Abstract
One of the hurdles to achieving personalized medicine has been implementing the laboratory processes for performing and reporting complex molecular tests. The rapidly changing test rosters and complex analysis platforms in molecular diagnostics have meant that many clinical laboratories still use labor-intensive manual processing and testing without the level of automation seen in high-volume chemistry and hematology testing. We provide here a discussion of design requirements and the results of implementation of a suite of lab management tools that incorporate the many elements required for use of molecular diagnostics in personalized medicine, particularly in cancer. These applications provide the functionality required for sample accessioning and tracking, material generation, and testing that are particular to the evolving needs of individualized molecular diagnostics. On implementation, the applications described here resulted in improvements in the turn-around time for reporting of more complex molecular test sets, and significant changes in the workflow. Therefore, careful mapping of workflow can permit design of software applications that simplify even the complex demands of specialized molecular testing. By incorporating design features for order review, software tools can permit a more personalized approach to sample handling and test selection without compromising efficiency.
Laboratory processes in molecular diagnostics are currently highly complex and have proven difficult to standardize and automate. Contributors to this complexity include the wide variety of tissue sample types that can be tested, the numerous steps involved in material preparation, frequent implementation of novel technical platforms leading to variable and unpredictable assay performance, and complex post-testing analysis methods. Another component of the complexity, especially in molecular oncology, is that different tumor samples require different order sets including multistep and reflex testing. Finally, the high cost of some molecular testing often requires prescreening or prioritization of limited samples. Therefore, new sample handling models that are applicable to the clinical laboratory are needed but not yet fully developed.1
We present here a discussion of a software design considerations related to the personalized molecular testing, and the results of implementation of applications that address some of these laboratory issues.
Materials and Methods
Project Specifications and Prior Laboratory Workflow Systems
This project was conducted over the years 2006 to 2008 in the Molecular Diagnostics Laboratory at The University of Texas M.D. Anderson Cancer Center, which performs RNA and DNA-based molecular testing for cancer and immunological applications (currently approximately 20,000 samples comprising 30,000 tests per year from cancer patients). The laboratory reports results into two different laboratory information systems (LIS), namely Cerner Classic (Cerner, Kansas City, MO) or Powerpath (IMPAC, Sunnyvale, CA), for blood and bone marrow samples and tissues and body fluids, respectively. The applications described were designed to provide internal laboratory management and workflow and to interface with the LIS reporting systems.
Before the development of these applications, the laboratory workflow included a manual system for tracking and processing of samples, processed materials and products, use of numerous different spreadsheets for run worksheets, and a referential database for recording testing results. The manual system for sample/specimen routing in our laboratory involved the generation of differently colored cards for each processed material type (eg, RNA, DNA, T-cell sort, etc). These paper cards then tracked along with each of the laboratory workstations until testing was completed and the report was generated.
Definitions of Data Elements
The initial biological materials received in the laboratory, including blood, bone marrow aspirate, other body fluids or paraffin-embedded tissues blocks/slides, are referred to herein as samples. The processed DNA, RNA, or protein lysates are referred to as processed materials and are sometimes obtained following further purification, microdissection, or cell separation of the samples. These are then used as the substrate(s) for testing, often following PCR amplification, with fragment analysis by capillary electrophoresis, DNA sequencing, microarrays, or other molecular methodologies.
Data elements from the paper test requisition captured at the time of sample accessioning include ordering physician, medical record number, sample ID registered in the LIS, sample type (eg, blood), and test(s) ordered. Data elements produced by the accessioning personnel include sample cell count and sample condition (eg, lysed or clotted blood). The initial status of any test (ie, “active,” “screen,” “hold,” or “cancel”) is determined by the accessioning personnel based on the sample type, quality or quantity, previous testing results and the laboratory's internal ordering rules/protocols.
Data elements captured during the sample processing steps are different for DNA, RNA, and protein lysates, as well as for cell sorting and plasma preparation. Minimum data elements collected for most processed materials included the technician(s) doing the processing, quantity and concentration (usually determined by spectrophotometry), as well as quality and purity.
