamilton syringe were inserted. A small magnetic stir bar was placed at the bottom of the cuvette to mix the reagents. The temperature was maintained at 25 °C. The titration buffer (100 mM phosphate pH 7.4) was made anaerobic by flushing with ultrapure argon while stirring. The final experimental volume was 1.3 ml with an optical density ~1.0 near the main CPR visible band (near 450 nm). The following redox mediators were used; methyl viologen (−430 mV), benzyl viologen (−374 mV), 2-hydroxy-1,4-naphthoquinone (−145 mV), and anthraquinone 2-sulfonate (−230 mV) to a final concentration of 4 μM each. All potentials are reported against the standard hydrogen electrode (SHE).
Protein and mediator mixtures were deoxygenated under the flow of argon gas for several minutes. The protein was reduced by the addition of a small excess of anaerobically prepared sodium dithionite solution (concentration determined using a molar absorption coefficient 315 = 8.05 mM−1 cm−1) (34). The reduced protein spectrum was recorded to confirm complete reduction. Redox titrations followed Dutton's method (35). A small aliquot of oxidant/reductant (ferricyanide/dithionite) was added and the solution was stirred until equilibration was reached ~15–20 min, and the spectrum (350–800 nm) recorded using a CARY 300 UV-Visible spectrophotometer. The electrochemical potential was monitored using a Hewlett Packard (HP) 34401A Multimeter (input resistance was 10 GΩ) coupled to the gold electrode and the Ag/AgCl reference electrode (Bioanalytical Systems, Inc). Prior to modification, the gold working electrode was cleaned as described previously (36-37) and the electrodes immersed in saturated 4,4′-dithiodipyridine solution for 1 h. The electrode system was calibrated against a standard calomel electrode, and observed potential obtained relative to the Ag/AgCl reference. In order to control for potential interaction of CPR-Nanodiscs with the mediators, we analyzed the spectral changes occurring upon addition of a mediator mixture to Nanodiscs. The spectra of the mixture is well represented by the sum of the spectra of the components (mediators, proteins, oxidants and reductants) with no evidence for any significant spectrally manifested interaction at the concentrations of components used.
Analysis of titrations
Redox data was evaluated using Origin (OriginLab) software as described previously (25) and were fit to a modified Nernst equation describing four electron redox process that relates the absorbance changes at a specific wavelength to the redox potential of the four different redox couples: .
| (Equation 1) |
Here A is the absorption at a specific wavelength, a-c are the component extinction coefficient values contributed by one flavin in the oxidized, semiquinone and hydroquinone states, respectively. d-f are the corresponding values for the second flavin. E is the potential at the working electrode and E1, E2, E3 and E4 correspond to the midpoint potentials of the oxidized/semiquinone (ox/sq) and the semiquinone/hydroquinone (sq/hq) redox couples of the two flavins. Although, it is difficult to spectrally distinguish the FMN and FAD prosthetic groups, the redox potential of each cofactor are cleanly separated allowing deconvolution during titration.
From the spectra of individual cofactors in Equation (1) a=d and b=e, but c is not equal to f, and these constraints were used during spectral fitting. The coefficients a-f are wavelength dependent but are independent of the midpoint potential. The linear combination of the coefficients a-f gives rise to the absorbance changes of the whole system at a given potential. Therefore analysis yields the coefficients a-f which are unique for that wavelength. The data was also evaluated at the isosbestic wavelengths at 501 nm and 429 nm. Isosbestic points are defined as wavelengths corresponding to near-zero absorbance change for the associated redox couples. At 429 nm, the redox potential of oxidized/semiquinone for both FMN and FAD can be determined and we use the constraints b=c and e=f (as the absorbance values for semiquinone and reduced states of one flavin are equal at this wavelength). At 501 nm, the redox potential of semiquinone and reduced can be determined, and a=b and d=e as the absorbance of the oxidized and semiquinone states are equal.
Global fitting used a Levinthal-Marquardt subroutine with the constraints a=d and b=e at wavelengths 450-460 nm, chosen to reflect oxidized species of FMN and FAD with minimal interference from mediator absorbances and 580-605 nm, correspond to the semiquinone absorbance wavelengths, using b=c and e=f at 429 nm and a=d and d=e at 501 nm. The coefficients a-f for each wavelength were given limits using the maximal value of absorbance at that wavelength. From the non-linear curve fitting we obtain unique redox potential values, which are the same for all wavelengths, and unique values for the parameter a-f.
In order to verify the deconvolution analysis, a cross-validation approach was used. First, 10% of the data points are removed and the remaining 90% used as training data set to analyze the first 10%. Secondly, the coefficients a-f represent the basis spectra for the flavin intermediates and therefore can be used to determine the protein concentration. To demonstrate the compatibility of the data analysis we can calculate the total protein concentration from these derived parameters. The concentrations calculated from the a(501) and a(429) coefficients are 8.3 μM (ε 501 = 9.2 × 10 3 M −1 cm −1) and 9.8 μM (ε 429 = 18.2 × 103 M −1 cm −1) for 100% POPC and16 μM (ε 501 = 9.2 × 10 3 M −1 cm −1) and 12 μM (ε 429 = 18.2 × 103 M −1 cm −1) for 50% POPS. The coefficients a-f relate to the intrinsic spectral properties of cofactors and are independent of the external conditions such as the lipid composition.
RESULTS
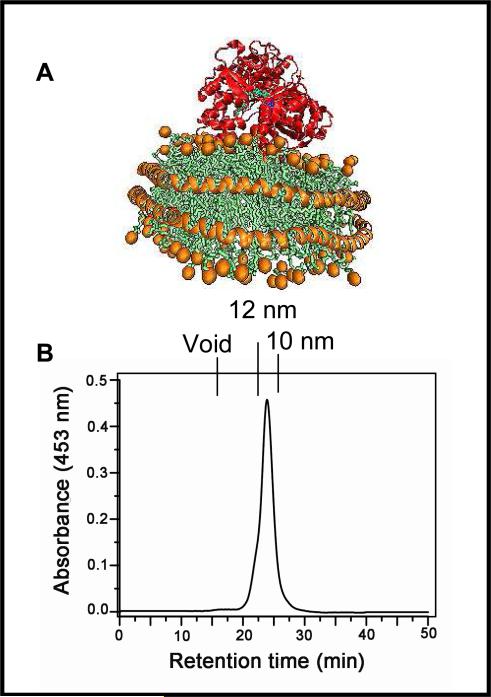
In order to study the effect of the membrane environment on the redox potential of CPR we use Nanodiscs (29, 38). Nanodiscs are discoidal self-assembled, water soluble, lipid bilayers encirculated by an amphipathic protein belt (Figure 1A). Nanodiscs have been used to solubilize and functionally stabilize many membrane proteins for investigations in solution and on surfaces (39-40). Nanodiscs provide an ideal environment for generating active monodisperse integral membrane proteins which avoids potential aggregation that could make redox potential measurements difficult (41). Nanodiscs have advantages over liposomes in this study as the local membrane environment around the protein in the Nanodisc can be rigorously controlled (42) and the number of CPR per Nanodisc can be defined which ensures a homogenous population of protein without higher order aggregation.
