Abstract
MicroRNAs (miRNAs) pair with target sequences in the 3’UTR of mRNAs to post-transcriptionally repress gene expression. Here we report that TNF-mediated induction of endothelial adhesion molecules can be regulated by miRNAs that are induced by TNF. Specifically, E-selectin and ICAM-1 are targets of TNF-induced miRNAs miR-31 and miR-17-3p, respectively. Specific antagonism of these TNF-induced miRNAs increased neutrophil adhesion to cultured ECs. Conversely, transfections with mimics of these miRNAs decreased neutrophil adhesion to ECs. These data suggest that miRNAs provide negative feedback control of inflammation.
Keywords: HUVECs, endothelial cell activation, neutrophils, adhesion molecules
INTRODUCTION
TNF, a pro-inflammatory cytokine, induces de novo expression of E-selectin, ICAM-1 and VCAM-1, endothelial cell (EC) surface proteins that bind leukocytes (1-3). miRNAs control gene expression by pairing with partially complementary target sites in mRNA 3’ untranslated regions (3’UTRs), resulting in translational repression and/or mRNA destabilization (4-6). Most miRNAs are constitutively expressed in a lineage-specific manner that peaks during embryological development, but some miRNAs are regulated dynamically in the adult organism. Vascular endothelial growth factor (VEGF) induces a subset of EC miRNAs that regulate cell growth, survival, and angiogenesis (7-9). Other miRNAs are induced in different cell types by a variety of pro-inflammatory stimuli (10-15). A constitutively expressed, EC-restricted miRNA (miR-126) modulates TNF-induced VCAM-1 expression (16). We report here that TNF increases miR-155, -31, -17, and -191 without changing miR-20a, -222 and -126 in human ECs, and that E-selectin and ICAM-1 are targets of TNF-induced miR-31 and miR-17-3p, respectively, regulating neutrophil binding to ECs. These data reveal how inducible miRNAs comprise a negative feedback loop to control inflammation.
MATERIALS AND METHODS
Cell culture
Human cells were obtained from discarded tissues or peripheral blood of de-identified donors under protocols approved by the Yale Human Investigation Committee. HUVECs and human dermal fibroblasts (HDFs) were isolated and serially cultured as described (8, 17, 18). Neutrophils were isolated from whole blood of healthy adult donors (19).
miRNA array analyses
HUVEC miRNA expression was assessed following stimulation for 2 or 24 h with 10ng/ml TNF or buffer control using Exiqon miRCURY™ LNA Arrays as described (8). The data have been deposited in MIAMExpress with ArrayExpress accession number: E-MTAB-150 and accessible through http://www.ebi.ac.uk/microarray-as/ae/. Array results were validated by Northern blot (8, 18) or by quantitative RT-PCR using a mirVanaTM qRT-PCR miRNA Detection Kit (Ambion) (8, 18).
Reporter gene assays
cDNAs encoding the entire 3’UTR of ICAM-1 (1.329 kb) or E-selectin (SELE) (1.883 kb) mRNAs were amplified by RT-PCR from total HUVEC RNA using XhoI and NotI linker/primers and directionally cloned downstream of the Renilla luciferase open reading frame in the psiCHECK2™ vector (Promega) that also contains a constitutively expressed firefly luciferase gene. ICAM-1 or SELE 3’UTRs were cloned in reverse orientation as controls lacking the miRNA target sequence (20). Additionally, the region complemetary to the miR-31 seed sequence in position 94-100 of the human E-selectin 3’UTR, TCTTGCC was scrambled to CTGCCTT (mSELE 3’UTR) and the region complementary to the miR-17-3p seed sequence in the positions 638-644 and 1148-1154 in human ICAM-1 3’UTR, ACTGCAG was scrambled to GTAAGCC (mICAM-1 3’UTR). All constructs were confirmed by sequencing. COS-7 cells (gift of Dr. William Sessa, Yale University) were co-transfected with 1μg of the indicated reporter construct and the indicated miRIDIAN miRNA mimic or negative control mimic sequences (CM) (Dharmacon) using Lipofectamine 2000 (Invitrogen). Renilla luciferase activity was normalized firefly luciferase activity using a Dual-Glo Luciferase Assay System (Promega) and reported as the percentage of control cells co-transfected with the same concentration of CM.
HUVEC transfection
HUVECs were transfected with 40 nM miRIDIAN miRNA mimics or with 60 nM miRIDIAN miRNA inhibitors (Dharmacon) utilizing Oligofectamine (Invitrogen) (8, 18). Control samples were transfected with an equal concentration of CM or inhibitor negative control sequence (CI) as described (8). The effects of transfections with miR-mimics/Inhibitors was assessed by qRT-PCR. miR-31 and miR-17-3p levels were efficiently increased after 18h and persisted after 36 h following transfection and transfection with miR-31 did not affect the endogenous levels of miR-17-3p and vice versa (not shown). Transfection with miRNA inhibitors for miR-31 or miR-17-3p efficiently reduced the TNF stimulated levels of miR-31 and miR-17-3p below levels in untreated cells indicating an efficient inhibition of indicated miRNAs (not shown).
