Abstract
AIM: To investigate the pathophysiology of irritable bowel syndrome (IBS) by comparing the global mucosal metabolic profiles of IBS patients with those of healthy controls.
METHODS: Fifteen IBS patients fulfilling the Rome II criteria, and nine healthy volunteers were included in the study. A combined lipidomics (UPLC/MS) and metabolomics (GC × GC-TOF) approach was used to achieve global metabolic profiles of mucosal biopsies from the ascending colon.
RESULTS: Overall, lipid levels were elevated in patients with IBS. The most significant upregulation was seen for pro-inflammatory lysophosphatidylcholines. Other lipid groups that were significantly upregulated in IBS patients were lipotoxic ceramides, glycosphingolipids, and di- and triacylglycerols. Among the metabolites, the cyclic ester 2(3H)-furanone was almost 14-fold upregulated in IBS patients compared to healthy subjects (P = 0.03).
CONCLUSION: IBS mucosa is characterised by a distinct pro-inflammatory and lipotoxic metabolic profile. Especially, there was an increase in several lipid species such as lysophospholipids and ceramides.
Keywords: Functional gastrointestinal diseases, Irritable bowel syndrome, Histopathology
INTRODUCTION
Irritable bowel syndrome (IBS) is a functional bowel disorder characterised by abdominal pain or discomfort and an irregular bowel habit[1]. The prevalence is up to 20% in Western adults, which makes IBS the most common diagnosis in gastroenterology. The precise aetiology and pathophysiology of IBS are incompletely understood, despite extensive interest and investigation. The current knowledge does, however, suggest that altered gut motility, visceral hyperalgesia, and dysregulation of the brain-gut axis are central to IBS.
IBS is diagnosed by the presence of symptoms according to the Rome criteria[2], with concomitant exclusion of organic diseases; hence, there is no specific biological, radiological, endoscopic, or physiological marker for IBS. Medical treatment options for IBS are limited, possibly due to the lack of biomarkers and data about the pathophysiology of the condition. Different immune markers are among the most studied putative biomarkers in IBS. Increased mast cell counts, mast cells in close proximity to nerves, and mast cell mediators that are able to stimulate murine visceral sensory nerves appear to be characteristic of IBS[3-6]. Elevated plasma levels of pro-inflammatory interleukin (IL)-6 and IL-8 have been observed in IBS[7], and peripheral blood mononuclear cells obtained from IBS patients produce higher amounts of tumour necrosis factor (TNF)-α, IL-1β, IL-6 and IL-12 than cells from healthy controls[8,9]. Furthermore, an increased number of immunocytes have frequently been observed in mucosa from IBS patients[10,11]. In addition to immune markers, other markers, such as 5-hydroxytryptamine (5-HT, serotonin) and gut hormones, have also been associated with IBS. Decreased expression of 5-HT has been associated with constipation-predominant IBS[12]. Elevated plasma 5-HT concentrations have been observed in a mixed IBS population[13] and in post-infectious IBS[14], while the opposite was demonstrated in constipation IBS[14]. Moreover, IBS patients have also been demonstrated to have decreased 5-HT levels and turnover, and lower 5-HT transporter mRNA concentrations[14,15].
The recent technological development of analytical instruments combined with rapid progress in bioinformatics has opened novel opportunities to quickly and simultaneously measure and model huge numbers of molecular metabolites in biological samples[16,17]. This metabolomic approach is considered a powerful tool for characterising complex phenotypes and developing biological markers for specific physiological states. Thus, metabolomics provides an interesting platform to investigate the pathophysiology of a complex syndrome like IBS at the molecular level. Studies on the molecular abnormalities in IBS are needed to understand the mechanisms behind the emergence of symptoms, and to enable the development of novel therapies.
The aim of the current study was to compare the global metabolic profiles of mucosal biopsies from IBS patients with those from healthy subjects using a high-throughput approach comprising lipidomics and metabolomics.
MATERIALS AND METHODS
Subjects
Sixteen adult IBS patients fulfilling the Rome II criteria[18] and without organic intestinal diseases were recruited to participate in the study. One statistical outlier IBS patient was left out of the analyses after an initial quality check of the results, and thus a total of 15 patients were analysed (Table 1). Nine healthy subjects (mean age 49 years, SD 14; 4 male) without organic intestinal diseases or gastrointestinal symptoms consistent with IBS and undergoing colonoscopy for clinical reasons were recruited as controls. The healthy subjects were either polyp control patients having a minimum of 3 years since the previous polyp finding or anaemic patients. Inclusion criteria for all subjects were: an age between 20 and 65 years; normal blood count (erythrocytes, haemoglobin, haematocrit, MCV, MCH, MCHC, platelets, leukocytes); serum creatinine, alanine aminotransferase (ALT) and alkaline phosphatase (ALP) within reference values; and normal gut histology as evaluated by an experienced pathologist (PS). Subjects were excluded if they had a history of major or complicated gastrointestinal surgery, severe endometriosis, complicated abdominal adhesions, malignant tumours, were pregnant or lactating, or had received antimicrobials during the previous month. Patients with lactose intolerance were allowed to participate if they followed a continuous low-lactose or lactose-free diet.
Table 1.