Figure 1.
(A) Schematic representation of CPR docking in the lipid bilayer membrane of Nanodiscs (B) The HPLC elution profiles of CPR-Nanodisc assemblies following the flavin absorbances at 453 nm. A Stokes diameter scale is shown on the top axis.
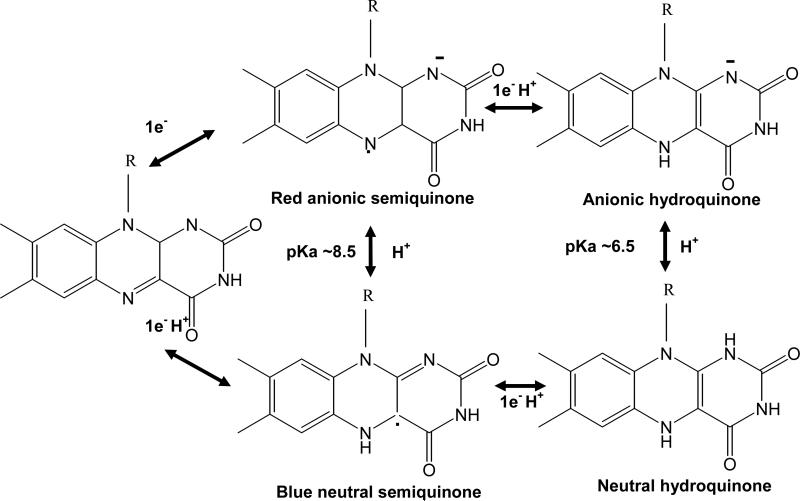
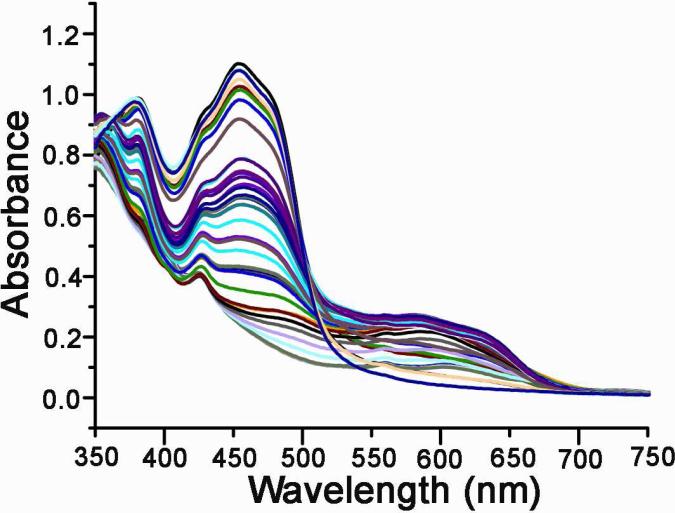
Full length CPR was purified to homogeneity as described (27), incorporated into his-tagged Nanodiscs and purified. The homogeneity of CPR-Nanodiscs is evident from the HPLC profile (Figure 1B). SDS-PAGE analysis demonstrates the lack of proteolysis during the Nanodisc assembly process. CPR-Nanodiscs were made with two different membrane compositions, 100% POPC (palmitoyl oleoyl phosphatidyl choline) and with 50% POPS (palmitoyl oleoyl phosphatidyl serine) and 50% POPC. CPR has three redox phases which are spectrally distinguishable as it cycles from fully oxidized through semiquinone to the fully reduced hydroquinone form (Schematic 1). These spectral characteristics of CPR-Nanodiscs are similar to CPR in solution (27) as shown in Figure 2. The visible absorbance maxima for the fully oxidized CPR are at 455 and 382 nm, with a shoulder at approximately 484 nm. A characteristic of the semiquinone is a broad band centered at 587 nm, with a shoulder at approximately 630 nm. For the oxidized to semiquinone transition, there is an isosbestic point at 501 nm. In the transition from semiquinone to hydroquinone, however, there is mixture of species and no clear isosbestic point is evident. At very negative potentials, the blue semiquinone signal decreases as the FAD semiquinone is reduced and there is a second isosbestic point at approximately 429 nm in this phase of titration. Redox titrations and concomitant spectral changes are initiated from the fully oxidized protein and proceed to full flavin reduction by addition of small aliquots of sodium dithionite and then back to oxidized flavin by addition of ferricyanide (Figure 2). Full equilibration was confirmed after the addition of each reducing/oxidizing aliquot by observation of a stable electrode voltage and spectra. No hysterisis was observed in the redox titrations and spectra recorded at similar potentials during oxidative and reductive titrations were identical.
Scheme 1.
Structure of flavin redox states.
Figure 2.
Spectral changes during a redox titration of full-length CPR in Nanodiscs.
Previous redox titrations of CPR utilized the solubilized trypsinized form of the enzyme (25-26, 43). In order to provide a consistent comparison, we measured the redox potential of full length rat CPR which is soluble and does not show any turbidity in 100 mM phosphate buffer pH 7.4 in the absence of glycerol. Figure 2 shows a typical set of spectra obtained for a redox titration of CPR. It has been previously shown that the FAD domain has a lower reduction potential than the FMN domain (44) and this was used to deconvolute the potentials using the Nernst equation as described in Materials and Methods. The redox potentials obtained are close to the literature values for soluble human CPR as shown in Table 1 and Table 2 for pH 7.5 (26).
Table 1.