Western blotting
Western blots were performed (8, 18) using mouse mAbs reactive with human ICAM-1 and E-selectin (R&D Systems) under non-reducing conditions or with Tie2 and VE-cadherin polyclonal Abs (Santa Cruz) or Hsp-90 mAb (BD Biosciences) under reducing conditions. Binding of fluorophore-conjugated secondary Abs (Rockland) were visualized using an Odyssey Infrared Imaging System (LI-COR Biotechnology).
Cell surface immunostaining
HUVECs were tansfected and/or TNF-treated as indicated, stained with directly fluor-conjugated mAbs as described (18) and analyzed on a FACScalibur (Becton Dickinson) using CellQuest analysis software collecting 5,000 gated cells per sample. Specific antibodies used in these analyses were FITC or PE directly conjugated and reactive with human E-selectin (R&D Systems) or ICAM-1 (BD Bioscience, Pharmingen), respectively.
Neutrophil adhesion assays
Adhesion of neutrophils to confluent EC monolayer was measured as described (19) with slight modifications. Neutrophils were labeled with calcein-AM (Molecular Probes) for 10 min at 37°C and suspended in PBS containing 1mM magnesium chloride, 0.5mM calcium chloride and 0.1 g/L glucose. Transfected HUVECs were treated with 10ng/ml TNF for 6 h, washed twice in PBS plus and then incubated with 1 ml of labeled neutophils (105) for 15 min at 37°C under static conditions. Samples were fixed and bound neutrophils were counted in five randomly selected 10× fields. Where indicated, HUVECs were incubated with anti-E-selectin and/or anti-ICAM-1 blocking antibodies (R&D Systems) at 50 or 10 μg/ml, respectively, for 1 h before the addition of the neutrophils.
Statistical analyses
Statistical differences between groups were assessed by two-tailed paired Student's t test. Data from qRT-PCR and reporter assays were expressed relative to the control of each experiment and 95.0% confidence intervals (C.I.) were calculated; differences were judged as significant when the C.I. interval did not include the “1” or “100” value.
RESULTS AND DISCUSSION
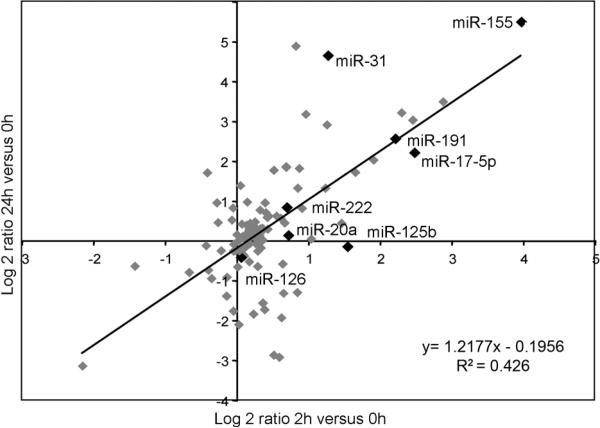
We characterized miRNA expression in HUVECs stimulated for 2 or 24 h with 10ng/ml TNF using microarrays. Several miRNAs are induced after 2 h; some of these are further increased after 24 h whereas others declined (Fig 1A and Table S1). Levels of selected miRNAs species identified in the arrays (Fig. 1A, highlighted in black) were confirmed by Northern blotting (Fig S1A) and/or by qRT-PCR (Fig. S1B). Levels of miR-155, -31, -17-5p, -191 and -125b are increased by TNF but not miR-222, -20a and -126 (Fig. S1A and S1B). Several TNF-induced HUVEC miRNAs were not induced in HDFs, suggesting at least partial specificity of the EC response. miR-155 is highly induced by TNF in both HUVECs and HDFs (Fig. S1C). TNF also induces miR-155 in immune cells (12), in synovial fibroblasts (21), and in lymphomas (9, 22). miR-155 is also upregulated in ECs by VEGF (8).
FIGURE. 1.
TNF regulation of miRNA levels in human ECs. RNA was isolated from ECs that were treated or not for 2 or 24 h with TNF (10 ng/ml). Data are presented as the Log2 ratio of miRNA expression of average treated (2 h or 24 h) versus average untreated (0 h). miR-155, 31, 17-5p, -191, -126, -222, -20a are highlighted in black.
Other miRNAs induced by TNF also overlap with those induced by VEGF (Fig. S2). VEGF induces several components of the c-myc/E2F regulated oncogenic cluster miR-17/92 (including miR-17-5p, -18a, and -20a) (8), but only miR-17-5p is upregulated by TNF (Fig.1B and S1A). While a single primary RNA transcript encodes the entire cluster, differences in efficiency of post-transcriptional processing (23, 24) or selective blockade of pri-miRNA processing (25) may result in differential expression of mature miRNAs. miR-17-5p is upregulated in psoriasis, atopic eczema and systemic lupus erythematosus (26, 27). In human pulmonary aortic ECs, the miR-17/92 cluster is modulated by IL-6 via STAT3 (28). Secretion of IL-6 is induced in ECs by TNF (29) raising the possibility of an indirect autocrine/paracrine effect. In HUVECs, both guide and passenger strands of miR-17 (5p and 3p, respectively) are induced by TNF (Fig. S1A).