Sociodemographic and clinical characteristics of IBS patients (n = 15) (mean ± SD)
| Age (yr) | 42 ± 16 |
| Sex (n): F/M | 11/4 |
| BMI (kg/m2) | 23.3 ± 5.0 |
| Predominant bowel habit1: (n) | |
| Diarrhoea | 2 |
| Constipation | 3 |
| Alternating | 10 |
According to the Rome II criteria; BMI: Body mass index.
The Human Ethics Committee of the Hospital District of Pirkanmaa, Finland, approved the study protocol. All subjects provided written informed consent.
Sample collection and preparation
Mucosal biopsies (mean weight 5.2 mg/sample; SD 1.5) from the ascending colon were obtained from each subject during colonoscopy after bowel cleansing. The samples were immediately frozen at -20°C, and stored at -70°C until required for analysis. The samples for lipidomic analysis were weighed into Eppendorf tubes, and 10 μL of 0.9% sodium chloride and 10 μL of an internal standard mixture (10 lipid compounds, 0.1 μg each) were added. The samples were extracted with 100 μL of chloroform:methanol (2:1; two min vortexing, one h extraction time) and centrifuged (7800 g, 3 min). Of the lower organic phase, 60 μL aliquots were transferred into vial inserts and 10 μL of a standard mixture containing three labelled lipid compounds was added. The internal standard mixture contained the following lipid compounds with heptadecanoic acid (C17:0) as the esterified fatty acid: D-erythro-Sphingosine-1-Phosphate (C17 Base, Avanti Polar Lipids, Alabaster, AL, USA), LysoPC (Avanti Polar Lipids), MG (17:0)[rac] (Larodan Fine Chemicals, Malmö, Sweden), PG (17:0/17:0)[rac] (Avanti Polar Lipids), Cer (d18:1/17:0) (Avanti Polar Lipids), PS (17:0/17:0) (Avanti Polar Lipids), PC (17:0/17:0) (Avanti Polar Lipids), PA (17:0/17:0) (Avanti Polar Lipids), PE (17:0/17:0) (Avanti Polar Lipids), DG (17:0/17:0)[rac] (Larodan Fine Chemicals), and TG (17:0/17:0/17:0) (Larodan Fine Chemicals). The labeled standard mixture consisted of LysoPC (16:0-D3) (Larodan Fine Chemicals), PC (16:0/16:0-D6) (Larodan Fine Chemicals), and TG (16:0/16:0/16:0-13C3) (Larodan Fine Chemicals).
For water-soluble compounds, the samples were weighed into Eppendorf tubes and 10°C of 1000 ppm (mg/mL) labelled palmitic acid (16:0-16, 16, 16D3) was added as an internal standard. The samples were extracted with 500 μL methanol (two min vortexing, 0.5 h extraction time) and centrifuged (7800 g, 3 min). The separated supernatants were evaporated to dryness under nitrogen, and the residues were derivatised with 2% methoxyamine HCl in pyridine (MOX; 25 μL, 90 min at 30°C) and N-Methyl-N-trimethylsilyltrifluoroacetamide (MSTFA; 50 μL, 30 min at 37°C). All samples were run in duplicate.
Analysis of lipids by UPLC/MS
Characterisation of lipid molecular species in colonic mucosa was performed by a lipidomics strategy using ultra performance liquid chromatography coupled to mass spectrometry (UPLC/MS, Waters Micromass Q-Tof Premier). The column (50°C) was a Waters Acquity UPLC™ BEH C18 (Waters Inc., Milford, MA, USA) 1 mm × 50 mm with 1.7 μm particles. The solvent system included: (A) ultra pure water (1% 1 mol/L NH4Ac, 0.1% HCOOH) and (B) LC/MS grade acetonitrile:isopropanol (5:2, 1% 1 mol/L NH4Ac, 0.1% HCOOH). The gradient started from 65% A and 35% B, reached 100% B in 6 min and remained there for the next 7 min. There was a 5 min re-equilibration step before the next run. The flow rate was 0.200 mL/min and the injected amount 1.0 μL. Lipid profiling was carried out using ESI+ mode, and the data were collected at a mass range of m/z 300-2000 with a scan duration of 0.08 s.
Lipids were identified using an internal spectral library or with tandem mass spectrometry. The normalisation of lipidomics data was performed as follows: all monoacyl lipids, except cholesterol esters (such as monoacylglycerols and monoacylglycerophospholipids), were calibrated with lysophosphatidylcholine lysoPC (17:0) internal standard; all diacyl lipids, except ethanolamine phospholipids, were normalised with phosphatidylcholine PC (17:0); the diacyl ethanolamine phospholipids were calibrated with phosphatidylethanolamine PE (17:0); and the triacylglycerols and cholesterol esters were calibrated with triacylglycerol TG (17:0). Other molecular species were normalised by lysoPC (17:0) for retention time < 310 s, PC (17:0) for retention time between 310 and 450 s, and TG (17:0) for higher retention times. Data was processed using the MZmine software, version 0.60[19], and metabolites were identified using an internal spectral library or with tandem mass spectrometry (UPLC/MS/MS).
