Abstract
As part of efforts to develop improved methods for NMR protein sample preparation and structure determination, the Northeast Structural Genomics Consortium (NESG) has implemented an NMR screening pipeline for protein target selection, construct optimization, and buffer optimization, incorporating efficient microscale NMR screening of proteins using a micro-cryoprobe. The process is feasible because the newest generation probe requires only small amounts of protein, typically 30–200 μg in 8–35 μl volume. Extensive automation has been made possible by the combination of database tools, mechanization of key process steps, and the use of a micro-cryoprobe that gives excellent data while requiring little optimization and manual setup. In this perspective, we describe the overall process used by the NESG for screening NMR samples as part of a sample optimization process, assessing optimal construct design and solution conditions, as well as for determining protein rotational correlation times in order to assess protein oligomerization states. Database infrastructure has been developed to allow for flexible implementation of new screening protocols and harvesting of the resulting output. The NESG micro NMR screening pipeline has also been used for detergent screening of membrane proteins. Descriptions of the individual steps in the NESG NMR sample design, production, and screening pipeline are presented in the format of a standard operating procedure.
Electronic supplementary material
The online version of this article (doi:10.1007/s10858-009-9386-z) contains supplementary material, which is available to authorized users.
Keywords: NMR screening, Micro-cryoprobe, Structural genomics, Construct optimization
Introduction
NMR spectroscopy is a powerful method for providing qualitative and quantitative information about biophysical properties of proteins in solution, including the tertiary structure, the secondary structure distribution, rotational correlation times, internal dynamics, and amide proton exchange rates. A large amount of information can be extracted from a few very simple experiments using a natural abundance or 15N-enriched sample. NMR sample screening provides a valuable approach for identifying protein constructs and solution conditions providing the best quality data, enabling resonance assignments and more extensive biophysical studies. As a result, NMR screening of multiple protein target constructs and solution conditions can greatly impact the efficiency, accuracy, speed, cost and ultimately the success of NMR research in structural biology and structural genomics. As NMR investigations shift more and more towards challenging larger proteins, multi-domain systems, and membrane proteins in detergent solutions, where many parameters and conditions require testing before a suitable sample is obtained, the role and value of screening NMR samples will become even more significant.
The Northeast Structural Genomics Consortium (NESG) (www.nesg.org) has implemented a largely “automated pipeline” for target selection, construct optimization, protein sample production, and efficient microscale screening of protein NMR targets using a micro-cryoprobe requiring only small amounts of protein, typically 10–200 μg of protein sample in 8–35 μl volume for each experiment. A remarkable degree of automation has been made possible by the combination of ad-hoc database tools, mechanization of key process steps and the use of a micro-cryoprobe that provides excellent data while requiring little optimization and manual setup thanks to the small coil diameter and enhanced mass sensitivity. During an initial exploratory phase, screening was conducted using a room temperature 600 MHz 1-mm probe (Bruker TXI 1 Microprobe). Subsequently, we switched to a more advanced 600 MHz 1.7-mm probe (Bruker TCI 1.7 MicroCryoprobe). This probe provides a mass sensitivity (S/N per μg of solute) that is one order of magnitude higher than conventional 5-mm probes. This impressive figure translates into a 1.7-mm probe that is as sensitive as a 5-mm room temperature probe with the sample volume requirement reduced by about an order of magnitude (~30 versus ~300 μl). Using this 1.7-mm micro NMR cryoprobe, a 600 MHz spectrometer system is used seamlessly for target screening, the acquisition of data necessary for backbone and side-chain chemical shift assignments and structure determination of [U-13C,15N] proteins up to 20 kDa.
Microcoil probe technology has been demonstrated to be very valuable for protein NMR studies, particularly for proteins for which only limited quantities are available or for which many conditions need screening. Wüthrich and co-workers have shown that microcoil probes are useful for NMR screening in a miniaturized high throughput structural genomics pipeline (Peti et al. 2005), and have also demonstrated the value of micro probes in screening detergent conditions for membrane protein structure determination by NMR (Zhang et al. 2008). Peti et al. (2004) have determined backbone and simultaneous aliphatic and aromatic side chain resonance assignments on <500 μg quantities of [U-13C, 15N]-labelled proteins using a flow-through HCN z-gradient CapNMR probe (MRM/Prostasis Inc.), and Aramini et al. (2007) have demonstrated complete 3D structure determination of small proteins using <100 μg of protein sample with a 1-mm Bruker Microprobe. In other applications, microcoil NMR probes have been combined with a micromixer to investigate solvent induced conformational transitions in ubiquitin (Kakuta et al. 2003) and capillary HPLC to characterize tryptic fragments of a protein kinase (Hentschel et al. 2005).
This article describes the general overall process of initial NMR sample characterization in the NESG, emphasizing the role of 1D and simple 2D NMR screening in selecting for optimal construct and solvent conditions, as well as determining oligomerization state, in order to validate protein targets for subsequent preparation with double- or triple-labeling (15N, 13C, 2H) for structure determination. We also outline the database infrastructure that has been put in place to allow for flexible implementation of our screening protocols and for harvesting and archiving of the resulting data. Descriptions of the individual steps in the NESG sample design, production and screening pipeline are presented in the following sections.