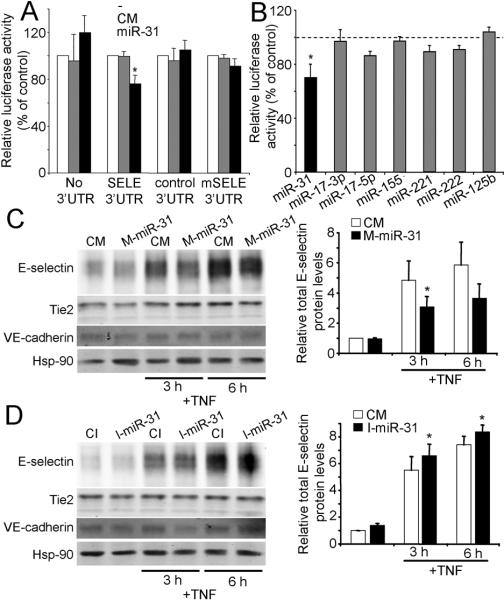
We used miRNA target prediction algorithms (5) to identify a putative binding site in the 3’UTR of E-selectin for miR-31 (Fig. S3A), a highly TNF-induced miRNA (Fig. 1), and an additional site for miR-221 and miR-222 (Fig. S3A), two highly homologous miRNAs, derived from the same pri-microRNA transcript that contain identical seed sequences and are not TNF-regulated. (Fig. 1). The miR-31 predicted site is a canonical 7mer-m8 site supported by an additional 3’ pairing optimally centered on miRNA nt 13-16 (5). The human “seed” region of miR-31 complementary to the sequence in E-selectin 3’UTR is conserved across species (Fig. S3A). The miR-221/222 predicted site is a non-conserved 6mer in the center of the E-selectin 3’UTR (Fig. S3A). We tested these predicted miRNA/mRNA interactions using an E-selectin 3 -UTR luciferase reporter plasmid in COS cells. The relative luciferase activity was significantly reduced (≈ 25%) by co-transfection with miR-31 but not with CM (Fig. 2A). miR-31 did not inhibit reporter vectors lacking E-selectin 3’UTR, with reverse oriented 3’UTR (control 3’ UTR) or with a mutational change in the sequence complementary to the miR-31 seed sequence (mSELE 3’UTR) (Fig. 2A). Luciferase activity of SELE 3’UTR was significantly reduced with as little as 10 nM of miR-31 (Fig. S4A). Luciferase activity of SELE 3’UTR was not significantly reduced by miR-221 and -222 (Fig. 2B). Positioning in the center of a long 3’UTR and lesser efficacy of 6mer sites (5) may explain this lack of effect. SELE 3’UTR luciferase activity was also not affected by TNF-induced miR-17-3p, -17-5p, -155, or -125b (Fig. 2B), none of which are predicted to bind to the E-selectin 3’UTR.
FIGURE. 2.
miR-31 regulation TNF-induced E-selectin expression by targeting the E-selectin 3’UTR. COS-7 cells were co-transected with the indicated constructs and with 40 nM of M-miR-31 or CM (A) or with SELE 3’UTR construct and 40 nM of the indicated miRNA mimics or CM (B). Data are expressed as relative luciferase activity to control samples co-transfected with an equal concentration CM (mean ± SEM of three experiments performed in duplicate). *Significantly different from cells transfected with CM and with control 3’UTR and with the indicated miRNAs, p≤0.05. C and D, Western blot analysis of E-selectin protein levels in ECs, (C) transfected for 12 h with M-miR-31 or CM or (D) with I-miR-31 or CI, in both cases 24 h post-transfection the cells were treated or not with TNF. Hsp-90 was used as loading control. VE-cadherin served as a control protein that is not targeted by miR-31. Graphs in the right show the relative total E-selectin protein levels compared to non-treated CM or CI-transfected controls (mean ± SEM of four experiments). *Significantly different from TNF treated control, p≤0.05.
To analyze the effect of miR-31 on E-selectin protein expression, HUVECs were transfected miR-31 mimic (M-miR-31) for 12 h and and, after a 24 h recovery period, stimulated with TNF for 3 or 6 h, the peaks of E-selectin mRNA and protein expression, respectively (1, 17). By immunoblotting, M-miR-31 reduced TNF-induced E-selectin levels by ≈ 35% at 3h (Fig 2C). Furthermore, E-selectin levels increased by 25 and 20% at 3 and 6h, respectively, when cells were transfected with an antisense miR-31 inhibitor (I-miR-31) prior to TNF stimulation (Fig. 2D). Two other EC proteins, VE-cadherin and Tie2, were not changed by these treatments (even though a putative binding site for miR-31 was predicted in Tie2). By FACS, TNF-induced E-selectin cell surface expression increased when miR-31 was inhibited (Fig S5A).