Redox potential of full length CPR in solution and in Nanodiscs. All potential are reported in mV vs. SHE in 100 mM phosphate buffer at pH 7.4.
| Enzyme | EFMN (ox/sq) mV | EFMN (sq/hq) mV | EFMN (ox/hq) mV | EFAD (ox/sq) mV | EFAD (sq/hq) mV | EFAD (ox/hq) mV |
|---|---|---|---|---|---|---|
| Soluble CPR | −68 ± 5 | −246 ± 5 | −157 | −325 ± 5 | −372 ± 2 | −348 |
| 100%-PC-CPR-Nanodisc | −16 ± 10 | −127 ± 2 | −71 | −309 ± 9 | −246 ± 6 | −277 |
| 50%PS-50%PC-CPR-Nanodisc | − 30 ± 5 | − 207 ± 0 | −118 | −332 ± 12 | −277 ± 10 | −304 |
| ΔE1 = E°100% PC-CPR-Nanodisc - E°Soluble-CPR | + 52 ± 15 | +119 ± 7 | +86 | +16 ± 14 | +126 ± 8 | +71 |
| ΔE2 = E°50% PC-CPR-Nanodisc - E°Soluble-CPR | + 38 ± 10 | +39 ± 5 | +39 | −7 ± 17 | +95 ± 12 | +44 |
| ΔE3 = ΔE2 - ΔE1 | −14 ± 25 | −80 ± 12 | −63 | −23 ± 31 | −31 ± 30 | −27 |
Table 2.
Redox potential of di-flavin reductases
| Enzyme | EFMN (ox/sq) mV | EFMN (sq/hq) mV | EFAD (ox/sq) mV | EFAD (sq/hq) mV | Conditions | Reference |
|---|---|---|---|---|---|---|
| Human t-CPR* | −66 | −269 | −283 | −382 | pH 7.0 | (25) |
| Human t-CPR* | −89 | −246 | −328 | −381 | pH 7.5 | (26) |
| Human t-CPR* | −114 | −261 | −366 | −385 | pH 8.0 | (26) |
| Human t-CPR* | −133 | −251 | −380 | −419 | pH 8.5 | (26) |
| Rabbit CPR | −110 | −270 | −290 | −365 | pH 7.0 | (43) |
| Rat CPR | −68 | −246 | −325 | −372 | pH 7.4 | This Work |
| Rat CPR-100% PC-Nanodisc | −16 | −127 | −309 | −246 | pH 7.4 | This Work |
| Rat CPR-50%PS50%PC-Nanodisc | −30 | −207 | −332 | −277 | pH 7.4 | This Work |
| NOS (−calmodulin)$ | −49 | −274 | −232 | −280 | pH 7.1 | (57) |
| NOS (+calmodulin)$ | −30 | −267 | −234 | −284 | pH 7.1 | (57) |
| Endothelial NOS | −105 | −240 | −230 | −260 | pH 7.0 | (58) |
| Neuronal NOS | −120 | −220 | −250 | −260 | pH 7.0 | (58) |
| Inducible NOS | −105 | −245 | −240 | −270 | pH 7.0 | (58) |
| P450-BM3 | −213 | −193 | −292 | −372 | pH 7.0 | (54) |
| Sulfite reductase | −152 | −327 | −382 | −322 | pH 7.7 | (59) |
t CPR, truncated CPR with its membrane spanning domains missing
NOS, nitric oxide synthase
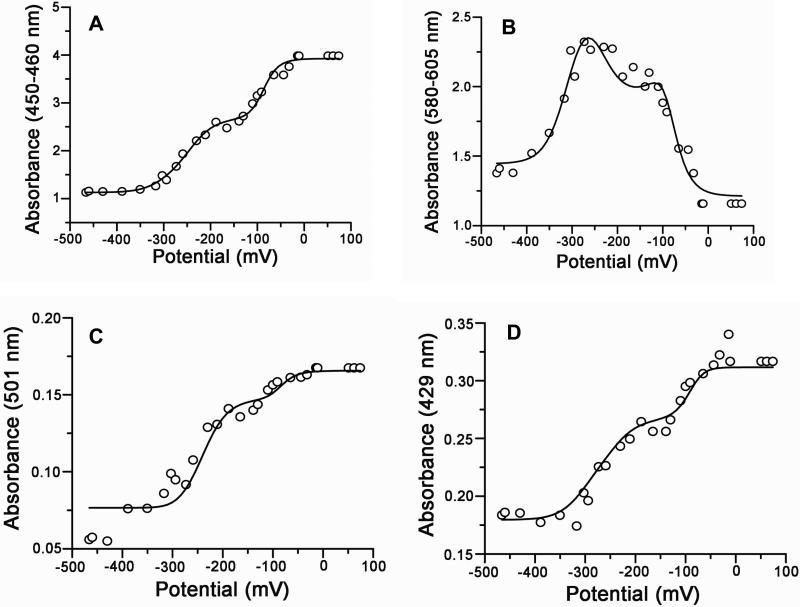
In order to examine the effect of the membrane environment on CPR, the redox potentials were determined when self-assembled into 100% POPC-Nanodiscs. Figure 3 shows the redox titration of CPR-Nanodiscs in 100 mM phosphate buffer at pH 7.4. As with the soluble CPR, the redox potential of the CPR-Nanodiscs also showed three phases. However, due to the presence of the membrane, the redox potentials of all the redox couples shift to more positive values in 100% POPC-CPR-Nanodiscs (Table 1). Therefore, CPR incorporated into a membrane environment shows significant differences in redox potential from those of full length CPR. In order to understand if the redox potentials are changing due to the membrane composition, anionic lipids in the form of POPS were incorporated in the membrane bilayer of Nanodiscs by self-assembly. Figure 4 shows the redox titration of CPR-Nanodiscs in a 50% POPS-50% POPC Nanodisc preparation.
Figure 3.