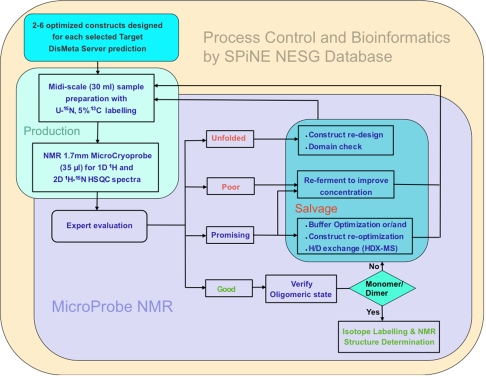
NESG high-throughput pipeline flow chart
Standard operating protocols are fundamental to platforms for high throughput (HTP) data collection. The schematic of Fig. 1 describes the standard operating protocol (SOP) used by the NESG Protein Sample Production group to rank NMR samples as potential targets for structural studies. The SPiNE database software (Bertone et al. 2001; Goh et al. 2003) developed for the NESG project, is the central switchboard that monitors and records information transferred between the bioinformatics, protein production and NMR screening groups. NMR screening runs are set up by software menus, and data is collected and partially processed in an unsupervised fashion. Evaluation of the NMR data and binning of the samples into “good”, “promising”, “poor” or “unfolded” categories is done by expert protein NMR spectroscopists using 1D and 2D spectra generated in a largely automated fashion by the screening pipeline. Samples not suitable for structure determination follow distinct “salvage pathways” based on the sensitivity, solubility, stability (with respect to slow precipitation) of the samples, degree of disorder or ‘unfoldedness’, and oligomerization state. The targets that return for screening under optimized conditions are monitored more closely and screened with a higher degree of scrutiny. This may involve acquiring data at several temperatures, or conducting experiments to characterize backbone dynamics (Farrow et al. 1994). Details of the individual steps in the pipeline are described in the following sections of the manuscript.
Fig. 1.
Schematic representation of standard operating protocol for protein NMR sample screening. The main components of the pipeline are grouped and highlighted (production, salvage, NMR). The NESG SPiNE laboratory information management system is used to oversee the entire process from disorder prediction calculations to data archival
Disorder prediction with dismeta server
The overall process begins when the targeted protein family sequences to be studied undergo a bioinformatics-based analysis that evaluates the protein sequence for regions that are likely to be disordered, and for structural features that may complicate NMR structure analysis such as signal peptides characteristic of secreted proteins, transmembrane helical regions characteristic of membrane proteins, and potential metal binding sites. NESG has developed a web-based tool (DisMeta) to aid HTP construct design. The DisMeta server (www-nmr.cabm.rutgers.edu/bioinformatics/disorder), illustrated in Fig. 2, employs a wide range of disorder prediction tools and several sequence-based structural prediction tools. Genomes encode for many disordered or natively unfolded proteins or protein regions that are very important in protein–protein interactions, modulating signaling, trafficking, and transport of proteins in the cell (Dunker et al. 2002; Iakoucheva et al. 2002; Liu et al. 2002; Plaxco and Gross 2001; Wright and Dyson 1999). However, when approaching a novel protein of unknown structure, the mission of the NESG project is to identify stable folded regions of the protein, obtain the structure by either X-ray or NMR methods, and to gain functional insights based on surface biophysical properties or other bioinformatics analyses. These structures form the basis for further studies of the complete protein, potentially including the structural and functional characterization of disordered regions. Disorder often complicates the goal of obtaining a structure of the ordered core of the protein by suppressing crystal formation or by creating large peaks in NMR spectra that overwhelm the lower intensity peaks originating from the folded portion of the protein.
Fig. 2.
Construct optimization of Staphylococcus saprophyticus SSP0609 protein (NESG target SyR11) and identification of a large disordered segment in the N-terminal region of the protein. a DisMeta report showing disorder in the N-terminal 55 residues of the sequence, b 1H-15N HSQC recorded at 30°C of full length SSP0609 (res. 1-155), c 1H-15N HSQC of the best truncated SSP0609 construct (res. 50-155), d difference spectrum shows the disordered amino-terminal region of the full-length SyR11 protein. NMR structure was solved (PDB ID, 2K3A)
Bioinformatics methods provide means for rapid identification of disordered regions in proteins. As the several disorder prediction software packages that have been developed each approach the problem from a slightly different point of view, we have found it useful to combine a number of these programs under a server and to extract a more robust consensus disorder prediction. The DisMeta Server (www-nmr.cabm.rutgers.edu/bioinformatics/disorder) runs a wide range of disorder prediction software, including DISEMBL (Linding et al. 2003a), DISOPRED2 (Ward et al. 2004), DISPro (Cheng et al. 2005), DRIP-PRED (MacCallum 2006), FoldIndex (Prilusky et al. 2005), FoldUnfold (Galzitskaya et al. 2006), GlobPlot2 (Linding et al. 2003b), IUPred (Dosztanyi et al. 2005), Prelink (Coeytaux and Poupon 2005), RONN (Yang et al. 2005), and VSL2 (Peng et al. 2006). The server has been designed to run standalone or interfaced directly with our target database for batch prediction and parsing of all NESG targets. Fig. 2 shows a representative DisMeta output for the Staphylococcus saprophyticus SSP0609 protein (Rossi et al. 2009) (NESG ID: SyR11), a secreted bacterial antigen with an intrinsically disordered amino-terminal signal peptide that was identified and excluded by this approach.
Construct design
Construct design is carried out largely using automated tools developed by the NESG project. The software uses reports from DisMeta to identify the predicted secondary structure regions, signal peptides characteristic of secreted proteins, trans-membrane segments, and disordered regions. The construct design software will generate multiple alternative constructs for each ‘interest region’ of the structural core (at least 2 constructs per ‘interest region’). If either the N- or C- terminus of alternative constructs is predicted to be located in the middle of a helix or strand, it will be extended to the adjacent predicted loop region. Signal peptides, inter-membrane segments and large disordered regions predicted from the DisMeta report, are excluded from the construct design. For ‘interest regions’ with short disordered regions at the N or C-terminal ends, more constructs will be generated, excluding these flexible region(s) from the designed construct.
The standard E. coli expression systems used in the NESG project produce proteins inside of the cell, and are not generally suitable for producing secreted proteins that may contain disulfide bonds. However, proteins (or domains) containing zero or one Cys residue can be successfully made in intracellular E. coli expression systems, and these are also identified by the construct design software.