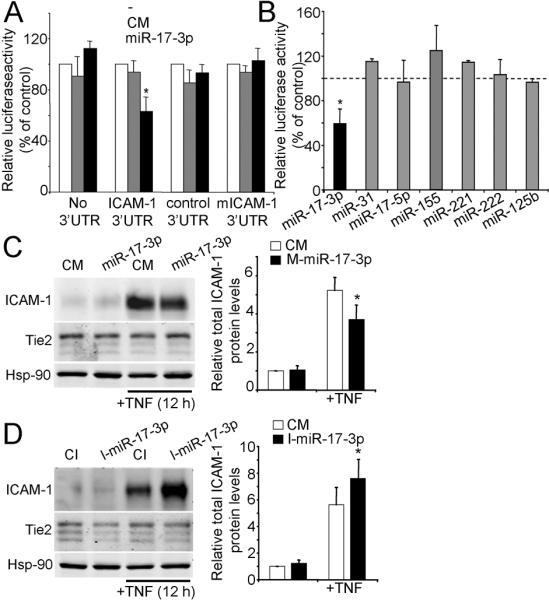
Two canonical 7mer-m8 and one 7mer-A1 putative binding sites (5) in the human 3’UTR of ICAM-1 were predicted for the TNF-induced miR-17-3p and for non-TNF-induced miR-221/222, respectively (Fig. S3B). None of these sites is conserved across species but were independently predicted by 3 different target prediction algorithms (TargetScan, miRBase and RegRNA) and a large fraction of non-conserved sites can be functional (30). In reporter gene assays, miR-17-3p efficiently repressed ICAM-1 3’UTR luciferase activity by ≈ 40% (Fig. 3A). Luciferase activity was unaffected by co-transfection of CM and miR-17-3p did not affect reporter vectors lacking ICAM-1 3’UTR, with reverse oriented 3’UTR (control 3’ UTR) or with a mutation in the sequence complementary to the miR-17-3p seed sequence (mICAM-1 3’UTR). Luciferase activity of ICAM-1 3’UTR was significantly reduced at 40nM (Fig. S4B). ICAM-1 3’UTR luciferase activity was unaffected by miR-221 or -222 (Fig. 3B), suggesting the predicted site is non-functional (5). It was recently reported that miR-222 can regulate expression of ICAM-1 in tumor cells by direct interaction with its 3’UTR (31). A key difference from our experiments is that Kohanbash et al. tested the isolated target sequence rather than testing the sequence in the context of the entire 3’UTR. Contextual features of the 3’UTR, such as secondary structures or local AU-rich regions among others, can govern miRNA/mRNA interactions (5). ICAM-1 3’UTR luciferase activity was not affected by other TNF-induced miRNAs (miR-31, -17-5p, -155, -125b) (Fig. 3B) not predicted to bind to the ICAM1 3’UTR. In general, the miRNA strand which has the lowest thermodynamic stability at its 5’-terminus acts as the mature miRNA (guide strand), and the other (passenger) strand (miRNA*) is degraded. However, in some cases both miRNA strands may accumulate in tissues at significant levels (32) and there are validated examples of trans-regulatory RNAs with demonstrable activities (33). While basal miR-17-3p levels are very low compared to miR-17-5p levels, miR-17-3p is clearly stimulated upon TNF treatment (Fig. S6).
FIGURE 3.
miR-17-3p regulation TNF-induced ICAM-1 expression by targeting the ICAM-1 3’UTR. COS-7 cells were co-transected with the indicated constructs and with 40 nM of M-miR-17-3p or CM (A) or with ICAM-1 3’UTR construct and 40 nM of the indicated miRNA mimics or CM (B). Data are expressed as relative luciferase activity to control samples co-transfected with an equal concentration CM (mean ± SEM of three experiments performed in duplicate). *Significantly different from cells transfected with CM and with control 3’UTR and with the indicated miRNAs, p≤0.05. C and D, Western blot analysis of ICAM-1 protein levels in ECs, (C) transfected for 12 h with M-miR-17-3p or CM or (D) with I-miR-17-3p or CI and, in both cases, cells were treated or not with TNF 24 h post-transfection. Hsp-90 was used as loading control. Tie2 served as a control protein that is not targeted by miR-17-3p. Graphs in the right show the relative total ICAM-1 protein levels compared to non-treated CM or CI-transfected controls (mean ± SEM of four experiments). *Significantly different from TNF treated control, p≤0.05.
To analyze the effect of miR-17-3p on ICAM-1 protein expression, HUVECs were transfected miR-17-3p mimic (M-miR-17-3p) for 12 h and, after a 24 h recovery period, stimulated with TNF for 12h. M-miR-17-3p reduced TNF-induced ICAM-1 expression by ≈ 30% (Fig. 3C). ICAM-1 levels increased up to 35% when cells were transfected with an antisense miR-17-3p inhibitor (I-miR-17-3p) prior to TNF stimulation (Fig. 3D). Tie2 levels were not affected by miR-17-3p (Fig. 3C and D). Although the total protein levels were consistently induced by miR-17-3p antagonism, increases in TNF-induced ICAM-1 cell surface expression observed by FACS did not reach statistical significance (Fig S5B). The increase was significant in some HUVEC donors, consistent with a polymorphic response.
We did not find predicted sites for regulation of VCAM-1 by TNF-induced miRNAs. miR-126 is an EC-specific miRNA that has been shown to regulate the expression of VCAM-1 and limit the adherence of leukocytes to EC (16); miR-126 is not regulated by TNF (Fig. 1 and ref. (16)). miRGen Target interface does not predict any sites for miR-126 in the 3’UTRs of either E-selectin or ICAM-1.