Analysis of water-soluble metabolites by GC × GC-TOF
A broad screening of water-soluble metabolites was conducted by a comprehensive two-dimensional gas chromatography coupled to a high speed time-of-flight mass spectrometry (GC × GC-TOF)[20]. The instrument used was a Leco Pegasus 4D GC × GC-TOF with Agilent 6890N GC from Agilent Technologies, USA and CombiPAL autosampler from CTC Analytics AG, Switzerland. Modulator, secondary oven, and time-of flight mass spectrometer were from Leco Inc., USA. The GC was operated in split mode (1:20) using helium as carrier gas at 1.5 mL/min constant flow. The first GC column was a relatively non-polar RTX-5 column, 10 m × 0.18 mm × 0.20 μm, and the second was a polar BPX-50, 1.10 m × 0.10 mm × 0.10 μm. The temperature programme was as follows: initial 50°C, 1 min -> 280°C, 7°C/min, one min. The secondary oven was set to +30°C above the primary oven temperature. The second dimension separation time was set to 3 s. The mass range used was 40 to 600 amu and the data collection speed was 100 spectra/s. A commercial mass spectral library, Palisade Complete 600K, was used for identifying metabolites.
Statistical analysis
Partial least squares discriminant analysis (PLS/DA) was utilized as a supervised modelling method using the SIMPLS algorithm to calculate the model. The contiguous-blocks cross-validation method and Q2 scores were used to develop the models. Top loadings for latent variables associated with drug-specific effects were reported. The VIP (variable importance in the projection) values were calculated to identify the most important molecular species for clustering of specific groups. Multivariate analyses were performed using Matlab, version 7.2 (Mathworks Inc., Natick, MA, USA), and the PLS Toolbox, version 4.0, for the Matlab package (Eigenvector Research Inc., Wenatchee, WA, USA). One statistical outlier IBS patient was left out of the analyses after an initial quality check of the results. Univariate comparisons for individual metabolites between the groups were performed using the Wilcoxon rank-sum test. A P value < 0.05 was considered statistically significant. To account for multiple comparisons, the False Discovery Rate (FDR) Q-value is also reported[21].
RESULTS
Lipidomic analysis
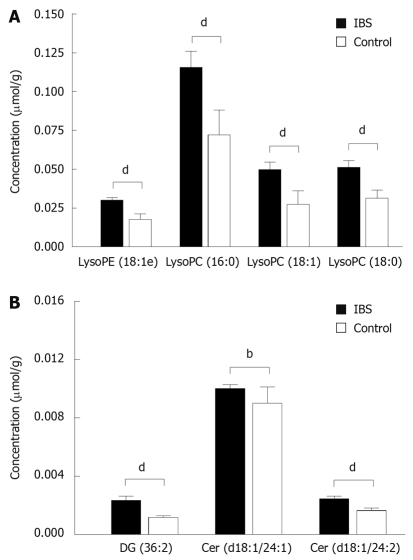
By applying UPLC/MS, a total of 651 lipid peaks were found, and 75 of them were identified using the internal spectral library, as described by Yetukuri et al[22], or with tandem mass spectrometry using UPLC/MS/MS. PLS/DA analysis of lipidomic data revealed significant differences in the mucosal lipid profiles of IBS patients and healthy controls. Overall, lipid species were upregulated in biopsies from IBS patients compared to those from healthy subjects. The 20 lipids with the largest differences between the groups by fold change appear in Table 2. A significant upregulation in the concentrations of typical cell membrane metabolites, lysophospholipids, in IBS patients was the most obvious finding (Figure 1A). Other lipid groups with a significant contribution to the distinction between IBS patients and healthy controls were ceramides (Figure 1B), glycosphingolipids, and di- and triacylglycerols. All of these showed upregulation in the IBS group.
Table 2.
The 15 lipids with the largest and most significant differences between patients and controls
| Lipid name | Fold (IBS/healthy control) | P value1 | FDR Q value |
| Cer (d18:1/24:1) | 1.3 | 0.001 | 0.022 |
| Cer (d18:1/24:2) | 1.4 | 0.00004 | 0.0018 |
| DG (36:2) | 1.9 | 0.000001 | 0.0003 |
| GlycoSL (m/z = 1199.805) | 1.9 | 0.001 | 0.018 |
| GlycoSL (m/z = 1195.851) | 2.0 | 0.0003 | 0.0069 |
| lysoPC (16:0) | 2.1 | 0.00006 | 0.0020 |
| lysoPC (18:0) | 1.9 | 0.0002 | 0.0049 |
| lysoPC (18:1) | 2.8 | 0.00002 | 0.0013 |
| lysoPE (18:1e) | 2.4 | 0.00002 | 0.0013 |
| 7TG (46:5) | 1.4 | 0.04 | 0.19 |
| TG (48:5) | 1.6 | 0.03 | 0.17 |
| TG (48:6) | 1.7 | 0.03 | 0.18 |
| TG (49:3) | 2.1 | 0.009 | 0.089 |
| TG (51:4) | 1.6 | 0.04 | 0.19 |
| TG (51:5) | 1.8 | 0.02 | 0.12 |
Wilcoxon rank sum test. Cer: Ceramide; DG: Diacylglycerol; GlycoSL: Glycosphingolipid; lysoPC: Lysophosphatidylcholine; lysoPE: Lysophosphatidylethanolamine; TG: Triacylglycerol.