Cloning, expression, and purification
Once boundaries of the ordered core of the protein targets are identified, a number of primers are designed using the automated primer design software Primer Prim’er (Everett et al. 2004) and cloned into a set of E. coli pET vectors containing short hexaHis tags at the N- or C-terminal regions. A detailed description of the robotic cloning and expression platform used for NMR protein sample production has been published (Acton et al. 2005). The primers generated for PCR amplification of the targeted coding sequences add 15 base pair regions on each end of the DNA fragment. These sequences overlap with the multiple cloning site of either our pET15 or pET21 T7 expression vector derivatives, allowing for high-throughput, high-efficiency Infusion-based ligation independent cloning (Clontech). Expression vectors are constructed in a high throughput fashion in 96-well format using a Qiagen BioRobot 8000 system (Acton et al. 2005).
The growth medium used for fermentation is MJ9 (Jansson et al. 1996), a modified minimal medium containing a stronger buffering system and supplemental vitamins and trace elements optimized for efficient isotopic-enrichment of proteins. We have found that MJ9 medium can support the same cell density and protein expression levels as rich media such as LB (data not shown), although not as high as rich media such as Terrific Broth (Tartoff and Hobbs 1987). For NMR screening, samples are prepared with 100% 15N and 5% 13C enrichment. The fermentation process begins with transformation of the target expression vector into the appropriate BL21(DE3) strain of E. coli, followed by an LB preculture. This preculture is then used to inoculate a 8 ml overnight culture which is grown to saturation. The entire volumes of each overnight culture is then used to inoculate a 67 ml fermentation in a 100 ml tube (Midi Scale Fermentation) containing MJ9 supplemented with uniformly U-15NH4-salts (1–2 g/l) and a mixture of 100% U-13C-glucose (5%) and unenriched glucose (95%) (3–4 g/l) the sole sources of nitrogen and carbon, respectively. The cultures are incubated with constant aeration with 100% O2 at 37°C until OD600 ~1.0–1.5 units, equilibrated at 17°C, and induced with IPTG. Incubation with vigorous aeration in a 17°C room continues overnight followed by harvesting through centrifugation. Aliquots of the induced cells are taken and SDS polyacrylamide gel electrophoresis analysis is performed on sonicated aliquots to assay for expression and solubility (Acton et al. 2005). The cell pellets are then stored at −20°C.
In the purification stage of sample prepartion, cell pellets are disrupted by sonication, and centrifuged to remove their insoluble portion. The resultant supernatant is then applied to an ÄKTAxpress™ (GE Healthcare) system using two-step protocol consisting of HisTrap HP affinity chromatography followed directly by HiLoad 16/60 Superdex 75 gel filtration chromatography. Samples are concentrated using an Amicon ultrafiltration concentrator (Millipore) to 0.3 to 1.0 mM in 95% H2O/5% 2H2O solution containing an appropriate screening buffer; e.g. 20 mM MES, 200 mM NaCl, 10 mM DTT, 5 mM CaCl2 at pH 6.5, or 20 mM NH4OAc, 200 mM NaCl, 10 mM DTT, 5 mM CaCl2 at pH 5.5 or pH 4.5 (see Supplementary Table S1 entries MJ001, MJ002 and MJ003 for complete reagent list). All buffers contain 50 μM DSS standard for internal referencing (Markley et al. 1998). Aliquots (8 or 35 μl) are then transferred to 1.0-mm or 1.7-mm SampleJet Tubes (Bruker) using Gilson 96-well liquid-handler.
Preparation of samples for NMR screening
Various protein samples to be screened are placed in 96-well plates, and 1.7-mm NMR tubes are robotically filled with 35 μl of protein. NMR spectra are then obtained using a 1.7-mm micro-cryoprobe on a Bruker 600 MHz instrument equipped with a Bruker B-ACS 60 sample handler. The device is controlled by Bruker IconNMR software. Integrated database tools have been developed to reduce operator intervention to a minimum during data acquisition and archival. The IconNMR run execution is scripted based on sample ID and conditions, which are manually entered only one time, upstream at the sample production stage. The robotic autosampler holds up to sixty samples and each sample is locked, tuned and shimmed prior to data acquisition (see image of the dedicated hardware in Supplementary Figure S1). The sample temperature is regulated at 20°C and in the first, completely automated step, 1D 1H NMR spectra with solvent presaturation are acquired using a standard template with optimized values for parameters such as the carrier position, proton sweep width, proton 90° pulse width, saturation power and delay for water-presaturation, and 256 scans for each spectrum. The only human involvement in running a set of 1D screening spectra is the loading of the samples into the autosampler.
1D NMR screening and scoring
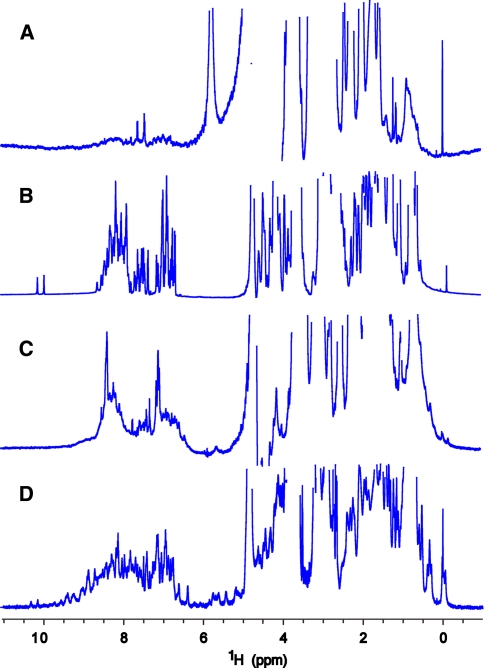
1D 1H NMR spectra with solvent presaturation are acquired for the entire block of 60 samples within about 24 h. The 1D spectra are then scored manually. Illustrative 1D proton spectra are shown in Fig. 3. The criteria for scoring are as follows: (a) signal to noise (b) upfield-shifted methyl protons indicating a folded core formed by aromatic and methyl stacking, and (c) dispersion of the amide protons. Each sample contains 50 μM DSS internal standard that is used for both referencing and evaluating the sensitivity as compared to the concentration measured by UV absorption. Aside from some subjective characteristics, most proteins are easily classified by 1D proton NMR and queued (or not) for 2D 1H-15N HSQC analysis.