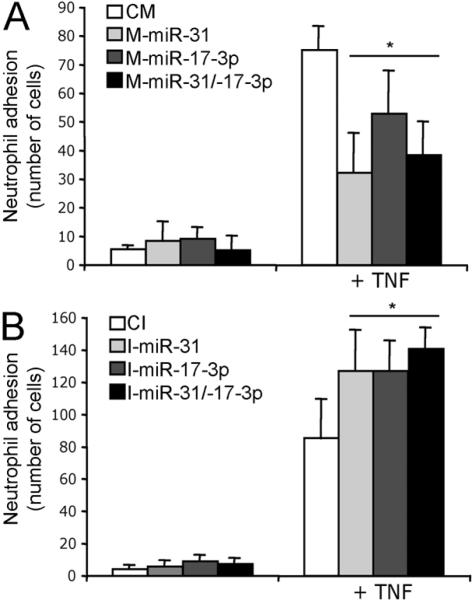
Finally, we tested if the miRNA effects on TNF-induced adhesion molecule expression are functional. As expected, TNF increased neutrophil adhesion to ECs. Exogenous overexpression of M-miR-31, M-miR-17-3p or the combination of the two significantly reduced neutrophil binding (Fig. 4A and Fig S7A) while inhibition of miRNAs miR-31 and/or miR-17-3p increased neutrophil adherence to TNF-stimulated ECs (Fig. 4B and Fig. S7B). It is likely that TNF-induced miRNAs, miR-31 and miR-17-3p, regulate neutrophil adhesion through the regulation of TNF-induced expression of E-selectin and ICAM-1, respectively, although other actions of these miRNAs could also affect neutrophil binding. To test this possibility, we performed experiments combining blocking antibodies with miRNA mimic or inhibitor transfections. As expected, the incubation with blocking antibodies alone profoundly reduced neutrophil binding (Fig S7C and D). Transfection of mimics did not cause any evidence of further reduction in TNF-induced neutrophil binding (Fig. S7C). Transfection of inhibitors produced a small increase in neutrophil binding in the presence of blocking antibodies that did not reach statistical significance (Fig S7D). These experiments do not rule out the possibility that these TNF-induced miRNAs affect neutrophil adhesion by actions on targets other than E-selectin or ICAM-1, but if such effects exist, they are too small to detect when interactions with E-selectin or ICAM-1 are blocked.
FIGURE 4.
Regulation of neutrophil adhesion by miR-31 and/or miR-17-3p. ECs transfected for 12 h with M-miR-31 and/or M-miR17-3p or CM (A) or with I-miR-31 and/or, I-miR-17-3 or CI (B) and 24 h after the transfection period treated or not with TNF. Calcein AM labeled neutrophils were added to the ECs, incubated at 37°C for 15 min. Graphs show the mean of number bound cells from triplicates ± SD of one experiment of three with similar results. *Significantly different from TNF treated control, p≤0.05.
In summary, we illustrate two examples of miRNA-mediated feedback control of expression of adhesion molecules (E-selectin and ICAM-1). This kind of regulation, in which miRNA-directed target repression acts to oppose the overall outcome of a biological response and the regulatory miRNAs are induced by the same signals that cause the response, is likely to be important for fine-tuning of the process (34). Antisense oligos (antimiRs) that target specific miRNAs have recently been shown to be very efficient in vivo (35). In the system we describe, miRNA delivery designed to simulate, rather than antagonize, the function of endogenous mature miRNAs could be useful as an anti-inflammatory therapy.
Supplementary Material
SUPPLEMENTAL DATA
Supplemental Figures and Tables Legends
Fig. S1. Confirmation of miRNA expression of the indicated miRNAs in ECs by qRT-PCR. (D) Examples of miRNA expression by qRT-PCR in HDF after treatment with TNF for the indicated times. (C) and (D), values are relative to the non-treated control and correspond to the mean ± SEM of three experiments performed in duplicate. 5SrRNA was use for normalization. *Significantly different from control, p≤0.05.
Fig. S2. Comparative analysis of VEGF- or TNF-regulated miRNAs. The plot show Log2 ratio of TNF regulated miRNAs (24 h vs 0 h) reported here versus VEGF regulate{Ambros, 2004 #32}d miRNA (9 h vs 0 h) reported in (11). For clarity, only the regulated miRNAs annotated in Sanger miRNA database are shown (predicted miRNA sequences: No known hsa target and miRPLUS-Exiqon sequences have been excluded). Black diamonds show similarly or differently regulated miRNAs. Examples (miR-155, 31, 17-5p, -191, -126, -222, -20a and 18a) are annotated. Grey diamonds show miRNAs solely regulated by TNF and white diamonds show miRNAs solely regulated by VEGF.
Fig. S3. Sequence complementarity of potential miRNA binding sites in human E-selectin and ICAM-1 3’UTR. A, miR-31 and miR-221 binding sites in human E-selectin 3’UTR. Predictions and type of binding site obtained from TargetScan and miRBase are depicted (left). Note that the seed binding site for miR-31 is conserved across species (right). B, miR-17-3p and miR-221 and -221 binding sites in human ICAM-1 3’UTR. Predictions and type of binding sites obtained from TargetScan are depicted. miRBase and RegRNA target databases predict the same binding sites (not shown)
Fig. S4. Dose responseA, COS-7 cells were co-transected for 24 h with the indicated constructs and with increasing concentrations of miR-31 mimic or the corresponding concentrations of CM. COS-7 cells were co-transected for 24 h with SELE 3’UTR construct and 40 nM of the indicated miRNA mimics or CM. B, COS-7 cells were co-transected for 24 h with the indicated constructs and with increasing concentrations of miR-17-3p mimic or the corresponding concentrations of CM. A and B, Data are expressed as relative luciferase activity to control samples co-transfected with an equal concentration CM and correspond to the mean ± SEM of three experiments performed in duplicate. *Significantly different from cells transfected with CM and with control 3’UTR, p≤0.05.