Software Design Process
To produce design specifications for an integrated computerized workflow solution, laboratory personnel were sequentially interviewed, including managers, supervisors, and bench technicians. This allowed us to develop, from the start, a parallel set of workflow diagrams from the perspective of the laboratory managers (Figure 1A), or the bench technicians (Figure 1B). The most experienced technicians in the processing and testing areas of the laboratory were interviewed to obtain information on the granular components of their activities. Finally, all of the findings from the interviews and observations were taken back to the laboratory director for final discussion and refinement before work on the system began. These diagrams were then translated into tools for each of these personnel roles.
Figure 1.
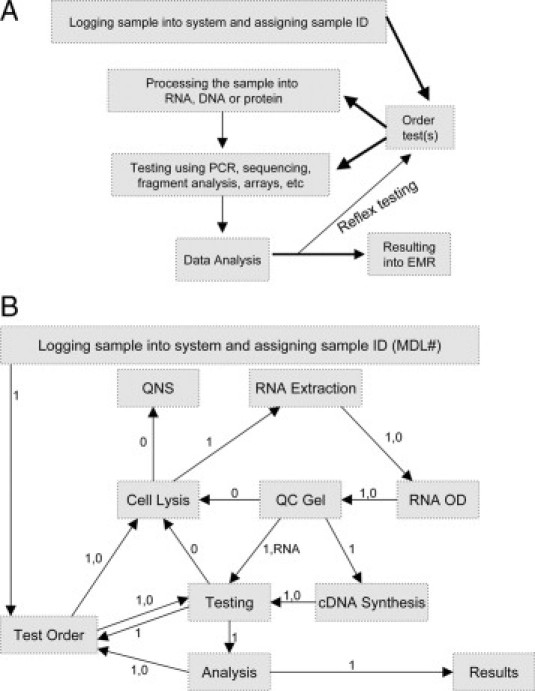
Overview of laboratory workflow. A: Design improvements were separately implemented for sample login, processing, and testing. B: Laboratory workflow for RNA from the perspective of the bench technician. Workflow at this level is similar to tiered structure necessary for programming, and includes workflow irregularities. Abbreviations: OD: optical density from spectrophotometry reading, QC: quality control, QNS: quantity not sufficient (cancel testing). In flow diagram, 1 indicates “proceed with next step” and 0 indicates “problem” with possible actions of trouble shoot (supervisor review), retest, or cancel.
Programming Tools and Network Architecture
The computer hardware included one database server and a separate application web server. We developed an overall container application composed of modular tools programmed using the .NET 2.0 platform (Microsoft, Bellevue, WA) that was accessed by end-users via a web server. Client machines across the laboratory received software updates automatically as changes were made and uploaded to the application server. These applications connected to a consolidated database hosted on a SQL Server 2000 platform (Microsoft). The database was designed to support complex testing algorithms and dynamic building of the testing library by a nested table structure. The format of results, accessed by external systems, was managed through stored procedure-based queries or by the open database connectivity interface.
Results
Redesign of the Sample Login Process: Implementing Screening Tools to Aid in Accessioning
The core functionality of the accessioning workstation in any molecular laboratory involves receipt of samples with their accompanying physician orders, log-in and cross-check of the patient demographics and sample data between the internal laboratory tracking system and the LIS and/or electronic medical record, and then allocation of appropriate portions of the sample to the various processed materials to be generated (Table 1). Since molecular test rosters are complex and require different sample types and amounts of processed material for DNA, RNA and protein methods, proper accessioning can be difficult, especially when material is limited.
Table 1.