Fig. 3.
1D 1H NMR spectra with H2O presaturation of representative NESG targets obtained with a 1.7-mm micro NMR cryoprobe at 20°C with corresponding NESG target IDs. a HR3159A spectrum scores as “poor” on account of broad poorly dispersed resonances. b LmR69A spectrum scores “unfolded” due to sharp and poorly dispersed peaks in all regions. c EwR71A spectrum scores as “promising” with the presence of upfield-shifted methyl peaks but crowding of the amide region (7–9 ppm), and relatively broad peaks. d NsR431C spectrum scores “good” with sharp uniform intensity and upfield-shifted methyls
2D NMR screening and scoring
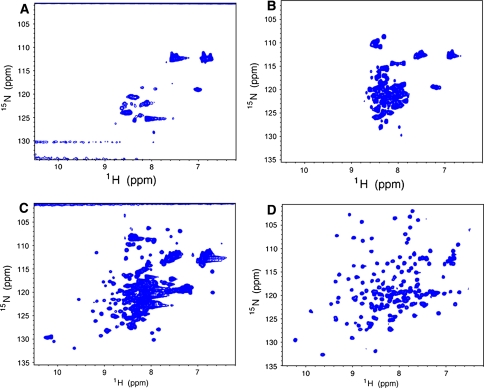
2D 1H-15N HSQC spectra are queued with the best “good” samples heading the queue. Each of the 2D spectra is typically run for about 2 h at 20°C. An attempt to acquire 2D spectra on all the samples is made unless the protein shows no signal (poor classification), but a maximum of 10 h is allotted for the least concentrated sample. In addition, digital resolution in the 15N dimension is adjusted for larger proteins (18–20 kDa) as deemed necessary to obtain a better spectrum for scoring. Conversely, weak samples are run with emphasis on sensitivity (e.g. 64 points in 15N evolution and 4× the number of scans).
The spectrometer routines allow for rapid 2D processing and peak-picking of the 2D spectra. Scoring is based upon dispersion and the number of resolved peaks. The dispersion and uniformity of backbone and side-chain amide intensities are considered in the subjective scoring criteria. While 1D 1H NMR provides key elements for scoring the protein, the presence of unfolded or exchanged broadened regions is determined much more accurately by 2D 1H-15N HSQC. Representative 2D spectra acquired using optimal parameters are shown in Fig. 4. Some targets show promising 1H-15N HSQC and good 1D dispersion, but have poor solubility or regions of disorder that complicate the structure determination. Such targets are further screened for more appropriate solvent using a standard set of 12 buffers (Supplementary Table S1) and(or) directed to amide hydrogen deuterium exchange with mass spectrometry (HDX-MS) (Sharma et al. 2009) to identify more precisely the locations of disordered N- or C-terminal segments which can be removed in order to improve the spectrum. Several problematic targets have yielded high quality NMR structures by this approach (Sharma et al. 2009). Rotational correlation times, based on 15N longitudinal and transverse relaxation time (T 1 and T 2) measurements made using 1D NMR spectra (described below), are also obtained with the 1.7-mm micro cryoprobe at this stage of screening, providing confirmation of the oligomerization state initially determined by gel filtration. For larger proteins and homodimers, these measurements also indicate whether or not structure determination can be carried out with a [U-13C,15N] sample, or if a triply-labeled [U-2H,13C,15N] sample will be required.
Fig. 4.
1H-15N HSQC spectra of NESG targets recorded with a 1.7-mm micro-cryoprobe at 20°C labeled with corresponding NESG target IDs. a NR26 scored as “poor” due to low sensitivity and low peak count. b HR5272A scored as “unfolded”, as all backbone amides are in the random coil region (∼7–9 ppm), and the side-chain amide resonances are largely overlapped. c GmR58A scored as “promising”, as the spectrum shows the simultaneous presence of folded regions and unfolded segments with higher peak intensity. d NsR431C scored as “good”, with uniform intensity, good dispersion, and correct peak count. In our pipeline, target GmR58A is a candidate for construct optimization using HDX-MS, while target NsR431C was promoted for uniform 15N,13C-enrichment and structure determination
Throughput and bottlenecks
Using the robotic sample changer with the 1.7-mm micro cryoprobe, about 60 1D spectra are acquired in ~24 h. These data are used to prioritize samples for 2D 1H-15N HSQC data collection. 2D spectra are each acquired for 2–6 h, depending on the sensitivity of the sample and size of the target molecule. Generally, 2D screening for a sixty-sample batch of targets is completed in about a week. For the NESG NMR structure production pipeline, scheduling on the spectrometer is adjusted to allow for the screening of about one hundred samples in each 2-week period of each month.
One of the limitations of the automatic screening is fluctuation of the sample volume, due to evaporation, that causes lock failure. Care must be taken to ensure that volume fluctuation be kept within 5 μl by acquiring data as quickly as possible given the available sample concentration or by sealing the microtube. The automation software is configured to send error notification via e-mail to the operator. One obvious solution can be to start with slightly larger sample volumes i.e., 40–45 μl samples. However, samples prepared with larger volumes have been observed to result in poorer performance of the autoshimming software that is run in setting up each sample.
Data management
The NESG has developed a dedicated, flexible and secure database, SPiNE that tracks the overall sample flow through all the stages from target selection to PDB deposition (Bertone et al. 2001; Goh et al. 2003). For the purpose of microscale NMR screening, the SPiNE database was expanded to handle purified protein samples in 96-well format. Sample tube identifiers (PST IDs) are defined in each position of the block. The hand-off of each block of protein samples for NMR screening also occurs through SPiNE. The database directly generates the instructions for the spectrometer in a simple text file that is downloaded to the instrument and controls data collection for each sample. This would otherwise be a laborious manual task to be conducted from within the IconNMR interface. The scripts are conveniently downloaded and saved in the appropriate directory on the 600 MHz NMR spectrometer, they are pre-filled with the (1) block label ID, (2) PST IDs, and (3) the 1D and 2D acquisition parameters. After NMR acquisition, the raw data and processed spectra are uploaded into SPiNE for viewing. Fig. 5 shows snapshots of the operations carried out in SPiNE for the automated run and the database upload. In addition to the initial screening data, the subsequent fate of the protein is also tracked by the database. The salvage pathway is defined and the new redesigned samples are re-screened and compared to the original data.