Fig. S5. Surface expression of E-Selectin or ICAM-1. ECs were transfected with I-miR-31 (A) or miR-17-3p and treated or not with TNF for 6 h (A) or 12h (B). E-Selectin or ICAM-1 surface expression was measured by FACS. (a) Plots are from one of three independent experiments with similar results. (B) Relative ICAM-1 surface expression compared to non-treated CI-transfected controls and correspond to the to the means ± SEM of four experiments.
Fig. S6. Relative miRNA expression of miR-17-3p. miRNA expression of miR-17-3p in ECs by qRT-PCR after treatment with TNF for the indicated times. Values are relative to the levels of miR-17-5p in the absence of TNF and correspond to the mean ± SD of three experiments performed in duplicate. 5SrRNA was use for normalization.
Fig. S7. Regulation of neutrophil adhesion by miR-31 and/or miR-17-3p. ECs transfected for 12 h with M-miR-31 and/or M-miR17-3p or CM (A, C) or with I-miR-31 and/or, I-miR-17-3 or CI (B, D) and 24 h after the transfection period treated or not with TNF. Calcein AM labeled neutrophils were added to the ECs, incubated at 37°C for 15 min, and then washed four times and fixed. Representative micrographs used for the quantification are shown. Scale bars represent 100 μm. (C and D) ECs treated as above were incubated with control Ig (50 μg/ml) anti-human E-Selectin (50 μg/ml) and/or anti-human ICAM-1 (10 μg/ml) blocking mAb 1 h before the addition of the neutrophils. Graphs show the relative neutrophil adhesion as a percentage of TNF treated control (mean ± SD) of two experiments performed in duplicate. ANOVA reveals a significant effect of both mimics and inhibitors in control Ig-treated samples, but neither mimics nor inhibitors have statistically significant effects in the presence of blocking mAb.
Table S1. TNF-regulated miRNAs in ECs. Background-subtracted median signals for pixels from each probe spot in both the Cy3 and the Cy5 channels were used for analysis. Data were normalized using spike-in controls. The ratio for treated vs. untreated was calculated based on the average ratio of Cy5/Cy3 of replicates samples from two different islolation of HUVECs. Every isolation consisted of cells from 3 different cords pooled together. Data are presented as the Log2 ratio of miRNA expression of average treated (2 h or 24 h) versus average untreated (0h).
AKNOWLEDGEMENTS
We thank Dr. Anjelica Gonzalez for helpful comments regarding neutrophil adhesion assays; Louise Benson, Gwen Davis-Arrington, Lisa Gras and Todd Jensen for help with cell isolation and culture; Penn Microarray Facility at University of Pennsylvania and the Keck Facility at Yale University for miRNA array analyses; Dr. William C. Sessa for providing COS-7 cells and access to LI-COR and Dr. Carlos Fernández-Hernando for critical discussions in preparation of this manuscript.
This work was supported by NIH grants R01-HL36003 and HL51014 to J.S. Pober and SDG-AHA (0835481N) to Y. Suárez. C. Wang was supported by an NIH Medical Scientist Training Program (T32-GM07205STP).
Abbreviations
- ECs
Endothelial cells
- HDF
human termal fibroblasts
- miRNAs
microRNAs
- UTR
untranslated region
- VEGF
vascular endothelial growth factor
- VE-cadherin
vascular endothelial cadherin
- CM
control mimic sequence
- CI
control inhibitor sequence
Footnotes
CONFLICT OF INTEREST DISCLOSURE
The authors declare no conflicts of interest.