Tasks for the Accessioning Station and How They Were Addressed
| Need | Sample task | Solution(s) |
|---|---|---|
| Accurate entry/cross-check of demographics and sample data | Logging a sample into both the lab system and LIS | Barcoding |
| Automatic matching of sample demographics against hospital records system | ||
| Algorithm-driven panel-based or individualized test ordering | Order a mutation test (alternatively) for one exon, a set of exons, or a set of genes based on diagnosis or pre-determined clinical indication(s) | Database design with granular definitions down to testable level |
| On-screen display of gene-exon-codon in a nested format | ||
| Allow either gene-specific or panel-based ordering based on rules | ||
| Prioritize testing when sample is limited | Decide which of the ordered tests to complete first | Supervisor reviews the queues, and the provisional (hold) status for tests |
| Eliminate duplicate ordering | Don't allow two mutation screens or translocation panels for the same patient | Display previous test results for each patient, with on-screen color highlighting duplicates |
| Choosing the correct followup tests | Ordering the correct assays for a tumor-associated translocation or mutation previously detected | MRD, prior results, and tumor-type database fields available at login |
| Rules-based population of data fields, whenever possible |
To facilitate these functions, design elements in the log-in tool included bar-code reading of LIS sample labels that was used to populate demographics in the laboratory database and to cross-check against the hospital admission database. These barcodes contained the institutional medical record number and a sample ID that are used to identify the sample within the electronic medical record. On-screen guidance on the sample requirements required for each test type and for those in clinical trials were implemented (Figure 2, and not shown). When sample quantity was limited, as indicated by highlighted values in the sample cell count field, supervisor review could be initiated for testing prioritization (Figure 3). Before moving samples to processing stations, they were tagged with another barcode containing a (hidden) internal numeric ID for tracking of all products generated from it.
Figure 2.
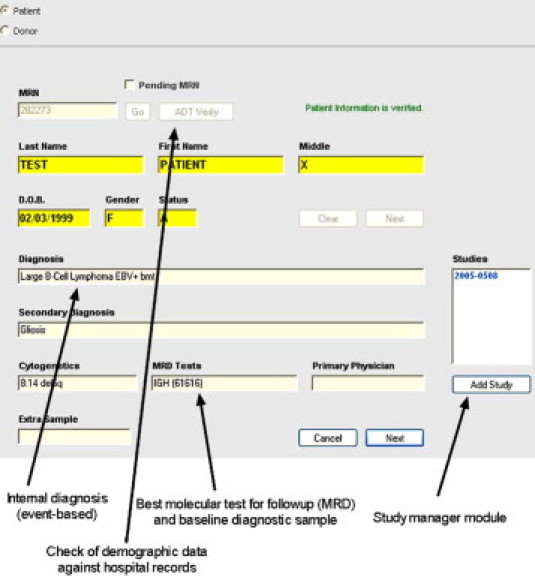
Login of samples: incorporating rules for order checking using internal laboratory data. Screenshot of a .NET software workflow management application covering the first step in the sample login process. Highlighted data fields relate to internal laboratory diagnosis, which accumulates disease-related and treatment events over time. (The example shown is an Epstein-Barr virus [EBV]-positive lymphoma that underwent bone marrow transplant [bmt]), with the appropriate test(s) for disease follow-up/minimal residual disease (MRD) monitoring indicated as quantitative immunoglobulin heavy chain (IGH) PCR. Also presented in the MRD field is the laboratory number of the prior baseline sample needed for quantitative comparison of disease levels with current sample. Rules for ordering can be tied to the values in these fields. On the right, a link to a separate module to control testing and notification for clinical trials involving this patient is also included.
Figure 3.
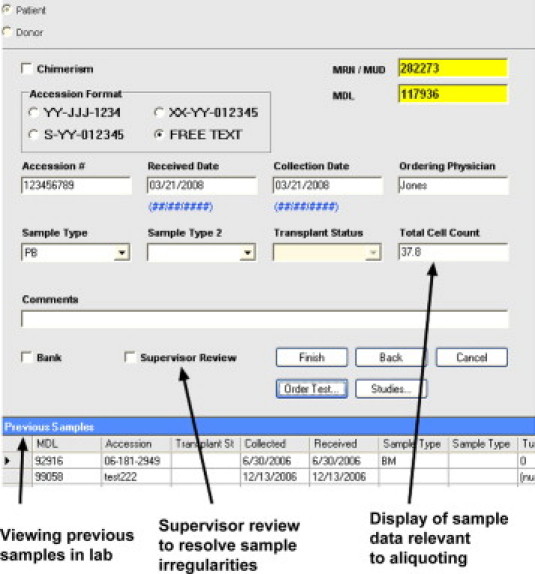
Login of samples: sample aliquoting and adequacy check before specific test ordering. Screenshot of a .NET software application covering the second step in the sample login process highlighting display of other recent samples from the same patient as a trigger to look for redundant testing. In the center of the field, an option for adding to supervisor review queue to resolve problems. On the right, display of data needed (eg, total cell count) for proper aliquoting of sample at this step to process into RNA, DNA, or protein materials or to bank the sample, for later use.