Fig. 5.
SPiNE database views showing a “Screening Block” page, push-button generation of IconNMR script for B-ACS 60 sample changer run, and view of the NMR screening record, including 1D and 2D spectra that have been uploaded to the database and made available for viewing and analysis. HSQC NMR spectra for hundreds of NESG target proteins that are stored in SPiNE are publicly available from the NESG Home Page (www.nesg.org) under the menu item Statistics
Salvage pathways
For proteins providing marginal quality (e.g. “Promising”) HSQC spectra, several “salvage” processes have been developed to provide improved solvent conditions and/or construct design. Some of the most effective strategies include sample buffer optimization by NMR and amide hydrogen deuterium exchange with mass spectrometry detection (HDX-MS) for construct optimization (Sharma et al. 2009).
Buffer optimization
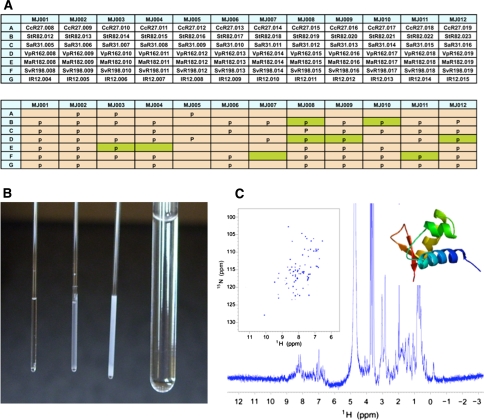
The need for buffer optimization is established following the first screening run carried out with a standard buffer at pH 6.5 (or pH modified to avoid proximity to the pI of the protein, where aggregation and precipitation can occur) and 200 mM ionic strength (see Supplementary Table S1 for exact descriptions of buffers). If the sample is deemed adequate for structure determination but not stable enough with respect to slow precipitation during the data acquisition time periods, 4–10 days, required for a structure determination, then the sample is directed to buffer optimization. The microprobe is particularly useful here because it reduces the sample requirement for the process. A stepwise description of sample preparation for buffer optimization is provided in the Supplementary Materials. Briefly, purified protein samples are exchanged into twelve (12) buffer conditions using a desalting column and transferred to micro NMR tubes for NMR screening. Supplementary Table S1 contains a list the twelve buffers commonly employed by the NESG project in buffer optimization, and which have a proven track record of success in our hands. Under special circumstances that depend on the pI of an individual protein, buffers with pH up to 8.0 may be evaluated. Fig. 6a illustrates how improved buffer conditions are found for a set of seven targets prepared as described above and screened in twelve different buffer conditions from our set (MJ001-012). Inspection of the sample tube for precipitation is often ineffective in establishing the viability of a buffer condition (Fig. 6b), and many samples that show some precipitation actually contain soluble protein in sufficient amounts for structure determination by NMR as shown for NESG target StR82 (Fig. 6c) (Aramini et. al DOI:10.2210/pdb2jt1/pdb).
Fig. 6.
a Buffer optimization block of seven NESG protein targets (NESG IDs: CcR27, StR82, SaR31, VpR162, MaR182, SvR196, IR12) in twelve buffers (MJ001 to MJ0012, Supplementary Table S1). Protein signal is detected (pale green) even though the tube shows abundant precipitation indicated by a ‘p’ in the grid. b From the left, 1-mm microtubes showing no or increasing degrees of precipitation of target StR82 in different buffers. Signal is detected in the center tube, but not in the clear (left) or the heavily precipitated (right) microtubes. c The best spectra for pefl from S. typhimurium (NESG target StR82) were recorded at 20°C in 450 mM NaCl at pH 6.5 (buffer MJ008). The insets show the 2D 1H-15N HSQC and the ribbon diagram of the structure solved using optimal conditions (PDB ID: 2JT1)
Amide hydrogen deuterium exchange with mass spectrometery detection (HDX-MS) for construct optimization
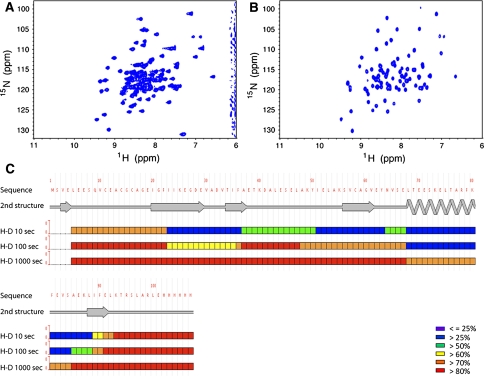
Disorder prediction methods described above using the DisMeta server have improved the outcome of our production pipeline tremendously. In some instances, however, more detailed biophysical analysis is required to gain the information necessary to solve the protein structure. The NESG NMR sample preparation pipeline employs HDX-MS (Englander 2006; Sharma et al. 2009; Woods and Hamuro 2001) for the identification of the exact boundaries of disordered N- and C-terminal segments for optimizing constructs scored as “Promising” in the microscale NMR screening pipeline. The technique, which requires ~50 μg of unlabeled protein, revolves around the concept that backbone amide protons in solvent-inaccessible ordered regions of a protein will exchange with solvent deuterium at a slower rate than those in flexible or disordered regions. The results are depicted in a so-called “heat map”, in which amide 1H/2H exchange rates are represented by colors ranging from blue (slow amide proton exchange) to red (fast amide proton exchange). HDX-MS has been applied to optimize design of protein constructs providing improved crystallization success compared to the full-length proteins (Pantazatos et al. 2004; Spraggon et al. 2004). In a recent pilot study on a small set of targets from the NESG (Sharma et al. 2009), we demonstrated the feasibility of using HDX-MS to design truncated constructs yielding NMR spectra that are more amenable for NMR structure determination, while maintaining the structural integrity of the remaining ordered region of the protein. More recently, the method has been used to improve construct design of promising samples, yielding good spectra and 3D structures for some dozen proteins. The HDX-MS analysis used in construct optimization of V. parahaemolyticus VPA0419 (NESG target VpR68) serves as a representative example of how the technique can yield dramatic improvements to the quality of 1H-15N HSQC NMR spectra, and provide structures of protein targets that could not otherwise be studied (Fig. 7). Note that, in contrast to the predicted disorder map provided by the consensus DisMeta server for this protein (not shown), which indicated a lack of consensus among the different disorder prediction methods and hence a low reliability for identifying disordered segments, the HDX-MS results reveal that both amino and carboxyl-terminal segments of VpR68 are disordered. The solution NMR structure of a HDX-MS -optimized construct, obtained by removing 27 disordered N-terminal residues, was subsequently solved by the NESG consortium (Singarapu et al., DOI: 10.2210/pdb2jz5/pdb).