Figure 1.
The concentrations (mean ± SE) of selected lysophospholipids in mucosal biopsies from irritable bowel syndrome (IBS) patients (n = 15) and healthy controls (n = 9) as measured by UPLC/MS. (A) Patients and controls differ significantly from each other for all presented lysophospholipids, as well as for (B) diacylglycerol and ceramides. LysoPE: Lysophosphatidylethanolamine; LysoPC: Lysophosphatidylcholine. P values are based on Wilcoxon rank sum test with bP < 0.01 and dP < 0.001.
Metabolomic analysis
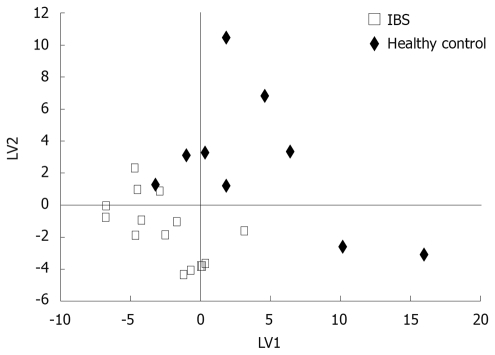
Broad metabolite screening by GC × GC-TOF resulted in several hundred mucosal metabolites, of which 107 were identified and kept in analyses. Based on PLS/DA analysis, a clear distinction between IBS cases and controls was obtained (Figure 2). Both upregulation and downregulation of metabolites were observed in IBS patients vs controls. The top ranked metabolites contributing to the distinction between the groups appear in Table 3.
Figure 2.
Partial least squares discriminant analysis (PLS/DA) of GC × GC-Tof-based metabolic profiles for IBS patients (n = 15) and healthy controls (n = 9). Two latent variables (LVs) were used (Q2 = 61%).
Table 3.
Major water soluble metabolites contributing to differentiation between patients and controls1
| Metabolite | Fold (IBS/healthy control) | P value2 | FDR Q value |
| 2(3H)-furanone | 13.7 | 0.03 | 0.25 |
| Ribitol | 3.6 | NS | 0.25 |
| Heptan | 2.9 | 0.02 | 0.25 |
| L-Mannose | 2.8 | NS | 0.25 |
| Creatinine | 1.7 | 0.04 | 0.25 |
| Dodecane | 1.5 | NS | 0.29 |
| Decanoic acid | 1.3 | NS | 0.52 |
| Dodecanoic acid | -1.5 | NS | 0.25 |
| n-Butylamine | -1.5 | 0.01 | 0.25 |
| D-ribose | -1.5 | NS | 0.25 |
| Glucopyranose | -1.6 | NS | 0.29 |
| Azelaic acid | -1.8 | 0.02 | 0.25 |
| Adipic acid | -2.7 | 0.0008 | 0.05 |
Separation is based on a variable importance projection (VIP) analysis with a cut-off value of 2;
Wilcoxon rank sum test. NS: Not significant.
The metabolite contributing most to the distinction was 2(3H)-furanone, a cyclic ester commonly produced in biochemical pathways, which was almost 14-fold upregulated in IBS patients compared to healthy subjects (P = 0.03). The fold changes for other top ranked metabolites were clearly lower (a 3.7 to -2.7 fold change). Other basic metabolites frequently found in biochemical pathways, such as the second messenger, D-ribose, were also among the major factors contributing to the distinction between cases and controls. Organic, carboxylic acids were found to be both slightly downregulated (dodecanoic, azelaic, and adipic acid) as well as slightly upregulated (decanoic acid) in IBS patients compared to healthy controls.
DISCUSSION
Data on the molecular abnormalities in IBS are urgently required, as the pathophysiology of the condition is largely unknown and current therapies are therefore also limited. In this study, the differences between colonic mucosa from IBS patients and healthy controls were investigated by employing two high-throughput metabolomic platforms, UPLC/MS based lipidomics and GC × GC-TOF based metabolomics. Data indicated multiple differences between IBS mucosa and healthy mucosa, including an increase in the IBS group of several lipid species, such as lysophospholipids and ceramides.
The present study is the first to utilise a metabolomic approach to investigate molecular differences between IBS patients and healthy controls. Metabolomics can be seen as an optimal tool for studying diseases with unknown or complex pathophysiology, as a global study of the metabolome is a non-targeted approach that requires no preselection of markers, in contrast to the traditional way of limiting the analysis to a predefined set of known compounds[23]. Recently, metabolomics has been utilised in the investigation of multiple diseases, e.g. inflammatory bowel disease[24], obesity[25], and cancer[26].
In the present study, lipids - particularly lysophosphatidylcholines (lysoPCs) and ceramides - were the most upregulated molecules in IBS patients. Mounting data suggest that certain lipids, including phospholipid derivatives and ceramides, play a role in modulating and enhancing pain sensitivity[27,28], which could be one explanation for their involvement in IBS pathophysiology. LysoPCs have not previously been associated with IBS, but studies indicate elevated levels of lysoPCs or phospholipase A2, an enzyme involved in lysoPC formation, in inflammatory bowel disease (IBD)[29,30]. A high lysoPC concentration has been suggested to impair mucosal barrier function and increase gastrointestinal permeability in vivo and in vitro[31-34]. The role of permeability defects in IBS is not fully elucidated, but a recent review by Camilleri and Gorman concludes that there appears to be at least one IBS subgroup with increased gut permeability[35]. Interestingly, lysoPCs have also been associated with vascular inflammation, endothelial dysfunction, and coronary atherosclerosis[36], implying that lysoPCs might also play a role in the subtle type of mucosal inflammation present in IBS.