REFERENCES
- 1.Bevilacqua MP, Nelson RM, Mannori G, Cecconi O. Endothelial-leukocyte adhesion molecules in human disease. Annu Rev Med. 1994;45:361–378. doi: 10.1146/annurev.med.45.1.361. [DOI] [PubMed] [Google Scholar]
- 2.Collins T, Read MA, Neish AS, Whitley MZ, Thanos D, Maniatis T. Transcriptional regulation of endothelial cell adhesion molecules: NF-kappa B and cytokineinducible enhancers. Faseb J. 1995;9:899–909. [PubMed] [Google Scholar]
- 3.Madge LA, Pober JS. TNF signaling in vascular endothelial cells. Exp Mol Pathol. 2001;70:317–325. doi: 10.1006/exmp.2001.2368. [DOI] [PubMed] [Google Scholar]
- 4.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 7.Dews M, Homayouni A, Yu D, Murphy D, Sevignani C, Wentzel E, Furth EE, Lee WM, Enders GH, Mendell JT, Thomas-Tikhonenko A. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38:1060–1065. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suarez Y, Fernandez-Hernando C, Yu J, Gerber SA, Harrison KD, Pober JS, Iruela-Arispe ML, Merkenschlager M, Sessa WC. Dicer-dependent endothelial microRNAs are necessary for postnatal angiogenesis. Proc Natl Acad Sci U S A. 2008;105:14082–14087. doi: 10.1073/pnas.0804597105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sonkoly E, Pivarcsi A. Advances in microRNAs: implications for immunity and inflammatory diseases. J Cell Mol Med. 2009;13:24–38. doi: 10.1111/j.1582-4934.2008.00534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bazzoni F, Rossato M, Fabbri M, Gaudiosi D, Mirolo M, Mori L, Tamassia N, Mantovani A, Cassatella MA, Locati M. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc Natl Acad Sci U S A. 2009;106:5282–5287. doi: 10.1073/pnas.0810909106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci U S A. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perry MM, Moschos SA, Williams AE, Shepherd NJ, Larner-Svensson HM, Lindsay MA. Rapid changes in microRNA-146a expression negatively regulate the IL-1beta-induced inflammatory response in human lung alveolar epithelial cells. J Immunol. 2008;180:5689–5698. doi: 10.4049/jimmunol.180.8.5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tili E, Michaille JJ, Cimino A, Costinean S, Dumitru CD, Adair B, Fabbri M, Alder H, Liu CG, Calin GA, Croce CM. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. 2007;179:5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 16.Harris TA, Yamakuchi M, Ferlito M, Mendell JT, Lowenstein CJ. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc Natl Acad Sci U S A. 2008;105:1516–1521. doi: 10.1073/pnas.0707493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pober JS, Bevilacqua MP, Mendrick DL, Lapierre LA, Fiers W, Gimbrone MA., Jr. Two distinct monokines, interleukin 1 and tumor necrosis factor, each independently induce biosynthesis and transient expression of the same antigen on the surface of cultured human vascular endothelial cells. J Immunol. 1986;136:1680–1687. [PubMed] [Google Scholar]
- 18.Suarez Y, Fernandez-Hernando C, Pober JS, Sessa WC. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ Res. 2007;100:1164–1173. doi: 10.1161/01.RES.0000265065.26744.17. [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez AL, El-Bjeirami W, West JL, McIntire LV, Smith CW. Transendothelial migration enhances integrin-dependent human neutrophil chemokinesis. J Leukoc Biol. 2007;81:686–695. doi: 10.1189/jlb.0906553. [DOI] [PubMed] [Google Scholar]
- 20.Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, Ivey KN, Bruneau BG, Stainier DY, Srivastava D. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15:272–284. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stanczyk J, Pedrioli DM, Brentano F, Sanchez-Pernaute O, Kolling C, Gay RE, Detmar M, Gay S, Kyburz D. Altered expression of MicroRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheum. 2008;58:1001–1009. doi: 10.1002/art.23386. [DOI] [PubMed] [Google Scholar]
- 22.Kluiver J, Poppema S, de Jong D, Blokzijl T, Harms G, Jacobs S, Kroesen BJ, van den Berg A. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J Pathol. 2005;207:243–249. doi: 10.1002/path.1825. [DOI] [PubMed] [Google Scholar]
- 23.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 24.Lund E, Dahlberg JE. Substrate selectivity of exportin 5 and Dicer in the biogenesis of microRNAs. Cold Spring Harb Symp Quant Biol. 2006;71:59–66. doi: 10.1101/sqb.2006.71.050. [DOI] [PubMed] [Google Scholar]
- 25.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dai Y, Huang YS, Tang M, Lv TY, Hu CX, Tan YH, Xu ZM, Yin YB. Microarray analysis of microRNA expression in peripheral blood cells of systemic lupus erythematosus patients. Lupus. 2007;16:939–946. doi: 10.1177/0961203307084158. [DOI] [PubMed] [Google Scholar]
- 27.Sonkoly E, Wei T, Janson PC, Saaf A, Lundeberg L, Tengvall-Linder M, Norstedt G, Alenius H, Homey B, Scheynius A, Stahle M, Pivarcsi A. MicroRNAs: novel regulators involved in the pathogenesis of Psoriasis? PLoS ONE. 2007;2:e610. doi: 10.1371/journal.pone.0000610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brock M, Trenkmann M, Gay RE, Michel BA, Gay S, Fischler M, Ulrich S, Speich R, Huber LC. Interleukin-6 modulates the expression of the bone morphogenic protein receptor type II through a novel STAT3-microRNA cluster 17/92 pathway. Circ Res. 2009;104:1184–1191. doi: 10.1161/CIRCRESAHA.109.197491. [DOI] [PubMed] [Google Scholar]
- 29.Jirik FR, Podor TJ, Hirano T, Kishimoto T, Loskutoff DJ, Carson DA, Lotz M. Bacterial lipopolysaccharide and inflammatory mediators augment IL-6 secretion by human endothelial cells. J Immunol. 1989;142:144–147. [PubMed] [Google Scholar]
- 30.Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 31.Ueda R, Kohanbash G, Sasaki K, Fujita M, Zhu X, Kastenhuber ER, McDonald HA, Potter DM, Hamilton RL, Lotze MT, Khan SA, Sobol RW, Okada H. Dicer-regulated microRNAs 222 and 339 promote resistance of cancer cells to cytotoxic T-lymphocytes by down-regulation of ICAM-1. Proc Natl Acad Sci U S A. 2009;106:10746–10751. doi: 10.1073/pnas.0811817106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ro S, Park C, Young D, Sanders KM, Yan W. Tissue-dependent paired expression of miRNAs. Nucleic Acids Res. 2007;35:5944–5953. doi: 10.1093/nar/gkm641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okamura K, Phillips MD, Tyler DM, Duan H, Chou YT, Lai EC. The regulatory activity of microRNA* species has substantial influence on microRNA and 3′ UTR evolution. Nat Struct Mol Biol. 2008;15:354–363. doi: 10.1038/nsmb.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsang J, Zhu J, van Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell. 2007;26:753–767. doi: 10.1016/j.molcel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, Berger U, Gullans S, Kearney P, Sarnow P, Straarup EM, Kauppinen S. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPLEMENTAL DATA
Supplemental Figures and Tables Legends
Fig. S1. Confirmation of miRNA expression of the indicated miRNAs in ECs by qRT-PCR. (D) Examples of miRNA expression by qRT-PCR in HDF after treatment with TNF for the indicated times. (C) and (D), values are relative to the non-treated control and correspond to the mean ± SEM of three experiments performed in duplicate. 5SrRNA was use for normalization. *Significantly different from control, p≤0.05.