Fig. 7.
HDX-MS -based construct optimization of VPA0419 protein from V. parahaemolyticus (NESG target, VpR68). a 1H-15N HSQC spectra (20°C) of full length (res. 1-109) and b construct optimized (res. 17-99) solved by NMR (PDB ID: 2JZ5). c HDX-MS results for full-length of V. parahaemolyticus VPA0419 (10, 100, and 1,000 s 1H/2H exchange durations), the legend indicates the color-coded H/D exchange rate
Using NMR to determine oligomerization states of proteins
A priori knowledge of the oligomerization state of a protein is critical to sample labeling choice and to accurate protein structure determination by solution NMR techniques. The principal approaches employed in the NESG for elucidation of the oligomerization state of targets selected for NMR structure determination include: (1) analytical gel filtration chromatography, (2) static light scattering, and (3) 1D 15N NMR relaxation measurements. Our standard protocol for analytical gel filtration with static light scattering detection has been described elsewhere (Acton et al. 2005). Here we discuss our standard procedure for measurements of rotational correlation times from 1H-detected 1D 15N relaxation measurements, which are executed on the NMR sample to be studied as part of the microscale NMR screening.
The rotational correlation time of a protein in solution is the time for a protein to rotate one radian. For an approximately spherical globular protein, the rotational correlation time (τc), is related to its effective hydrodynamic radius (a), and thus its oligomerization state, according to the Stokes–Einstein equation (Eq. 1), where η is viscosity and T is temperature.
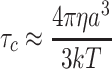 |
1 |
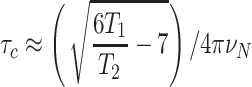 |
2 |
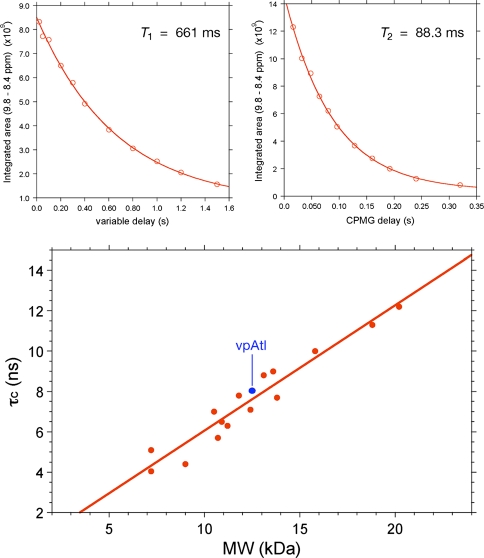
In the limit of slow molecular motion (τc > > 0.5 ns), the correlation time of a protein is related to the ratio of the longitudinal (T 1) and transverse (T 2) 15N relaxation times, and nuclear frequency (νN) according to Eq. 2, which is derived from Eq. 8 in Kay et al. (Kay et al. 1989) by considering only J(0) and J(ω) spectral densities and neglecting higher frequency terms. In practice, global 15N T 1 and T 2 relaxation times for an unknown protein target can be obtained quickly (ca. 1 h) on a 1.7-mm MicroCyroprobe using 1D 1H-detected 15N-edited relaxation experiments (Farrow et al. 1994) by fitting the integrated signal in the backbone amide 1H region of the spectrum as a function of delay time to an exponential decay (Fig. 8). One then computes the correlation time using Eq. 2, and compares it to a standard curve of τc versus protein molecular weight (MW) obtained at the same temperature on a series of known monomeric proteins of varying size (Fig. 8). Supplementary Table S2 provides the values used for τc versus MW plot determined for protein samples studied by the NESG consortium. As a general rule of thumb, the τc of a monomeric protein in solution at 20°C, in nanoseconds, is approximately 0.6 times its molecular weight (kDa). This approach is reliable up to MW ≈25 kDa, where accurate measurement of the diminishing 15N T 2 becomes more error prone. The 15N -1H TRACT NMR approach (Lee et al. 2006) expands the range of measurable τc by measuring the 15N Rα and Rβ fast and slow relaxing components arising from the TROSY effect. The NESG uses TRACT less routinely and primarily for larger proteins, homodimers, or membrane-associated proteins.
Fig. 8.