Lipids of the ceramide/sphingomyelin pathway are another lipid type that we observed to be altered in IBS. Previous studies indicate that ceramides might be involved in the pathology of IBD[37,38], whereas no reports on ceramides in IBS are available. Ceramides have, however, been shown to be toxic in several cell types. Current data reveal a possible role for ceramides in the damage of cells and tissues, and ultimately in the development of chronic metabolic diseases, such as diabetes and cardiovascular disease[39]. The toxic effects of ceramides might be partly mediated via the production of reactive oxygen species in cells[40]. It has been proposed that, similar to lysophosphatidylcholines, epithelial oxidative stress might also contribute to gut barrier dysfunction[41]. Our results thus suggest that the molecular mechanism behind increased permeability could, to some extent, be similar in IBS and IBD.
Based on a global analysis of water-soluble metabolites, IBS cases and healthy controls were rather well separated into two distinct groups. The physiological relevance of the main molecules contributing to the separation is, on the other hand, less evident than in the case of lipid species. Differences were seen in basic metabolites, such as 2(3H)-furanone (also known as lactone) and D-ribose, both of which are produced in common biochemical pathways in cells. A recent study investigating a Trichinella spiralis infected mouse model of post-infectious IBS and utilizing metabolomics, demonstrated an increase in molecules involved in energy metabolism (lactate, citrate, and alanine) in the IBS group[42]. The authors suggest that this might reflect a muscular hypercontractility possibly present in IBS, though it should be underlined that this was an experimental model. Concerning organic acids, our results are in line with the study by Martin et al[42], in that we found that organic acids contributed to the separation between cases and controls. Specifically, we did observe increased concentrations of alanine in the IBS group, but the difference between IBS and control groups was not significant (fold = 1.3, P = 0.14). Organic acids are known to be produced by the intestinal microbiota, and a disruption in the acid profile could reflect a possible deviance in microbiota previously reported in IBS[43,44]. In further support of findings by Martin et al[42], we detected elevated levels of creatinine in the IBS group. Creatine and creatinine are tightly interlinked with the energy metabolism in smooth muscle, and a raised creatinine concentration might be a sign of increased energy consumption and muscle contractility[45].
The field of metabolomics is evolving rapidly, and it is already considered a sensitive analytical tool for investigating the health-disease continuum[17]. Like any method, however, it has its own limitations. As large numbers of metabolites are included in the studies, caution is necessary in the interpretation of results[16]. The relevance of a single identified biomarker might not be high, but it could be that systematic up- or downregulation in specific groups of molecules (such as certain lipids in the current study) indicates a biologically relevant metabolite type. Another drawback of metabolomics is that a large proportion of spectral peaks are still unknown, and consequently more effort has to be placed on the compilation of standardised metabolite libraries[16,23]. Considering the current study setting, one obvious weakness is the small number of subjects. On the other hand, it is highly encouraging to see that IBS patients and healthy controls were fairly well differentiated, even with this limited sample size. Moreover, it would have been interesting to investigate whether differences between IBS and healthy subjects could also be observed in metabolic profiles from non-invasive tissues, such as faecal material or blood, as these are more easily obtained in clinical settings.
Taken together, our results suggest significant differences in the global mucosal metabolic profile between IBS patients and healthy controls. The current study is the first to attempt to identify colonic mucosal metabolites typical for IBS using a high-throughput metabolomic approach. In this study, IBS was particularly characterised by an upregulation of specific lipid groups, such as lysophosphatidylcholines and ceramides. These lipid species have been associated with the modulation of pain sensitivity and gut permeability, and our data thus indicate that these molecules might be involved in the pathophysiology of visceral pain and gut barrier dysfunction associated with IBS.
COMMENTS
Background
Irritable bowel syndrome (IBS) is the most common diagnosis in gastroenterology. The syndrome is classified as functional, and no biological marker exists for IBS. The precise aetiology and pathophysiology is not fully known, and this might partly explain why pharmacotherapy is considered rather ineffective.
Research frontiers
More data on the molecular abnormalities in IBS are required to better understand the mechanism behind the emergence of symptoms, and to be able to treat the patients in a safe and efficient way.
Innovations and breakthroughs
This study characterises the differences between colonic mucosa from IBS patients and healthy controls using two high-throughput metabolomic platforms, UPLC/MS based lipidomics and GC × GC-TOF based metabolomics. Metabolomics is a useful tool for investigating diseases with complex or unknown backgrounds, because it is possible to simultaneously measure and model a huge number of metabolites. Data indicated multiple differences between IBS mucosa and healthy mucosa, thus providing novel information about the pathophysiology of IBS. An increase in the IBS group of several lipid species, such as lysophospholipids and ceramides, was the major difference observed.