Since many laboratories (including our own) do not have fully-implemented online ordering, the accessioning station is also required to map the varied physician requests (often hand-written on a paper requisition) onto the laboratory's testing menu. This mapping is further complicated by the introduction of personalized medicine algorithms (especially in oncology) that have increasingly generated branch points for molecular testing that demand either tumor-specific test panels, or sequential (reflex) testing of a range of different genes, exons or even particular codons. A well-known example includes the sequential testing in the workup of a new chronic myeloproliferative neoplasm for BCR-ABL gene fusion, followed by mutation detection at codon 617 of JAK2, then codon 515 of MPL, or exon 12 of JAK2, until a positive result is obtained.
These testing algorithms are also subject to change based on evolution in the underlying clinical data, shifts in laboratory functionalities, and or cost considerations so the particulars of the algorithm are best not coded into the software. Instead, a flexible design is preferred that allows implementation of rules for ordering based on a variety of different data fields such as diagnosis (eg, codon 12/13 KRAS mutation for colon cancer versus both KRAS and NRAS testing for leukemia), clinical indication (eg, treatment-naïve versus therapy-resistant sample), or the previous result of testing performed (eg, confirmation of a previously identified mutation).
For these reasons, we sought to provide personnel at the accessioning station with the maximum amount of relevant data on diagnosis and the flexibility to implement panel-based, sequential algorithms, or individualized orders. Therefore, in our database design, all tests were regarded as components of potential panels to permit flexible display of hierarchical order sets but also to permit manual selection of individual components (Figure 4). For example, when mutation screening of a gene was requested, the current clinical and laboratory protocols could allow either ordering of the entire panel of exons or one or several particular exons, instead. Testing for any particular gene/exon/codon that was ordered but not in compliance with the current algorithms could also be placed on hold, and the request reviewed by the supervisor for cancellation or approval. Furthermore, the selection of particular order sets could be driven by rules-based algorithms derived from the data entered into the fields on the first two accessioning screens.
Figure 4.
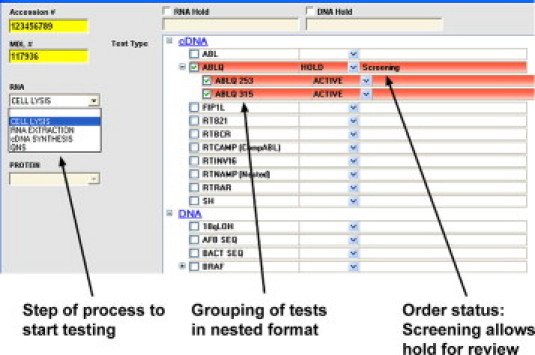
Login of samples: test ordering at a granular level. Screenshot of a .NET software application covering the third step in the sample login process. Shown on the left is a functionality to advance testing to any step to be used if partially or completely processed materials are received. On the right, the ordering sets are hierarchical to allow easy selection of any mix of components in a panel, including an entire gene or only specific exons. Shown are two tested codons (253 and 315) of the ABL1 kinase. The ordering status allows material to be put on hold for supervisor review before testing begins, but still allows processed material to be generated.
For followup samples, the accessioning workstation must also ensure that duplicate testing is not done (in accordance with laboratory ordering rules) and that the correct monitoring or minimal residual disease (MRD) test is ordered. For example, processing of a request for B-cell clonality studies could trigger an order for the (less-sensitive) non-quantitative PCR method or a more sensitive tumor-specific quantitative PCR method. The appropriate test for any particular patient is thus dependent on the particular diagnosis (eg, lymphoblastic leukemia versus lymphoma), whether or not the laboratory had previously received an initial (baseline) tumor sample, and whether specific primers to detect that tumor had already been designed.