15N T 1 and T 2 relaxation data for [U-5%-13C, U-15N] vpAtl (NESG ID, VpR247). The data were acquired on a Bruker AVANCE 600 MHz spectrometer with 1.7 MicroCryoprobe at 25°C using pseudo-2D 15N T 1 and T 2 gradient experiments. T 1 spectra were acquired with delays, T = 20, 50, 100, 200, 300, 400, 600, 800, 1,000, 1,200 and 1,500 ms, and a relaxation delay of 3 s. T 2 spectra were acquired with CPMG delays, T = 16, 32, 48, 64, 80, 96, 128, 160, 192, 240 and 320 ms, and with a relaxation delay of 1.5 s. (Top): 15N T 1 and T 2 values were extracted by plotting the decay of integrated 1HN intensity between δ ≈ 8.4 to 9.8 ppm and fitting the curves with standard exponential equations using the program ‘t1guide’ within Topspin2.1 (Bruker BioSpin). (Bottom): Plot of τc versus protein molecular weight for known monomeric NESG targets of ranging size (taking into account isotope enrichment as well as affinity tags in the sequence). 15N T 1/T 2 data for all monomeric proteins used for the τc versus MW plot (red) were obtained on the same Bruker 600 MHz spectrometer at 25°C, and analyzed as described above. Using this approach, we obtain a τc of 8.0 ns for vpAtl (blue), which is consistent with a monomer
Discussion
In this paper we have described, the salient aspects of our microscale NMR screening pipeline. Sample production, automatic data setup and acquisition, data analysis and data archiving have been optimized and streamlined. The aim was to obtain the most accurate results in the shortest possible time and in a cost effective manner, while minimizing operator intervention and error. This was made possible by (1) expanding the SPiNE database tools to oversee and coordinate the the microscale NMR screening pipeline, (2) application of state-of-the-art 600 MHz 1.7-mm micro cryoprobe technology adopted to reduce sample requirements and to better utilize limited NMR resources, (3) introduction of bioinformatics (DisMeta and other construct optimization software), and experimental techniques (HDX-MS) to identify and remove disordered N- or C-terminal segments from the protein construct. In its current form, the NESG microscale NMR screening pipeline is a combination of dedicated in-house database development, commercially available hardware, established proteomics techniques, and optimal expert input. The NESG micro NMR screening pipeline has also been used for detergent screening of membrane proteins (Mao et al. 2009). The strategy has proven essential for the success of the NESG consortium NMR structure production, and may provide a useful template for structural biology programs exploring samples and conditions suitable for studies of complex system that may require construct and/or buffer optimization.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We thank A. Eletski, K. Singarapu, Y. Tang and R. Mani for helpful discussions, and for datasets used in the production of this manuscript. This work was supported by the National Institutes of General Medical Science Protein Structure Initiative program, grant U54 GM074958.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
The authors Paolo Rossi and G. V. T. Swapna contributed equally to this work.
References
- Acton TB, Gunsalus KC, Xiao R, Ma LC, Aramini J, Baran MC, Chiang YW, Climent T, Cooper B, Denissova NG, Douglas SM, Everett JK, Ho CK, Macapagal D, Rajan PK, Shastry R, Shih LY, Swapna GV, Wilson M, Wu M, Gerstein M, Inouye M, Hunt JF, Montelione GT. Robotic cloning and protein production platform of the northeast structural genomics consortium. Methods Enzymol. 2005;394:210–243. doi: 10.1016/S0076-6879(05)94008-1. [DOI] [PubMed] [Google Scholar]
- Aramini JM, Rossi P, Anklin C, Xiao R, Montelione GT. Microgram-scale protein structure determination by NMR. Nat Methods. 2007;4:491–493. doi: 10.1038/nmeth1051. [DOI] [PubMed] [Google Scholar]
- Bertone P, Kluger Y, Lan N, Zheng D, Christendat D, Yee A, Edwards AM, Arrowsmith CH, Montelione GT, Gerstein M. SPINE: an integrated tracking database and data mining approach for identifying feasible targets in high-throughput structural proteomics. Nucleic Acids Res. 2001;29:2884–2898. doi: 10.1093/nar/29.13.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Sweredoski MJ, Baldi P (2005) Accurate prediction of protein disordered regions by mining protein structure data. Data Mining and Knowledge Discovery 11
- Coeytaux K, Poupon A. Prediction of unfolded segments in a protein sequence based on amino acid composition. Bioinformatics. 2005;21:1891–1900. doi: 10.1093/bioinformatics/bti266. [DOI] [PubMed] [Google Scholar]
- Dosztanyi Z, Csizmok V, Tompa P, Simon I. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J Mol Biol. 2005;347:827–839. doi: 10.1016/j.jmb.2005.01.071. [DOI] [PubMed] [Google Scholar]
- Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Intrinsic disorder and protein function. Biochemistry. 2002;41:6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- Englander SW. Hydrogen exchange and mass spectrometry: a historical perspective. J Am Soc Mass Spectrom. 2006;17:1481–1489. doi: 10.1016/j.jasms.2006.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett JK, Acton TB, Montelione GT. Primer prim’er: a web based server for automated primer design. J Struct Funct Genom. 2004;5:13–21. doi: 10.1023/B:JSFG.0000029238.86387.90. [DOI] [PubMed] [Google Scholar]
- Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15 N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- Galzitskaya OV, Garbuzynskiy SO, Lobanov MY. FoldUnfold: web server for the prediction of disordered regions in protein chain. Bioinformatics. 2006;22:2948–2949. doi: 10.1093/bioinformatics/btl504. [DOI] [PubMed] [Google Scholar]
- Goh CS, Lan N, Echols N, Douglas SM, Milburn D, Bertone P, Xiao R, Ma LC, Zheng D, Wunderlich Z, Acton T, Montelione GT, Gerstein M. SPINE 2: a system for collaborative structural proteomics within a federated database framework. Nucleic Acids Res. 2003;31:2833–2838. doi: 10.1093/nar/gkg397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschel P, Krucker M, Grynbaum MD, Putzbach K, Bischoff R, Albert K. Determination of regulatory phosphorylation sites in nanogram amounts of a synthetic fragment of ZAP-70 using microprobe NMR and on-line coupled capillary HPLC-NMR. Magn Reson Chem. 2005;43:747–754. doi: 10.1002/mrc.1628. [DOI] [PubMed] [Google Scholar]