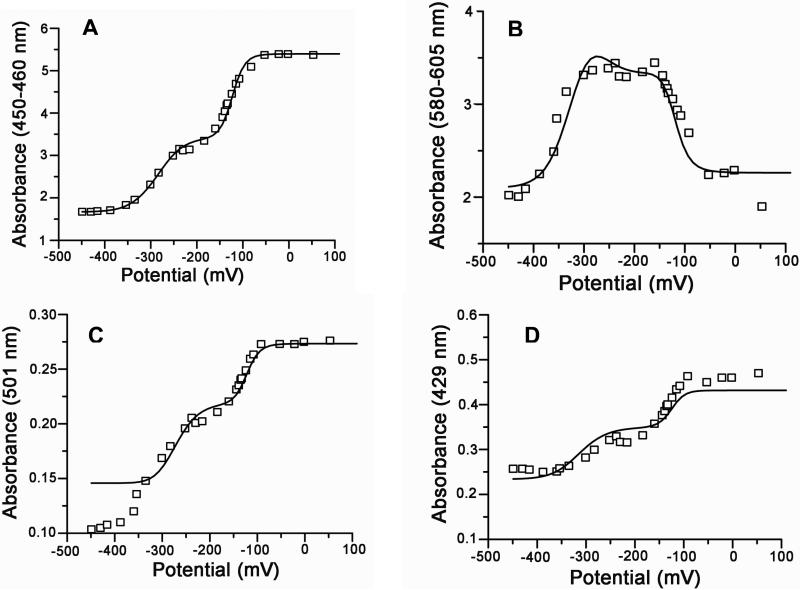
Multiple wavelength analysis of the redox titration of CPR-Nanodiscs assembled with POPC. The absorbance is plotted versus the solution potential (mV vs. SHE) at several wavelength regions: (A) 450-460 nm, (B) the isosbestic of semiquinone/reduced couple at 429 nm, (C) 580-605 and (D) the isosbestic of the oxidized/semiquinone at 501 nm. The solid lines represent fits to the four electron Nernst equation (Methods).
Figure 4.
Multiple wavelength analysis of the redox titration of CPR-Nanodiscs with 50% POPS and 50% POPC. (A-D) correspond to the wavelengths of absorption measurements as described in Figure 3.
Figure 3A and 4A, shows the dependence of the summed absorbance from 450-460 nm, corresponding to oxidized species of the FMN and FAD and having minimal interference from mediator absorbances. Figure 3B and 4B, presents the dependence of summed absorbances from 580-605 (corresponding to the semiquinone absorbance wavelengths). The asymmetric bell shape curve in figure 3B and 4B shows the presence of semiquinone contributions from both FMN and FAD. The isosbestic point at 429 and 501 was used to increase the validity of the redox potential analysis for the two redox couples (ox/sq and sq/hq). Figure 3C and 4C shows the redox potential dependence at the isosbestic point absorbance at 501 nm. At 501 nm, the redox potential of semiquinone and reduced can be determined and a=b and d=e, as the absorbance of the oxidized and semiquinone states are equal, (Equation 1). In Figure 3D and 4D, the absorbance at 429 is plotted against the potential which yields the redox potential of oxidized/semiquinone for both FMN and FAD by setting b=c and e=f (as the absorbance values for semiquinone and reduced states of one flavin are equal at this wavelength) in Equation 1. All data from each experiment shown in Figure 3A-D and Figure 4A-D were fitted to the four electrons Nernst equation (1) simultaneously using a global fitting algorithm (Methods) and the solid lines in each figure represent the best fit to these observed data sets. This approach also yields unique values for the spectral parameters a-f . These parameters are intrinsic for the protein and are not dependent on external conditions such as the lipid composition. These values are the absorbance of the pure species of oxidized, semiquinone and hydroquinone and are used to calculate the concentration of the pure species (see Methods).
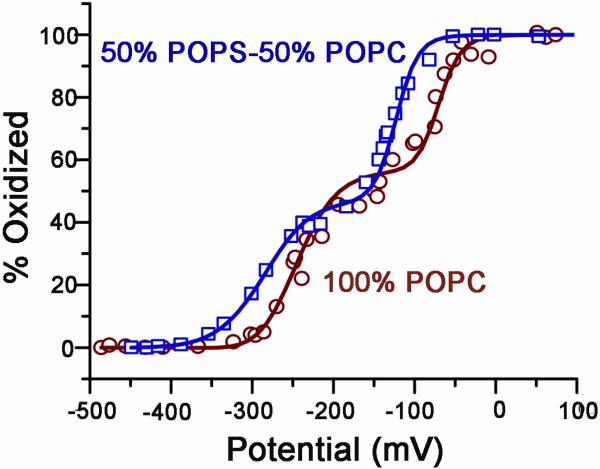
In Figure 5, we plot the electrode potential versus the percentage of oxidized CPR at wavelength 478 nm for 50% POPS and 100% POPC membrane composition. Clearly, the potentials of the redox couples of both FMN and FAD are shifted to more negative values in 50% POPS Nanodiscs as compared to 100% POPC, showing that the membrane composition has a major affect on the redox potential of CPR.
Figure 5.
Effect of anionic lipids on the CPR redox potentials. Potential CPR-Nanodiscs with 100% POPC (open circles) is shifted to higher values as compared to CPRNanodiscs containing 50% POPC/50% POPS (open squares).
DISCUSSION
The redox potentials of di-flavin proteins that couple to heme centers have been determined for a variety of enzyme systems (Table 2). There is variation in the redox potentials of the flavin couples and also differences in the identity of the redox state of the flavin that delivers electrons to the accepting redox partner. For example, with microsomal CPRs, the hydroquinone donates electrons to the heme during P450 catalysis and the di-flavins cycle between the “1-3-2-1” reduction states (4). In contrast, in bacterial CYP102, the short lived anionic semiquinone is the main species transferring electrons to the heme group and the enzyme circulates between the “0-2-1-0” redox states during P450 catalysis (4). Clearly, flavin redox potentials in these reductases are one of the key factors which control electron transfer in these systems.
In this work, we have determined the redox potential of full length CPR in a membrane environment. The reduction potential of the soluble full length CPR is found to be similar to the cleaved soluble human CPR, Tables 1 and 2 for pH 7.4 (26). However, the redox potentials of CPR in Nanodiscs demonstrate that the lipid bilayer can affect the redox potential of the flavin domains. The redox potentials of both FMN and FAD are more positive (ΔE1 and ΔE2, Table 1) when inserted into or in close proximity to the lipid bilayer. Theoretically, it is been shown that a shift of up to ~100 mV with respect to bulk solution can be realized by positioning the redox couples within 5-10 Å region of a membrane-solvent interface (45-46). The shift to more positive potential of an NADPH oxidase system was observed when the FAD redox potential was determined in a plasma membrane (47). In other experiments, cyt b-558 has been shown to have a more positive redox potential in membrane preparations as compared to when it is solubilized (48). In addition, we have shown previously that redox potential of CYP3A4 in a lipid bilayer shifts by ~ +100 mV compared to CYP3A4 in solution (41). Therefore, the membrane environment can effect the redox potentials of membrane proteins, generally shifting them to more positive values (49).