To accomplish these test mapping functions for followup samples, the first two accessioning screens (Figures 2 and 3) provided the laboratory's internal tracking diagnosis and previous testing done on other samples from the same patient, including a “MRD” data field that listed the best monitoring test(s) for that particular patient. For those tests requiring comparison with a baseline level (eg, BCR-ABL), the sample ID of an appropriate patient-specific baseline sample already available within the laboratory was also displayed.
Redesign of Sample Processing: Allowing Redirection of Samples Based on Common Problems
The most complicated aspect of the workflow in any molecular laboratory is related to material processing and test setup. Although for each sample type there is a standardized sequence of work (eg, Figure 5 for RNA extraction), a significant number of cases require re-extraction, re-purification, or other adjustments due to variations in sample quality or quantity. Furthermore, due to the increase in retrospective and reflex testing in most laboratories, materials often reenter the sample processing stream at various points.
Figure 5.
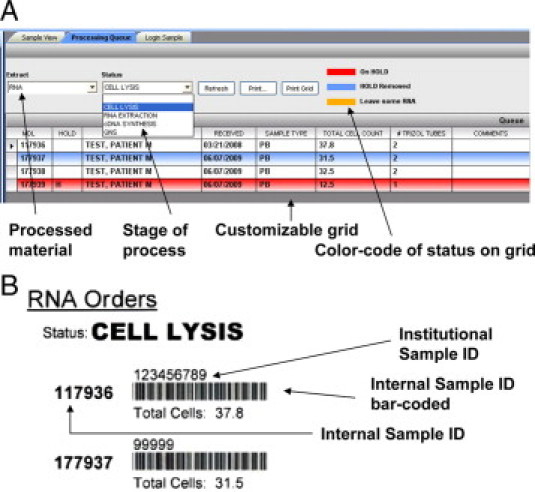
Materials processing queues: incorporating visual queues for irregularities. A: Software application covering sample conversion into DNA, RNA. and protein materials allows advancing or returning samples to specific points in the technical process; illustrated are the lysis, extraction and cDNA synthesis steps for RNA, or termination of the testing due to “quantity not sufficient” (QNS). Color-coding is used to highlight particular handling requirements for specific samples in the queue (eg, a sample on hold is illustrated). B: Printout of bar-coded labels for cases in queue for use at benchtop to mark completion status of cases or rapidly advance their status.
Therefore, we began our redesign of these steps with a careful mapping of workflow (eg, Figure 1b for RNA extraction), highlighting all possible branch points, and an assessment of the frequency of particular variances and problems encountered. This review revealed that the most common need was to repeat later steps in extraction due to inadequate or degraded starting material, technical failures or receipt of materials that were already partially processed. This led to a screen design for the processing workstations that placed only the most critical decision points as dropdown menu items at the top of the queue forms. This allowed technicians to quickly move an individual sample through the process manually, or to barcode groups of samples from one step to the next (Figure 5).
The tools developed required recording and viewing data only for those decision points in the workflow rather than modeling every step in the process. This led to display grids containing data elements that were essential and customized for a specific task, with a color key provided to allow rapid visual queuing of the current step in processing and any sample variances. For example, the tool for RNA extraction from blood displayed only four decision points in the dropdown menu: cell lysis, RNA extraction, cDNA synthesis, and completed versus quantity/quality not sufficient for testing (QNS) status (Figure 5A). Although our goal for this application was a completely paperless operation, a print feature was also included based on technician feedback given that the work often involved moving samples away from the computer monitor and thus barcode scanning samples off to the next step at any point (Figure 5B).
Redesign of Testing Queues: Accommodating Prioritization, and Sequential and Repeat Testing
Similar to the processing workflow, sortable order queues were designed for each of the laboratory workstations that allowed samples to be added to testing worksheets either singly or as a group (Figure 6). Process and workflow controls were also available on the testing queue to permit repeating the testing from any point in the laboratory process or removing the test due to inadequate material available.
Figure 6.