Applications
By better understanding the mucosal abnormalities behind IBS, it might be possible to improve the diagnosis and therapy of patients.
Peer review
This article is surely innovative, not only in the hypothesis, but also in the methodology. This could be a hot article if could be popularized appropriately. A review of the current literature indicates that the present article is a pioneer in its field.
Acknowledgments
The authors would like to express their deepest gratitude to Aino Oksanen, MD, PhD, Kristiina Söderlund, MD, Lea Veijola, MD, PhD and Ms. Anne-Maria Riihimäki for their invaluable help during the conduct of the study.
Footnotes
Supported by Valio Ltd and the Finnish Funding Agency for Technology and Innovation (TEKES); the preparation of this manuscript was funded in part by the Academy of Finland
Peer reviewer: Dr. Peyman Adibi, MD, Vice Chancellor for Research, Associate Professor of Medicine, Isfahan University of Medical Sciences, Isfahan 81767, Iran
S- Editor Tian L L- Editor Stewart GJ E- Editor Ma WH
References
- 1.Drossman DA, Camilleri M, Mayer EA, Whitehead WE. AGA technical review on irritable bowel syndrome. Gastroenterology. 2002;123:2108–2131. doi: 10.1053/gast.2002.37095. [DOI] [PubMed] [Google Scholar]
- 2.Longstreth GF, Thompson WG, Chey WD, Houghton LA, Mearin F, Spiller RC. Functional bowel disorders. Gastroenterology. 2006;130:1480–1491. doi: 10.1053/j.gastro.2005.11.061. [DOI] [PubMed] [Google Scholar]
- 3.O'Sullivan M, Clayton N, Breslin NP, Harman I, Bountra C, McLaren A, O'Morain CA. Increased mast cells in the irritable bowel syndrome. Neurogastroenterol Motil. 2000;12:449–457. doi: 10.1046/j.1365-2982.2000.00221.x. [DOI] [PubMed] [Google Scholar]
- 4.Guilarte M, Santos J, de Torres I, Alonso C, Vicario M, Ramos L, Martinez C, Casellas F, Saperas E, Malagelada JR. Diarrhoea-predominant IBS patients show mast cell activation and hyperplasia in the jejunum. Gut. 2007;56:203–209. doi: 10.1136/gut.2006.100594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbara G, Stanghellini V, De Giorgio R, Cremon C, Cottrell GS, Santini D, Pasquinelli G, Morselli-Labate AM, Grady EF, Bunnett NW, et al. Activated mast cells in proximity to colonic nerves correlate with abdominal pain in irritable bowel syndrome. Gastroenterology. 2004;126:693–702. doi: 10.1053/j.gastro.2003.11.055. [DOI] [PubMed] [Google Scholar]
- 6.Barbara G, Wang B, Stanghellini V, de Giorgio R, Cremon C, Di Nardo G, Trevisani M, Campi B, Geppetti P, Tonini M, et al. Mast cell-dependent excitation of visceral-nociceptive sensory neurons in irritable bowel syndrome. Gastroenterology. 2007;132:26–37. doi: 10.1053/j.gastro.2006.11.039. [DOI] [PubMed] [Google Scholar]
- 7.Dinan TG, Quigley EM, Ahmed SM, Scully P, O'Brien S, O'Mahony L, O'Mahony S, Shanahan F, Keeling PW. Hypothalamic-pituitary-gut axis dysregulation in irritable bowel syndrome: plasma cytokines as a potential biomarker? Gastroenterology. 2006;130:304–311. doi: 10.1053/j.gastro.2005.11.033. [DOI] [PubMed] [Google Scholar]
- 8.O'Mahony L, McCarthy J, Kelly P, Hurley G, Luo F, Chen K, O'Sullivan GC, Kiely B, Collins JK, Shanahan F, et al. Lactobacillus and bifidobacterium in irritable bowel syndrome: symptom responses and relationship to cytokine profiles. Gastroenterology. 2005;128:541–551. doi: 10.1053/j.gastro.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 9.Liebregts T, Adam B, Bredack C, Roth A, Heinzel S, Lester S, Downie-Doyle S, Smith E, Drew P, Talley NJ, et al. Immune activation in patients with irritable bowel syndrome. Gastroenterology. 2007;132:913–920. doi: 10.1053/j.gastro.2007.01.046. [DOI] [PubMed] [Google Scholar]
- 10.Chadwick VS, Chen W, Shu D, Paulus B, Bethwaite P, Tie A, Wilson I. Activation of the mucosal immune system in irritable bowel syndrome. Gastroenterology. 2002;122:1778–1783. doi: 10.1053/gast.2002.33579. [DOI] [PubMed] [Google Scholar]
- 11.Törnblom H, Lindberg G, Nyberg B, Veress B. Full-thickness biopsy of the jejunum reveals inflammation and enteric neuropathy in irritable bowel syndrome. Gastroenterology. 2002;123:1972–1979. doi: 10.1053/gast.2002.37059. [DOI] [PubMed] [Google Scholar]