The redox potential of FAD ox/hq, −304 mV in POPC:POPS (50:50) and −277 mV in pure POPC in CPR-Nanodiscs is more positive than the redox potential of NADPH (−320 mV) (50). Therefore, the reduction of CPR by NADPH is thermodynamically feasible when CPR is anchored to the membrane, resulting in a more favorable couple as compared to when CPR is in solution, where the ox/hq potential is −348 mV (4). On the contrary, it can be argued that the redox potential of NADPH changes on binding to CPR as the nicotinamide center is moved from an aqueous hydrophilic environment to a hydrophobic protein environment and also forms a π-π stacking interaction with the isoalloxazine ring of FAD. This has been shown to produce a shift to + 40 mV in the redox potential of NADPH upon binding to the protein (51). However, the affect of NADPH binding to the redox potential of FAD is varied for different proteins (26,51-52). Although it is possible that after hydride transfer the semiquinone form of FAD and FMN is stabilized by the proximity of a positively charged NADP+, in soluble CPR the redox potentials in the presence and absence of NADP+ and 2,5’-ADP are the same (26, 52). Therefore, the reduction of CPR by NADPH is thermodynamically feasible only when CPR is anchored to a membrane.
Upon reduction, the flavins go through three different redox states, oxidized (ox), semiquinone (sq) and hydroquinone (hq) as illustrated in Scheme 1. The sq and hq state can exist in both protonated and unprotonated forms depending on the pKa in the protein. From Figure 5 and Table 1, it can be seen that both the FMN and FAD sq/hq equilibria shift to higher redox potentials when in a membrane environment. The positive shift of the FMN ox/sq further stabilizes the semiquinone in the membrane bilayer which may provide an additional driving force for electron transfer from NADPH to P450. In the membrane environment of Nanodiscs, the redox potentials of the ox/sq and sq/hq couples redox potentials are reversed for FAD, thus destabilizing the semiquinone state of FAD. A similar inversion of the FAD has been reported in NADPH oxidase (53). The larger change in the redox potential of sq/hq redox couple (FADsq/hq ΔE = +126, FMNsq/hq ΔE = +119 mV) versus the ox/sq couple (FADox/sq ΔE = +16, FMNox/sq ΔE = +52 mV) in membranes compared to solution can be qualitatively understood via simple electrostatic considerations. As shown in Scheme 1, the pKa of semiquinone is ~ 8.5 and is possibly higher in the protein. Thus, at pH 7.4, the semiquinone is protonated and is charge neutral. This can be observed spectrally, as the neutral semiquinone has a characteristic peak at 585 nm while the anionic hydroquinone has a peak at 380 nm (54-55). The pKa of the hydroquinone is ~ 6.5 and therefore at pH 7.4 the anionic hydroquinone is formed, which has a net negative charge. Although the anionic semiquinone can be spectrally distinguished from the neutral semiquinone, the anionic hydroquinone and neutral hydroquinone are spectrally indistinguishable. Hence, at pH 7.4, it is expected that when reduction occurs from oxidized to semiquinone there is conversion from neutral oxidized to neutral semiquinone species and the ox/sq redox couple will not be altered as much as the sq/hq couple by membrane electrostatics. When reduction occurs from semiquinone to hydroquinone, the neutral to charged state conversion can be modulated by the membrane electrostatics. Therefore the insertion of the protein into a membrane bilayer perturbs the redox potential of the sq/hq couple to a larger extent than the ox/sq equilibrium. If true, then changing the pH to higher values should favor the formation of red anionic semiquinone so that the redox potential of ox/sq (neutral ox and anionic sq) should be more affected as compared to the sq/hq couple where both states are charged. Previously it has been reported that there is a disappearance of the CPR neutral semiquinone as the pH is raised (26). It was also observed that the redox couple, ox/sq, is more affected by changes in pH from 7.0 to 8.5 than the sq/hq couple, Table 2. Thus, there is differential effect of membrane electrostatics on the proton coupled redox states of the flavins which can be qualitatively explained on the basis of a charge stabilization in the membrane.
The redox potential of CPR-Nanodiscs with 50% POPS was measured in order to probe the effect of negatively charged lipids on the redox potential of CPR. The redox potential of both flavins become more negative compared to the Nanodiscs assembled with the zwitterionic POPC. We observe that the redox potential of the sq/hq couple of both FMN and FAD (ΔE3 in Table 1) are affected more than the corresponding ox/sq couples. Anionic lipids disfavor the formation of the anionic semiquinone and anionic hydroquinone. Hence the reduction from neutral semiquinone to anionic hydroquinone at pH 7.4 will be disfavored, leading to the redox potential of sq/hq couple becoming more negative when anionic lipids are in proximity. As the oxidized form and semiquinone form are both neutral at pH 7.4, the redox couple of ox/sq will not be affected to as great an extent. On the other hand, the changes in the redox potential of the ox/sq couple between 100% POPC to 50% POPS CPR-Nanodiscs cannot be explained on the basis of the simple argument.
CPR orchestrates the electron supply from NADPH to the P450 by stabilizing the one electron reduced form of FAD/FMNH* (56). In the presence of the membrane, the reduction potential of the FAD is more positive and becomes thermodynamically favorable for reduction by NADPH. The high midpoint potential of the FMN ox/sq couple provides the driving force for the interflavin electron transfer in full length CPR. In the presence of neutral lipids, although the redox potential of all the couples shift to more positive values, the difference between the FAD sq/hq couple and the FMN ox/sq couple decreases, hence the driving force for interflavin electron transfer decreases. On addition of anionic lipids, there is slight increase in this driving force. The FMN hydroquinone is thought to be the species which donates electrons to the P450 during enzyme catalysis. The redox potential of sq/hq couple of 100% POPC CPR-Nanodisc FMN is -127 mV, which is more positive than the redox potential of substrate bound CYP3A4-Nanodiscs -138 mV (41). This makes the electron transfer from the CPR FMN to CYP3A4 in neutral membranes slightly unfavorable thermodynamically. In the presence of anionic lipids, as in 50% POPS CPR-Nanodiscs, the redox potential of the FMN sq/hq is lowered (−207 mV) dropping below the redox potential of substrate bound CYP3A4, but more positive than substrate free CYP3A4, which thermodynamically favors electron transfer from FMN to CYP3A4. This may be the origin of an enhancement in substrate oxidation is observed in CPR dependent reaction in the presence of anionic lipids. Clearly, understanding the functional behavior of integral membrane redox proteins demands that experiment be conducted in a native-like membrane environment.