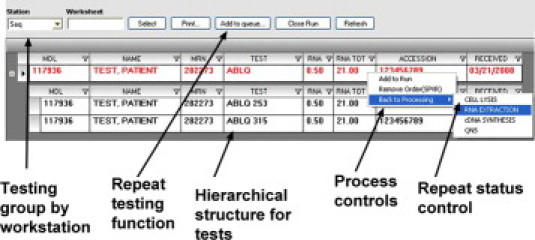
Testing queues: process controls and hierarchical organization of panels. Screenshot of a .NET software application demonstrating order queues for the DNA sequencing station shows the hierarchical structure of orders (a quantitative BCR-ABL mutation test is shown with condons 253 and 315 listed separately). Tests are added to the run worksheets by clicking on a panel or any individual component test. Other testing queues not diagrammed include quantitative PCR, PCR-fragment analysis, bead arrays, and comparative genomic hybridization/expression microarray.
One of the most critical aspects of performing expensive, complex molecular testing is grouping the orders into runs with the appropriate numbers of samples for maximum efficiency and ease of technician handling while maintaining adequate test turn-around times. By displaying all pending samples in the queues, the technicians could see which tests were in the pipeline, with those samples ready for testing indicated based on the presence or absence of adequate starting material indicated in the grid. The division of testing into workstations, namely fragment analysis/capillary electrophoresis, quantitative/real-time PCR, DNA sequencing, and microarrays, reflected test groups that share a common testing methodology and might be added together to a common run worksheet.
Another feature that was added was a supervisor comment field so that the bench technician could determine whether a sample had been approved for testing or not. Given the cost and complexity of testing, molecular diagnostic laboratories often need to review sample adequacy after materials have been made, but before proceeding with testing. We, therefore, included this feature so bench technicians could confirm for certain tests whether a sample was approved by the supervisor before proceeding to testing based on an order-level comment.
Outcomes Following Software Implementation
Over the period of implementation of these tools (May 2006 to early 2008), test volumes increased 36% across the laboratory, with only minimal increases in staffing levels. Overall turn-around times (TAT), measured in days from sample collection to report signout, were similar pre- and postimplementation for high-throughput PCR assays (on average 4–5 days), but TAT improved on average from 25 to 30% for the lower-volume more complex assays, particularly DNA sequencing. We believe that this improvement in TAT for DNA sequencing assays was related to: (1) having simplified pending queues with cases from both of the LIS worklists merged into a single universal pending queue for each workstation; (2) easier generation of hybrid runs with multiple tests assayed together; and (3) ease of generation of run worksheets. The availability of tools concisely listing prior results was also helpful to the technicians at the testing stations in identifying cases where testing was redundant or where the order needed to be modified.
Implementation of the workflow tools also led gradually to process changes throughout the laboratory. The most dramatic changes noted were improvements in the accuracy of test screening performed by the accessioning personnel. A metric reflecting this was the number of tests entered on samples at the login station that later needed to be canceled or credited. Comparing samples in the same month in the years before and after implementation, such misordering declined from 3% to 1.3%. The introduction, at the login step, of a “MRD” functionality for assisting in best test selection probably led to the most significant reductions in redundant or inappropriate testing. The elimination of paper cards for sample tracking and the increased use of bar coding led to a small decrease in lost or misdirected samples.
Although the discontinuation of the paper cards eliminated the need for card production, filing and re-filing (saving the work of one full clerical position), the goal of a totally paperless system was not completely achieved. For example, for ease of setup, bench technicians preferred to continue printing out their pending processing and testing work queues, and having that “run-level” paperwork track along with the run to the supervisor review step.
Discussion
We present here a model for molecular testing in the clinical laboratory that includes granular mapping of all steps in the processes, programming elements to reduce testing complexity, and design of a suite of tools to handle samples from receiving to reporting. The functions illustrated here focus mostly on workflow and sample management, which are the minimum essential requirements for testing. However, applications that facilitate rules-based reflex test selection, transformation of machine data into reportable values, and complex mapping of results from different platforms are now being added. Such modular toolkit-based applications are preferable over more structured LIS models for implementing personalized medicine informatics since they can quickly cover any gaps in laboratory workflow that arise when new tests, platforms, or clinical algorithms are introduced. Structured web-based applications also avoid the limitations of customized spreadsheets, including the tendency to improvise and change processes without vigorous validation, the propagation of data-entry errors, and the problems of maintaining a consistent, structured database.