- 12.Wang SH, Dong L, Luo JY, Gong J, Li L, Lu XL, Han SP. Decreased expression of serotonin in the jejunum and increased numbers of mast cells in the terminal ileum in patients with irritable bowel syndrome. World J Gastroenterol. 2007;13:6041–6047. doi: 10.3748/wjg.v13.45.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bearcroft CP, Perrett D, Farthing MJ. Postprandial plasma 5-hydroxytryptamine in diarrhoea predominant irritable bowel syndrome: a pilot study. Gut. 1998;42:42–46. doi: 10.1136/gut.42.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunlop SP, Coleman NS, Blackshaw E, Perkins AC, Singh G, Marsden CA, Spiller RC. Abnormalities of 5-hydroxytryptamine metabolism in irritable bowel syndrome. Clin Gastroenterol Hepatol. 2005;3:349–357. doi: 10.1016/s1542-3565(04)00726-8. [DOI] [PubMed] [Google Scholar]
- 15.Coates MD, Mahoney CR, Linden DR, Sampson JE, Chen J, Blaszyk H, Crowell MD, Sharkey KA, Gershon MD, Mawe GM, et al. Molecular defects in mucosal serotonin content and decreased serotonin reuptake transporter in ulcerative colitis and irritable bowel syndrome. Gastroenterology. 2004;126:1657–1664. doi: 10.1053/j.gastro.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 16.Orešič M, Vidal-Puig A, Hänninen V. Metabolomic approaches to phenotype characterization and applications to complex diseases. Expert Rev Mol Diagn. 2006;6:575–585. doi: 10.1586/14737159.6.4.575. [DOI] [PubMed] [Google Scholar]
- 17.Schnackenberg LK, Beger RD. Monitoring the health to disease continuum with global metabolic profiling and systems biology. Pharmacogenomics. 2006;7:1077–1086. doi: 10.2217/14622416.7.7.1077. [DOI] [PubMed] [Google Scholar]
- 18.Thompson WG, Longstreth GF, Drossman DA, Heaton KW, Irvine EJ, Muller-Lissner SA. Functional bowel disorders and functional abdominal pain. Gut. 1999;45 Suppl 2:II43–II47. doi: 10.1136/gut.45.2008.ii43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katajamaa M, Orešič M. Processing methods for differential analysis of LC/MS profile data. BMC Bioinformatics. 2005;6:179. doi: 10.1186/1471-2105-6-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Welthagen W, Shellie RA, Spranger J, Ristow M, Zimmermann R, Fiehn O. Comprehensive two-dimensional gas chromatography-time-of-flight mass spectrometry (GC × GC-TOF) for high resolution metabolomics: biomarker discovery on spleen tissue extracts of obese NZO compared to lean C57BL/6 mice. Metabolomics. 2005;1:65–73. [Google Scholar]
- 21.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yetukuri L, Katajamaa M, Medina-Gomez G, Seppänen-Laakso T, Vidal-Puig A, Orešič M. Bioinformatics strategies for lipidomics analysis: characterization of obesity related hepatic steatosis. BMC Syst Biol. 2007;1:12. doi: 10.1186/1752-0509-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schlotterbeck G, Ross A, Dieterle F, Senn H. Metabolic profiling technologies for biomarker discovery in biomedicine and drug development. Pharmacogenomics. 2006;7:1055–1075. doi: 10.2217/14622416.7.7.1055. [DOI] [PubMed] [Google Scholar]
- 24.Marchesi JR, Holmes E, Khan F, Kochhar S, Scanlan P, Shanahan F, Wilson ID, Wang Y. Rapid and noninvasive metabonomic characterization of inflammatory bowel disease. J Proteome Res. 2007;6:546–551. doi: 10.1021/pr060470d. [DOI] [PubMed] [Google Scholar]
- 25.Pietiläinen KH, Sysi-Aho M, Rissanen A, Seppänen-Laakso T, Yki-Järvinen H, Kaprio J, Orešič M. Acquired obesity is associated with changes in the serum lipidomic profile independent of genetic effects - a monozygotic twin study. PLoS One. 2007;2:e218. doi: 10.1371/journal.pone.0000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Denkert C, Budczies J, Kind T, Weichert W, Tablack P, Sehouli J, Niesporek S, Konsgen D, Dietel M, Fiehn O. Mass spectrometry-based metabolic profiling reveals different metabolite patterns in invasive ovarian carcinomas and ovarian borderline tumors. Cancer Res. 2006;66:10795–804. doi: 10.1158/0008-5472.CAN-06-0755. [DOI] [PubMed] [Google Scholar]
- 27.Park KA, Vasko MR. Lipid mediators of sensitivity in sensory neurons. Trends Pharmacol Sci. 2005;26:571–577. doi: 10.1016/j.tips.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 28.Malan TP Jr, Porreca F. Lipid mediators regulating pain sensitivity. Prostaglandins Other Lipid Mediat. 2005;77:123–130. doi: 10.1016/j.prostaglandins.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 29.Minami T, Tojo H, Shinomura Y, Matsuzawa Y, Okamoto M. Increased group II phospholipase A2 in colonic mucosa of patients with Crohn's disease and ulcerative colitis. Gut. 1994;35:1593–1598. doi: 10.1136/gut.35.11.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haapamäki MM, Grönroos JM, Nurmi H, Irjala K, Alanen KA, Nevalainen TJ. Phospholipase A2 in serum and colonic mucosa in ulcerative colitis. Scand J Clin Lab Invest. 1999;59:279–287. doi: 10.1080/00365519950185643. [DOI] [PubMed] [Google Scholar]