ACKNOWLEDGEMENTS
We acknowledge Dr. Ilia Denisov, Yelena Grinkova and Dr. Mark A. McLean at University of Illinois and Dr. Nathan Baker at Washington University for their helpful discussions. Editorial assistance from Aretta Weber is greatly appreciated.
Abbreviations
- CPR
cytochrome P450 reductase
- POPC
1-Palmitoyl-2-Oleoyl-sn-Glycero-3-Phosphocholine
- POPS
1-Palmitoyl-2-Oleoyl-sn-Glycero-3-Phosphoserine
- Ox
oxidized
- Sq
Semiquinone
- Hq
hydroquinone
Footnotes
This research was supported by the National Science Foundation (EEC-0647560), NIH GM 31756 and GM 33775.
REFERENCES
- 1.Denisov IG, Makris TM, Sligar SG, Schlichting I. Structure and chemistry of cytochrome P450. Chem. Rev. 2005;105:2253–2277. doi: 10.1021/cr0307143. [DOI] [PubMed] [Google Scholar]
- 2.Iyanagi T. Structure and function of NADPH-cytochrome P450 reductase and nitric oxide synthase reductase domain. Biochem. Biophy. Res. Comm. 2005;338:520–528. doi: 10.1016/j.bbrc.2005.08.043. [DOI] [PubMed] [Google Scholar]
- 3.Hannemann F, Bichet A, Ewen KM, Bernhardt R. Cytochrome P450 systems--biological variations of electron transport chains. Biochim. Biophys. Acta (BBA) - General Subjects. 2007;1770:330–344. doi: 10.1016/j.bbagen.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 4.Murataliev MB, Feyereisen R, Walker FA. Electron transfer by diflavin reductases. Biochim.Biophys. Acta (BBA) - Proteins & Proteomics. 2004;1698:1–26. doi: 10.1016/j.bbapap.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 5.Guengerich FP. Cytochrome p450 and chemical toxicology. Chem. Res. Toxicol. 2008;21:70–83. doi: 10.1021/tx700079z. [DOI] [PubMed] [Google Scholar]
- 6.Miller WL. Minireview: Regulation of Steroidogenesis by Electron Transfer. Endocrinology. 2005;146:2544–2550. doi: 10.1210/en.2005-0096. [DOI] [PubMed] [Google Scholar]
- 7.Porter TD, Wilson TE, Kasper CB. Expression of a functional 78,000 dalton mammalian flavoprotein, NADPH-cytochrome P-450 oxidoreductase, in Escherichia coli. Arch.Biochem.Biophy. 1987;254:353–367. doi: 10.1016/0003-9861(87)90111-1. [DOI] [PubMed] [Google Scholar]
- 8.Leclerc D, Wilson A, Dumas R, Gafuik C, Song D, Watkins D, Heng HHQ, Rommens JM, Scherer SW, Rosenblatt DS, Gravel RA. Cloning and mapping of a cDNA for methionine synthase reductase, a flavoprotein defective in patients with homocystinuria. Proc. Nat. Acad. Sci.(USA) 1998;95:3059–3064. doi: 10.1073/pnas.95.6.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paine MJI, Garner AP, Powell D, Sibbald J, Sales M, Pratt N, Smith T, Tew DG, Wolf CR. Cloning and Characterization of a Novel Human Dual Flavin Reductase. J. Biol. Chem. 2000;275:1471–1478. doi: 10.1074/jbc.275.2.1471. [DOI] [PubMed] [Google Scholar]
- 10.McMillan K, Bredt DS, Hirsch DJ, Snyder SH, Clark JE, Masters BS. Cloned, expressed rat cerebellar nitric oxide synthase contains stoichiometric amounts of heme, which binds carbon monoxide. Proc. Nat. Acad. Sci.(USA) 1992;89:11141–11145. doi: 10.1073/pnas.89.23.11141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, Das A, Sibhatu H, Jamal J, Sligar SG, Poulos TL. Exploring the Electron Transfer Properties of Neuronal Nitric-oxide Synthase by Reversal of the FMN Redox Potential. J. Biol. Chem. 2008;283:34762–34772. doi: 10.1074/jbc.M806949200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Narhi L, Fulco A. Characterization of a catalytically self-sufficient 119,000-dalton cytochrome P-450 monooxygenase induced by barbiturates in Bacillus megaterium. J. Biol. Chem. 1986;261:7160–7169. [PubMed] [Google Scholar]
- 13.Ostrowski J, Barber M, Rueger D, Miller B, Siegel L, Kredich N. Characterization of the flavoprotein moieties of NADPH-sulfite reductase from Salmonella typhimurium and Escherichia coli. Physicochemical and catalytic properties, amino acid sequence deduced from DNA sequence of cysJ, and comparison with NADPH-cytochrome P-450 reductase. J. Biol. Chem. 1989;264:15796–15808. [PubMed] [Google Scholar]
- 14.Yasukochi Y, Masters BS. Some properties of a detergent-solubilized NADPH-cytochrome c(cytochrome P-450) reductase purified by biospecific affinity chromatography. J. Biol. Chem. 1976;251:5337–5344. [PubMed] [Google Scholar]
- 15.Lamb DC, Warrilow AGS, Venkateswarlu K, Kelly DE, Kelly SL. Activities and Kinetic Mechanisms of Native and Soluble NADPH-Cytochrome P450 Reductase. Biochem. Biophy. Res. Comm. 2001;286:48–54. doi: 10.1006/bbrc.2001.5338. [DOI] [PubMed] [Google Scholar]
- 16.Lamb DC, Kim Y, Yermalitskaya LV, Yermalitsky VN, Lepesheva GI, Kelly SL, Waterman MR, Podust LM. A Second FMN Binding Site in Yeast NADPH-Cytochrome P450 Reductase Suggests a Mechanism of Electron Transfer by Diflavin Reductases. Structure. 2006;14:51–61. doi: 10.1016/j.str.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 17.Wang M, Roberts DL, Paschke R, Shea TM, Masters BSS, Kim J-JP. Three-dimensional structure of NADPH cytochrome P450 reductase: Prototype for FMN- and FAD-containing enzymes. Proc. Nat. Acad. Sci.(USA) 1997;94:8411–8416. doi: 10.1073/pnas.94.16.8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Louërat-Oriou B, Perret A, Pompon D. Differential redox and electron-transfer properties of purified yeast, plant and human NADPH-cytochrome P-450 reductases highly modulate cytochrome P-450 activities. Eur. J. Biochem. 1998;258:1040–1049. doi: 10.1046/j.1432-1327.1998.2581040.x. [DOI] [PubMed] [Google Scholar]