Much previous work has gone into design of complex software tools to manage the large-scale and highly complex genomic, proteomic, and gene expression data emerging from new array-based/multiplexed testing platforms.2,3 However, most of this type of testing remains in the research setting with different goals and different personnel time and cost constraints than routine clinical laboratory testing.4,5 Conversely, most of the attention in LIS software design has focused on improving the efficiency of relatively simple, large-scale automated high-volume chemistry and hematology testing,6,7 or on integration of molecular imaging data with clinical data.8,9 Comparatively little attention has been given to the smaller scale, mostly manual, but still complex processes that exist today in molecular diagnostics laboratories where most clinical-grade molecular testing is currently done.10,11 Furthermore, applications that plug the gaps in the informatic needs and flexibly bridge the instrument output data with the LIS and electronic medical record have not been developed with any degree of sophistication.
Before the implementation of the newly designed workflow system described here, complex manual processes already existed in our laboratory that were repeatedly being changed by individual bench technologists. As a result, workflow was haphazard and shifted with each personnel change. Such individualized paper-based or spreadsheet solutions, which took place in multiple stages over the 15 years that the laboratory had been in operation, encompassed many innovative ideas and process improvements and some of these were retained in the new design. Others that proved inefficient or redundant were eliminated, when the processes were more precisely mapped onto the responsibilities of lab managers, station supervisors, and bench technicians.
For example, the previous simple system of having paper cards follow the sample through the lab processes had several advantages including being able to capture serial retesting and processing irregularities that constantly occur in molecular testing by simply moving the card back to previous workstations. Also, a stack of cards provides an easy index of pending cases for setting up worksheets and tube labeling. For these reasons, cards had proven difficult to displace, despite years of incremental improvements in automation and computerization. The downside of such a manual system was that neither could it accommodate multiple tests easily nor support complex, multistep test branching. Placing tests “on hold” before supervisor review was also difficult. Needless to say, tracking and filing these cards was tedious and labor-intensive. The tools designed here allowed the laboratory to retain the back-and-forth simplicity of the card while permitting automation, bar-coding, and computerization of worklist and worksheet production. The chief benefit of a formalized, comprehensive software suite is that the prior individualized approaches to managing work in the laboratory were now standardized and easier to track and integrate with middleware, testing platforms and clinical information systems.
An important secondary goal of our design was to reduce the apparent complexity of the information regarding previous testing and the new complex algorithms. We aimed to present bench technicians, supervisors, and mangers with the minimal relevant data necessary to complete each stage of the testing process. For example in the processing and order queues, the grids did not contain all of the data pertaining to the sample, but only the information relevant to the task at hand. This was so as to not crowd the screen and simple data presentation according to the usability heuristic that “less is more.”12
Our design model relied heavily on incorporating elements from the previous simple manual (but laborious) steps in the laboratory process before beginning the actual software development. As with the Lean processes for laboratory redesign,13 we did extensive data gathering and analysis from individuals at all levels of the laboratory who had different work roles to guide tool development through the entire process.14 This allowed us to greatly reduce the number of postrelease revisions needed. However, we received many process suggestions following initial implementation that were added to the second version. Feedback from signout pathologists led us to develop tools for them to more easily update diagnostic and MRD fields and to view previous results in multiple formats. Following supervisors' suggestions, we expanded the functionality of problem/hold queues to allow updating of the status of intermediate testing, and structured reporting of resolution of the problems.
The use of a modular structure was most helpful in allowing a seamless integration of these newly developed workflow tools without interrupting the existing operations of the laboratory. However, with introduction of each new application, we noted that adaptation by the bench and supervisory personnel ended up introducing changes to the original workflow that in turn led to enhancements in the manner of data presentation. This insight has led us to begin to develop more “test-building” functionality to allow laboratory personnel to adapt the tools in a more dynamic fashion. Such an approach is essential as the demands imposed on the clinical laboratories by personalized medicine multiply.
Acknowledgements
We thank the staff of the Molecular Diagnostics Laboratory, particularly Rajyalakshmi Luthra, Cindy Lewing, and Susan Biscanin, for their feedback, and Mark Routbort and Sanjivkumar Dave for help in database design.
Footnotes
Supported in part by a developmental grant from the Leukemia SPORE (1P50CA100632) awarded by the National Cancer Institute, Department of Health and Human Service. M.G. and D.J. have intellectual property interests in some of the data models underlying the applications described.