- 31.Tagesson C, Franzen L, Dahl G, Weström B. Lysophosphatidylcholine increases rat ileal permeability to macromolecules. Gut. 1985;26:369–377. doi: 10.1136/gut.26.4.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Otamiri T, Sjödahl R, Tagesson C. Lysophosphatidylcholine potentiates the increase in mucosal permeability after small-intestinal ischaemia. Scand J Gastroenterol. 1986;21:1131–1136. doi: 10.3109/00365528608996433. [DOI] [PubMed] [Google Scholar]
- 33.Karlqvist PA, Franzen L, Sjödahl R, Tagesson C. Lysophosphatidylcholine and taurodeoxycholate increase stomach permeability to different-sized molecules. Scand J Gastroenterol. 1986;21:1039–1045. doi: 10.3109/00365528608996417. [DOI] [PubMed] [Google Scholar]
- 34.Sawai T, Lampman R, Hua Y, Segura B, Drongowski RA, Coran AG, Harmon CM. Lysophosphatidylcholine alters enterocyte monolayer permeability via a protein kinase C/Ca2+ mechanism. Pediatr Surg Int. 2002;18:591–594. doi: 10.1007/s00383-002-0860-x. [DOI] [PubMed] [Google Scholar]
- 35.Camilleri M, Gorman H. Intestinal permeability and irritable bowel syndrome. Neurogastroenterol Motil. 2007;19:545–552. doi: 10.1111/j.1365-2982.2007.00925.x. [DOI] [PubMed] [Google Scholar]
- 36.Kougias P, Chai H, Lin PH, Lumsden AB, Yao Q, Chen C. Lysophosphatidylcholine and secretory phospholipase A2 in vascular disease: mediators of endothelial dysfunction and atherosclerosis. Med Sci Monit. 2006;12:RA5–R16. [PubMed] [Google Scholar]
- 37.Sakata A, Yasuda K, Ochiai T, Shimeno H, Hikishima S, Yokomatsu T, Shibuya S, Soeda S. Inhibition of lipopolysaccharide-induced release of interleukin-8 from intestinal epithelial cells by SMA, a novel inhibitor of sphingomyelinase and its therapeutic effect on dextran sulphate sodium-induced colitis in mice. Cell Immunol. 2007;245:24–31. doi: 10.1016/j.cellimm.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 38.Homaidan FR, El-Sabban ME, Chakroun I, El-Sibai M, Dbaibo GS. IL-1 stimulates ceramide accumulation without inducing apoptosis in intestinal epithelial cells. Mediators Inflamm. 2002;11:39–45. doi: 10.1080/09629350210313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Summers SA. Ceramides in insulin resistance and lipotoxicity. Prog Lipid Res. 2006;45:42–72. doi: 10.1016/j.plipres.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 40.Andrieu-Abadie N, Gouaze V, Salvayre R, Levade T. Ceramide in apoptosis signaling: relationship with oxidative stress. Free Radic Biol Med. 2001;31:717–728. doi: 10.1016/s0891-5849(01)00655-4. [DOI] [PubMed] [Google Scholar]
- 41.Assimakopoulos SF, Vagianos CE, Patsoukis N, Georgiou C, Nikolopoulou V, Scopa CD. Evidence for intestinal oxidative stress in obstructive jaundice-induced gut barrier dysfunction in rats. Acta Physiol Scand. 2004;180:177–185. doi: 10.1046/j.0001-6772.2003.01229.x. [DOI] [PubMed] [Google Scholar]
- 42.Martin FP, Verdu EF, Wang Y, Dumas ME, Yap IK, Cloarec O, Bergonzelli GE, Corthesy-Theulaz I, Kochhar S, Holmes E, et al. Transgenomic metabolic interactions in a mouse disease model: interactions of Trichinella spiralis infection with dietary Lactobacillus paracasei supplementation. J Proteome Res. 2006;5:2185–2193. doi: 10.1021/pr060157b. [DOI] [PubMed] [Google Scholar]
- 43.Malinen E, Rinttilä T, Kajander K, Mättö J, Kassinen A, Krogius L, Saarela M, Korpela R, Palva A. Analysis of the fecal microbiota of irritable bowel syndrome patients and healthy controls with real-time PCR. Am J Gastroenterol. 2005;100:373–382. doi: 10.1111/j.1572-0241.2005.40312.x. [DOI] [PubMed] [Google Scholar]
- 44.Kassinen A, Krogius-Kurikka L, Mäkivuokko H, Rinttilä T, Paulin L, Corander J, Malinen E, Apajalahti J, Palva A. The fecal microbiota of irritable bowel syndrome patients differs significantly from that of healthy subjects. Gastroenterology. 2007;133:24–33. doi: 10.1053/j.gastro.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 45.Clark JF. The creatine kinase system in smooth muscle. Mol Cell Biochem. 1994;133-134:221–232. doi: 10.1007/BF01267956. [DOI] [PubMed] [Google Scholar]