- 19.Aigrain L, Pompon D, Moréra S, Truan G. Structure of the open conformation of a functional chimeric NADPH cytochrome P450 reductase. EMBO reports. 2009;10:742–747. doi: 10.1038/embor.2009.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamdane D, Xia C, Im SC, Zhang H, Kim JJ, Waskell L. Structure and Function of an NADPH-Cytochrome P450 Oxidoreductase in an Open Conformation Capable of Reducing Cytochrome P450. J. Biol. Chem. 2009;284:11374–11384. doi: 10.1074/jbc.M807868200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghosh DK, Holliday MA, Thomas C, Weinberg JB, Smith SME, Salerno JC. Nitric-oxide Synthase Output State: Design and propertied of nitric-oxide synthase oxygenase/FMN domain constructs. J. Biol. Chem. 2006;281:14173–14183. doi: 10.1074/jbc.M509937200. [DOI] [PubMed] [Google Scholar]
- 22.Ingelman-Sundberg M, Hagbjörk A-L, Ueng Y-F, Yamazaki H, Guengerich FP. High Rates of Substrate Hydroxylation by Human Cytochrome P450 3A4 in Reconstituted Membranous Vesicles: Influence of Membrane Charge. Biochem. Biophy.Res. Comm. 1996;221:318–322. doi: 10.1006/bbrc.1996.0593. [DOI] [PubMed] [Google Scholar]
- 23.Kim KH, Ahn T, Yun CH. Membrane properties induced by anionic phospholipids and phosphatidylethanolamine are critical for the membrane binding and catalytic activity of human cytochrome P450 3A4. Biochemistry. 2003;42:15377–15387. doi: 10.1021/bi035280k. [DOI] [PubMed] [Google Scholar]
- 24.Ahn T, Guengerich FP, Yun CH. Membrane Insertion of Cytochrome P450 1A2 Promoted by Anionic Phospholipids. Biochemistry. 1998;37:12860–12866. doi: 10.1021/bi980804f. [DOI] [PubMed] [Google Scholar]
- 25.Munro AW, Noble MA, Robledo L, Daff SN, Chapman SK. Determination of the redox properties of human NADPH-cytochrome P450 reductase. Biochemistry. 2001;40:1956–1963. doi: 10.1021/bi001718u. [DOI] [PubMed] [Google Scholar]
- 26.Brenner S, Hay S, Munro AW, Scrutton NS. Inter-flavin electron transfer in cytochrome P450 reductase; effects of solvent and pH identify hidden complexity in mechanism. FEBS Journal. 2008;275:4540–4557. doi: 10.1111/j.1742-4658.2008.06597.x. [DOI] [PubMed] [Google Scholar]
- 27.Shen A, Porter T, Wilson T, Kasper C. Structural analysis of the FMN binding domain of NADPH-cytochrome P-450 oxidoreductase by site-directed mutagenesis. J. Biol. Chem. 1989;264:7584–7589. [PubMed] [Google Scholar]
- 28.Vermilion JL, Coon MJ. Purified liver microsomal NADPH-cytochrome P-450 reductase. Spectral characterization of oxidation-reduction states. J. Biol. Chem. 1978;253:2694–2704. [PubMed] [Google Scholar]
- 29.Denisov IG, Grinkova YV, Lazarides AA, Sligar SG. Directed self-assembly of monodisperse phospholipid bilayer Nanodiscs with controlled size. J. Am. Chem. Soc. 2004;126:3477–3487. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- 30.Denisov IG, Baas BJ, Grinkova YV, Sligar SG. Cooperativity in cytochrome P450 3A4: linkages in substrate binding, spin state, uncoupling, and product formation. J. Biol. Chem. 2007;282:7066–7076. doi: 10.1074/jbc.M609589200. [DOI] [PubMed] [Google Scholar]
- 31.Aliverti A, Curti B, Vanoni MA. Identifying and quantitating FAD and FMN in simple and in iron-sulfur-containing flavoproteins. Methods Mol. Biol. 1999;131:9–23. doi: 10.1385/1-59259-266-X:9. [DOI] [PubMed] [Google Scholar]
- 32.Faeder EJ, Siegel LM. A rapid micromethod for determination of FMN and FAD in mixtures. Anal.Biochem. 1973;53:332–336. doi: 10.1016/0003-2697(73)90442-9. [DOI] [PubMed] [Google Scholar]
- 33.Munro AW, Noble MA. Fluorescence Analysis of Flavoproteins. Methods Mol. Biol. 1999;131:25–48. doi: 10.1385/1-59259-266-X:25. [DOI] [PubMed] [Google Scholar]
- 34.McKenna CE, Gutheil WG, Song W. A method for preparing analytically pure sodium dithionite. Dithionite quality and observed nitrogenase-specific activities. Biochim. Biophy. Acta (BBA) - Bioenergetics. 1991;1075:109–117. doi: 10.1016/0304-4165(91)90082-r. [DOI] [PubMed] [Google Scholar]
- 35.Dutton PL. Redox Potentiometry: Determination of Midpoint Potentials of Oxidation-Reduction Components of Biological Electron-Transfer Systems. Methods Enzymol. 1978;54:411–435. doi: 10.1016/s0076-6879(78)54026-3. [DOI] [PubMed] [Google Scholar]
- 36.Das A, Trammell SA, Hecht MH. Electrochemical and ligand binding studies of a de novo heme protein. Biophy. Chem. 2006;123:102–112. doi: 10.1016/j.bpc.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 37.Das A, Hecht MH. Peroxidase activity of de novo heme proteins immobilized on electrodes. J. Inorg. Biochem. 2007;101:1820–1826. doi: 10.1016/j.jinorgbio.2007.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bayburt TH, Grinkova YV, Sligar SG. Self-Assembly of Discoidal Phospholipid Bilayer Nanoparticles with Membrane Scaffold Proteins. Nano Letters. 2002;2:853–856. [Google Scholar]
- 39.Nath A, Atkins WM, Sligar SG. Applications of phospholipid bilayer nanodiscs in the study of membranes and membrane proteins. Biochemistry. 2007;46:2059–2069. doi: 10.1021/bi602371n. [DOI] [PubMed] [Google Scholar]