References
- 1.Lippi G, Guidi GC, Mattiuzzi C, Plebani M. Preanalytical variability: the dark side of the moon in laboratory testing. Clin Chem Lab Med. 2006;44:358–365. doi: 10.1515/CCLM.2006.073. [DOI] [PubMed] [Google Scholar]
- 2.Ohmann C, Kuchinke W. Future developments of medical informatics from the viewpoint of networked clinical research. Interoperability and integration. Methods Inf Med. 2009;48:45–54. [PubMed] [Google Scholar]
- 3.Burren OS, Healy BC, Lam AC, Schuilenburg H, Dolman GE, Everett VH, Laneri D, Nutland S, Rance HE, Payne F, Smyth D, Lowe C, Barratt BJ, Twells RC, Rainbow DB, Wicker LS, Todd JA, Walker NM, Smink LJ. Development of an integrated genome informatics, data management and workflow infrastructure: a toolbox for the study of complex disease genetics. Hum Genomics. 2004;1:98–109. doi: 10.1186/1479-7364-1-2-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan Q. The integration of personalized and systems medicine: bioinformatics support for pharmacogenomics and drug discovery. Methods Mol Biol. 2008;448:1–19. doi: 10.1007/978-1-59745-205-2_1. [DOI] [PubMed] [Google Scholar]
- 5.Amos J, Patnaik M. Commercial molecular diagnostics in the U.S.: The Human Genome Project to the clinical laboratory. Hum Mutat. 2002;19:324–333. doi: 10.1002/humu.10061. [DOI] [PubMed] [Google Scholar]
- 6.Bossuyt X, Verweire K, Blanckaert N. Laboratory medicine: challenges and opportunities. Clin Chem. 2007;53:1730–1733. doi: 10.1373/clinchem.2007.093989. [DOI] [PubMed] [Google Scholar]
- 7.Bartel M. Case study: improving efficiency in a large hospital laboratory. Clin Leadersh Manag Rev. 2004;18:267–272. [PubMed] [Google Scholar]
- 8.Yang JY, Yang MQ, Arabnia HR, Deng Y. Genomics, molecular imaging, bioinformatics, and bio-nano-info integration are synergistic components of translational medicine and personalized healthcare research. BMC Genomics. 2008;9 Suppl 2:I1. doi: 10.1186/1471-2164-9-S2-I1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sarachan BD, Simmons MK, Subramanian P, Temkin JM. Combining medical informatics and bioinformatics toward tools for personalized medicine. Methods Inf Med. 2003;42:111–115. [PubMed] [Google Scholar]
- 10.Guidi GC, Lippi G. Will “personalized medicine” need personalized laboratory approach? Clin Chim Acta. 2009;400:25–29. doi: 10.1016/j.cca.2008.09.029. [DOI] [PubMed] [Google Scholar]
- 11.Lippi G, Banfi G, Buttarello M, Ceriotti F, Daves M, Dolci A, Caputo M, Giavarina D, Montagnana M, Miconi V, Milanesi B, Mosca A, Morandini M, Salvagno GL. Recommendations for detection and management of unsuitable samples in clinical laboratories. Clin Chem Lab Med. 2007;45:728–736. doi: 10.1515/CCLM.2007.174. [DOI] [PubMed] [Google Scholar]
- 12.Zhang J, Johnson TR, Patel VL, Paige DL, Kubose T. Using usability heuristics to evaluate patient safety of medical devices. J Biomed Inform. 2003;36:23–30. doi: 10.1016/s1532-0464(03)00060-1. [DOI] [PubMed] [Google Scholar]
- 13.Persoon TJ, Zaleski S, Frerichs J. Improving preanalytic processes using the principles of lean production (Toyota Production System) Am J Clin Pathol. 2006;125:16–25. [PubMed] [Google Scholar]
- 14.Johnson CM, Turley JP. The significance of cognitive modeling in building healthcare interfaces. Int J Med Inform. 2006;75:163–172. doi: 10.1016/j.ijmedinf.2005.06.003. [DOI] [PubMed] [Google Scholar]