- Iakoucheva LM, Brown CJ, Lawson JD, Obradovic Z, Dunker AK. Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol. 2002;323:573–584. doi: 10.1016/S0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- Jansson M, Li YC, Jendeberg L, Anderson S, Montelione BT, Nilsson B. High-level production of uniformly 15 N- and 13C-enriched fusion proteins in Escherichia coli. J Biomol NMR. 1996;7:131–141. doi: 10.1007/BF00203823. [DOI] [PubMed] [Google Scholar]
- Kakuta M, Jayawickrama DA, Wolters AM, Manz A, Sweedler JV. Micromixer-based time-resolved NMR: applications to ubiquitin protein conformation. Anal Chem. 2003;75:956–960. doi: 10.1021/ac026076q. [DOI] [PubMed] [Google Scholar]
- Kay LE, Torchia DA, Bax A. Backbone dynamics of proteins as studied by 15 N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- Lee D, Hilty C, Wider G, Wuthrich K. Effective rotational correlation times of proteins from NMR relaxation interference. J Magn Reson. 2006;178:72–76. doi: 10.1016/j.jmr.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Linding R, Jensen LJ, Diella F, Bork P, Gibson TJ, Russell RB. Protein disorder prediction: implications for structural proteomics. Structure. 2003;11:1453–1459. doi: 10.1016/j.str.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Linding R, Russell RB, Neduva V, Gibson TJ. GlobPlot: exploring protein sequences for globularity and disorder. Nucleic Acids Res. 2003;31:3701–3708. doi: 10.1093/nar/gkg519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Tan H, Rost B. Loopy proteins appear conserved in evolution. J Mol Biol. 2002;322:53–64. doi: 10.1016/S0022-2836(02)00736-2. [DOI] [PubMed] [Google Scholar]
- MacCallum RM (2006) Order/disorder prediction with self organising maps
- Mao L, Tang Y, Vaiphei T, Shimazu T, Kim SG, Mani REW, Montelione GT, Inouye M (2009) Production of membrane proteins for NMR studies without purification. J Struc Funct Genom. doi:10.1007/s10969-009-9072-0 [DOI] [PMC free article] [PubMed]
- Markley JL, Bax A, Arata Y, Hilbers CW, Kaptein R, Sykes BD, Wright PE, Wuthrich K. Recommendations for the presentation of NMR structures of proteins and nucleic acids. IUPAC-IUBMB-IUPAB inter-union task group on the standardization of data bases of protein and nucleic acid structures determined by NMR spectroscopy. J Biomol NMR. 1998;12:1–23. doi: 10.1023/A:1008290618449. [DOI] [PubMed] [Google Scholar]
- Pantazatos D, Kim JS, Klock HE, Stevens RC, Wilson IA, Lesley SA, Woods VL., Jr Rapid refinement of crystallographic protein construct definition employing enhanced hydrogen/deuterium exchange MS. Proc Natl Acad Sci U S A. 2004;101:751–756. doi: 10.1073/pnas.0307204101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng K, Radivojac P, Vucetic S, Dunker AK, Obradovic Z. Length-dependent prediction of protein intrinsic disorder. BMC Bioinform. 2006;7:208. doi: 10.1186/1471-2105-7-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peti W, Norcross J, Eldridge G, O’Neil-Johnson M. Biomolecular NMR using a microcoil NMR probe–new technique for the chemical shift assignment of aromatic side chains in proteins. J Am Chem Soc. 2004;126:5873–5878. doi: 10.1021/ja039779d. [DOI] [PubMed] [Google Scholar]
- Peti W, Page R, Moy K, O’Neil-Johnson M, Wilson IA, Stevens RC, Wuthrich K. Towards miniaturization of a structural genomics pipeline using micro-expression and microcoil NMR. J Struct Funct Genom. 2005;6:259–267. doi: 10.1007/s10969-005-9000-x. [DOI] [PubMed] [Google Scholar]
- Plaxco KW, Gross M. Unfolded, yes, but random? Never! Nat Struct Biol. 2001;8:659–660. doi: 10.1038/90349. [DOI] [PubMed] [Google Scholar]
- Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckmann JS, Silman I, Sussman JL. FoldIndex: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics. 2005;21:3435–3438. doi: 10.1093/bioinformatics/bti537. [DOI] [PubMed] [Google Scholar]
- Rossi P, Aramini JM, Xiao R, Chen CX, Nwosu C, Owens LA, Maglaqui M, Nair R, Fischer M, Acton TB, Honig B, Rost B, Montelione GT. Structural elucidation of the Cys-His-Glu-Asn proteolytic relay in the secreted CHAP domain enzyme from the human pathogen Staphylococcus saprophyticus. Proteins. 2009;74:515–519. doi: 10.1002/prot.22267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Zheng H, Huang YJ, Ertekin A, Hamuro Y, Rossi P, Tejero R, Acton TB, Xiao R, Jiang M, Zhao L, Ma LC, Swapna GV, Aramini JM, Montelione GT. Construct optimization for protein NMR structure analysis using amide hydrogen/deuterium exchange mass spectrometry. Proteins. 2009;76:882–894. doi: 10.1002/prot.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spraggon G, Pantazatos D, Klock HE, Wilson IA, Woods VL, Jr, Lesley SA. On the use of DXMS to produce more crystallizable proteins: structures of the T. maritima proteins TM0160 and TM1171. Protein Sci. 2004;13:3187–3199. doi: 10.1110/ps.04939904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartoff KD, Hobbs CA (1987) Bethesda research laboratory focus, vol 9, p 12
- Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Woods VL, Jr, Hamuro Y. High resolution, high-throughput amide deuterium exchange-mass spectrometry (DXMS) determination of protein binding site structure and dynamics: utility in pharmaceutical design. J Cell Biochem Suppl. 2001;37:89–98. doi: 10.1002/jcb.10069. [DOI] [PubMed] [Google Scholar]
- Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- Yang ZR, Thomson R, McNeil P, Esnouf RM. RONN: the bio-basis function neural network technique applied to the detection of natively disordered regions in proteins. Bioinformatics. 2005;21:3369–3376. doi: 10.1093/bioinformatics/bti534. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Horst R, Geralt M, Ma X, Hong WX, Finn MG, Stevens RC, Wuthrich K. Microscale NMR screening of new detergents for membrane protein structural biology. J Am Chem Soc. 2008;130:7357–7363. doi: 10.1021/ja077863d. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.