- 40.Das A, Zhao J, Schatz GC, Sligar SG, Van Duyne RP. Screening of Type I and II Drug Binding to Human Cytochrome P450-3A4 in Nanodiscs by Localized Surface Plasmon Resonance Spectroscopy. Anal. Chem. 2009;81:3754–3759. doi: 10.1021/ac802612z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Das A, Grinkova YV, Sligar SG. Redox potential control by drug binding to cytochrome P450 3A4. J. Am. Chem. Soc. 2007;129:13778–13779. doi: 10.1021/ja074864x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shaw AW, Pureza VS, Sligar SG, Morrissey JH. The local phospholipid environment modulates the activation of blood clotting. J. Biol. Chem. 2007;282:6556–6563. doi: 10.1074/jbc.M607973200. [DOI] [PubMed] [Google Scholar]
- 43.Iyanagi T, Makino N, Mason HS. Redox properties of the reduced nicotinamide adenine dinucleotide phosphate-cytochrome P-450 and reduced nicotinamide adenine dinucleotide-cytochrome b5 reductases. Biochemistry. 1974;13:1701–1710. doi: 10.1021/bi00705a023. [DOI] [PubMed] [Google Scholar]
- 44.Vermilion JL, Coon MJ. Identification of the high and low potential flavins of liver microsomal NADPH-cytochrome P-450 reductase. J. Biol. Chem. 1978;253:8812–8819. [PubMed] [Google Scholar]
- 45.Lee SJ, Song Y, Baker NA. Molecular Dynamics Simulations of Asymmetric NaCl and KCl Solutions Separated by Phosphatidylcholine Bilayers: Potential Drops and Structural Changes Induced by Strong Na+-Lipid Interactions and Finite Size Effects. Biophy. J. 2008;94:3565–3576. doi: 10.1529/biophysj.107.116335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nymeyer H, Zhou HX. A Method to Determine Dielectric Constants in Nonhomogeneous Systems: Application to Biological Membranes. Biophy. J. 2008;94:1185–1193. doi: 10.1529/biophysj.107.117770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kakinuma K, Kaneda M, Chiba T, Ohnishi T. Electron spin resonance studies on a flavoprotein in neutrophil plasma membranes. Redox potentials of the flavin and its participation in NADPH oxidase. J. Biol. Chem. 1986;261:9426–9432. [PubMed] [Google Scholar]
- 48.Hata A, Kirino Y, Matsuura K, Itoh S, Hiyama T, Konishi K, Kita K, Anraku Y. Assignment of ESR signals of Escherichia coli terminal oxidase complexes. Biochim. Biophy. Acta (BBA) - Bioenergetics. 1985;810:62–72. doi: 10.1016/0005-2728(85)90206-3. [DOI] [PubMed] [Google Scholar]
- 49.Krishtalik LI, Tae GS, Cherepanov DA, Cramer WA. The redox properties of cytochromes b imposed by the membrane electrostatic environment. Biophy. J. 1993;65:184–195. doi: 10.1016/S0006-3495(93)81050-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodkey FL, Donovan JA., Jr. Oxidation-Reduction Potentials of the Triphosphopyridine Nucleotide System. J. Biol. Chem. 1959;234:677–680. [PubMed] [Google Scholar]
- 51.Sancho J, Gómez-Moreno C. Interaction of ferredoxin-NADP+ reductase from Anabaena with its substrates. Arch .Biochem. Biophy. 1991;288:231–238. doi: 10.1016/0003-9861(91)90189-p. [DOI] [PubMed] [Google Scholar]
- 52.Gutierrez A, Munro AW, Grunau A, Wolf CR, Scrutton NS, Roberts GCK. Interflavin electron transfer in human cytochrome P450 reductase is enhanced by coenzyme binding. Eur. J. Biochem. 2003;270:2612–2621. doi: 10.1046/j.1432-1033.2003.03633.x. [DOI] [PubMed] [Google Scholar]
- 53.Kakinuma K, Kaneda M, Chiba T, Ohnishi T. Electron spin resonance studies on a flavoprotein in neutrophil plasma membranes. Redox potentials of the flavin and its participation in NADPH oxidase. J.Biol.Chem. 1986;261:9426–9432. [PubMed] [Google Scholar]
- 54.Daff SN, Chapman SK, Turner KL, Holt RA, Govindaraj S, Poulos TL, Munro AW. Redox Control of the Catalytic Cycle of Flavocytochrome P-450 BM3. Biochemistry. 1997;36:13816–13823. doi: 10.1021/bi971085s. [DOI] [PubMed] [Google Scholar]
- 55.Hanley SC, Ost TWB, Daff S. The unusual redox properties of flavocytochrome P450 BM3 flavodoxin domain. Biochem.Biophy.Res. Comm. 2004;325:1418–1423. doi: 10.1016/j.bbrc.2004.10.189. [DOI] [PubMed] [Google Scholar]
- 56.Paine MJI, Scrutton NS, Munro AW, Gutierrez A, Roberts CS, Wolf CR. In: Cytochrome P450: Structure, Mechanism and Biochemistry. Montellano P. R. O. d., editor. Kluwer Academic/Plenum; New York: 2005. pp. 115–148. [Google Scholar]
- 57.Noble MA, Munro AW, Rivers SL, Robledo L, Daff SN, Yellowlees LJ, Shimizu T, Sagami I, Guillemette JG, Chapman SK. Potentiometric Analysis of the Flavin Cofactors of Neuronal Nitric Oxide Synthase. Biochemistry. 1999;38:16413–16418. doi: 10.1021/bi992150w. [DOI] [PubMed] [Google Scholar]
- 58.Gao YT, Smith SME, Weinberg JB, Montgomery HJ, Newman E, Guillemette JG, Ghosh DK, Roman LJ, Martasek P, Salerno JC. Thermodynamics of oxidation-reduction reactions in mammalian nitric oxide synthase isoforms. J. Biol. Chem. 2004;279:18759–18766. doi: 10.1074/jbc.M308936200. [DOI] [PubMed] [Google Scholar]
- 59.Ostrowski J, Barber M, Rueger D, Miller B, Siegel L, Kredich N. Characterization of the flavoprotein moieties of NADPH-sulfite reductase from Salmonella typhimurium and Escherichia coli. Physicochemical and catalytic properties, amino acid sequence deduced from DNA sequence of cysJ, and comparison with NADPH-cytochrome P-450 reductase. J. Biol. Chem. 1989;264:15796–15808. [PubMed] [Google Scholar]