Fig. S2. Comparative analysis of VEGF- or TNF-regulated miRNAs. The plot show Log2 ratio of TNF regulated miRNAs (24 h vs 0 h) reported here versus VEGF regulate{Ambros, 2004 #32}d miRNA (9 h vs 0 h) reported in (11). For clarity, only the regulated miRNAs annotated in Sanger miRNA database are shown (predicted miRNA sequences: No known hsa target and miRPLUS-Exiqon sequences have been excluded). Black diamonds show similarly or differently regulated miRNAs. Examples (miR-155, 31, 17-5p, -191, -126, -222, -20a and 18a) are annotated. Grey diamonds show miRNAs solely regulated by TNF and white diamonds show miRNAs solely regulated by VEGF.
Fig. S3. Sequence complementarity of potential miRNA binding sites in human E-selectin and ICAM-1 3’UTR. A, miR-31 and miR-221 binding sites in human E-selectin 3’UTR. Predictions and type of binding site obtained from TargetScan and miRBase are depicted (left). Note that the seed binding site for miR-31 is conserved across species (right). B, miR-17-3p and miR-221 and -221 binding sites in human ICAM-1 3’UTR. Predictions and type of binding sites obtained from TargetScan are depicted. miRBase and RegRNA target databases predict the same binding sites (not shown)
Fig. S4. Dose responseA, COS-7 cells were co-transected for 24 h with the indicated constructs and with increasing concentrations of miR-31 mimic or the corresponding concentrations of CM. COS-7 cells were co-transected for 24 h with SELE 3’UTR construct and 40 nM of the indicated miRNA mimics or CM. B, COS-7 cells were co-transected for 24 h with the indicated constructs and with increasing concentrations of miR-17-3p mimic or the corresponding concentrations of CM. A and B, Data are expressed as relative luciferase activity to control samples co-transfected with an equal concentration CM and correspond to the mean ± SEM of three experiments performed in duplicate. *Significantly different from cells transfected with CM and with control 3’UTR, p≤0.05.
Fig. S5. Surface expression of E-Selectin or ICAM-1. ECs were transfected with I-miR-31 (A) or miR-17-3p and treated or not with TNF for 6 h (A) or 12h (B). E-Selectin or ICAM-1 surface expression was measured by FACS. (a) Plots are from one of three independent experiments with similar results. (B) Relative ICAM-1 surface expression compared to non-treated CI-transfected controls and correspond to the to the means ± SEM of four experiments.
Fig. S6. Relative miRNA expression of miR-17-3p. miRNA expression of miR-17-3p in ECs by qRT-PCR after treatment with TNF for the indicated times. Values are relative to the levels of miR-17-5p in the absence of TNF and correspond to the mean ± SD of three experiments performed in duplicate. 5SrRNA was use for normalization.
Fig. S7. Regulation of neutrophil adhesion by miR-31 and/or miR-17-3p. ECs transfected for 12 h with M-miR-31 and/or M-miR17-3p or CM (A, C) or with I-miR-31 and/or, I-miR-17-3 or CI (B, D) and 24 h after the transfection period treated or not with TNF. Calcein AM labeled neutrophils were added to the ECs, incubated at 37°C for 15 min, and then washed four times and fixed. Representative micrographs used for the quantification are shown. Scale bars represent 100 μm. (C and D) ECs treated as above were incubated with control Ig (50 μg/ml) anti-human E-Selectin (50 μg/ml) and/or anti-human ICAM-1 (10 μg/ml) blocking mAb 1 h before the addition of the neutrophils. Graphs show the relative neutrophil adhesion as a percentage of TNF treated control (mean ± SD) of two experiments performed in duplicate. ANOVA reveals a significant effect of both mimics and inhibitors in control Ig-treated samples, but neither mimics nor inhibitors have statistically significant effects in the presence of blocking mAb.
Table S1. TNF-regulated miRNAs in ECs. Background-subtracted median signals for pixels from each probe spot in both the Cy3 and the Cy5 channels were used for analysis. Data were normalized using spike-in controls. The ratio for treated vs. untreated was calculated based on the average ratio of Cy5/Cy3 of replicates samples from two different islolation of HUVECs. Every isolation consisted of cells from 3 different cords pooled together. Data are presented as the Log2 ratio of miRNA expression of average treated (2 h or 24 h) versus average untreated (0h).