Abstract
The somatic mutation JAK2 V617F is associated with BCR-ABL1-negative myeloproliferative neoplasms. Detection of this mutation aids diagnosis of these neoplasms, and quantification of JAK2 V617F may provide a method to monitor response to therapy. For these reasons, we designed a clinical assay that uses allele-specific PCR and real-time detection with hydrolysis probes for the quantification of JAK2 V617F, wild-type JAK2, and GAPDH transcripts. Mutant and wild-type JAK2 were quantified by using external plasmid standards that contain the relevant JAK2 V617F or JAK2 sequence, respectively. We tested 55 peripheral blood specimens from patients with suspected myeloproliferative neoplasms and 55 peripheral blood specimens from patients not known to have myeloproliferative neoplasms. Low-level, nonspecific amplification was detected in reactions containing a high copy number of plasmid standards and in specimens from patients not known to have myeloproliferative neoplasms, necessitating the use of a laboratory-established mutant to wild-type cutoff. The limit of detection established by using cell line dilutions is 0.1%, and this method identified three JAK2 V617F-positive patients who were not detected by a less sensitive method. The assay characteristics and our initial evaluation indicate this method can be used for the detection and quantification of JAK2 V617F, which should be useful for diagnosis of myeloproliferative neoplasms and potentially for monitoring minimal residual disease in future trials of therapies targeted to myeloproliferative neoplasms.
In early 2005, multiple independent groups described an acquired mutation in the Janus family kinase JAK2 gene associated with the classic BCR-ABL1-negative myeloproliferative neoplasms (MPNs).1,2,3,4,5 JAK2 is a cytoplasmic tyrosine kinase that mediates cytokine and growth factor receptor signaling. The mutation JAK2 NM_004972.2:c.1849G>T results in the substitution of a phenylalanine for a highly conserved valine (p.Val617Phe, V617F) within the pseudokinase domain (JH2), a protein domain that negatively regulates JAK2 catalytic activity.6 It is hypothesized that JAK2 V617F decreases the inhibition mediated by the pseudokinase domain and results in constitutive kinase activation.7
The JAK2 V617F mutation is detected in >95% of patients with polycythemia vera (PV) and ∼50% of patients with essential thrombocythemia (ET) and primary myelofibrosis (PMF).8,9 This mutation is also detected less frequently in cases of other BCR-ABL1-negative MPNs, myelodysplastic syndrome, and acute myeloid leukemia,10,11,12 and most published data indicated that JAK2 V617F is not found in healthy controls or individuals with reactive erythrocytosis or thrombocytosis.13
While numerous different methods have been used to detect and quantify JAK2 V617F allele and transcript levels (reviewed in Steensma14), we wanted to develop a method that was optimal for use in a molecular pathology laboratory. To this end, we identified three key characteristics that guided our assay design. First, we wanted to ensure that the method had adequate sensitivity to detect clinically relevant levels of JAK2 V617F. Baxter et al3 demonstrated that by using a sensitive allele-specific PCR method, they could detect JAK2 V617F alleles in ∼50% of specimens from patients with PV, ET, and PMF in which no mutant alleles were detected by a less sensitive DNA sequencing method. Further work by Verstovsek et al15 and Wang et al8 indicated that the JAK2 V617F mutation is present in virtually all cases of PV if a sufficiently sensitive assay is used. In addition, a lower JAK2 V617F allele burden may be present in some patients receiving cytoreductive therapy,8,16 as well as in many patients with ET,17 further underscoring the need for a highly sensitive assay.
Second, we chose to develop a quantitative assay because recent studies suggested there may be clinical utility in quantifying JAK2 V617F mutation allele and transcript levels both as a surrogate of therapeutic efficacy against the malignant MPN clone and as an independent prognostic factor. Selective small-molecule JAK2 antagonists inhibit growth of cell lines that express the JAK2 V617F protein,18 demonstrate therapeutic efficacy in mouse models of JAK2 V617F-induced MPNs,18,19 and inhibit primary hematopoietic cells derived from MPN patients with JAK2 V617F mutations.20 Given the promising in vitro and in vivo studies, clinical trials of JAK2 inhibitors have been initiated in patients with MPNs, so quantification of the JAK2 V617F levels may provide a method to monitor response to these and other treatments. In addition, studies of JAK2 V617F transgenic mice and MPN patients suggested that the ratio of JAK2 V617F to wild-type JAK2 expression determines the MPN phenotype.21,22 Furthermore, data from numerous studies indicated that JAK2 V617F allele burden correlates with a variety of hematological and clinical characteristics (reviewed in Vannucchi et al23), and these genotype-phenotype correlations may have prognostic significance.
The final assay characteristic we considered when developing our JAK2 V617F assay was whether to quantify allele or transcript levels. While the vast majority of current clinical assays detects or quantifies JAK2 V617F alleles, the quantification of JAK2 V617F transcript levels may provide several advantages over the measurement of allele burden. Work by Zhao et al5 demonstrated that the percentage of JAK2 V617F is usually higher in cDNA than in genomic DNA when PV mononuclear cell samples are analyzed. In addition, recent work by Ma et al24 demonstrated that analysis of mRNA from plasma samples of MPN patients resulted in a higher JAK2 V617F to wild-type JAK2 ratio when compared with analysis of DNA specimens. Furthermore, their work suggests that testing plasma mRNA results in increased sensitivity for detecting the JAK2 V617F mutation as compared with testing plasma DNA from paired samples; the JAK2 V617F mutation was detected by mRNA analysis in 37 samples and by DNA analysis in 33 samples. In addition, because the JAK2 V617F transcript level is likely to more directly reflect the JAK2 V617F protein expression level, this may be a more biologically relevant measurement than allele burden. Finally, our assay involves the analysis of RNA extracted from peripheral blood buffy coats, which therefore incorporates granulocytes as well as platelets. The inclusion of platelet RNA may be particularly relevant for ET, because prior studies have indicated that the analysis of platelet RNA may improve the clinical sensitivity of JAK2 V617F assays.25
Based on these considerations, we developed an assay that uses allele-specific PCR and real-time detection with hydrolysis probes for the quantification of JAK2 V617F and wild-type JAK2 transcript levels. Our initial evaluation indicates the method is sufficiently sensitive, specific, and reproducible to be used in clinical molecular pathology laboratories.
Materials and Methods
Patient Specimens
We performed quantitative testing on 55 peripheral blood specimens submitted to the Stanford Molecular Pathology Laboratory between August 2007 and January 2008 on which our standard JAK2 V617F mutation testing by PCR melting-curve analysis was ordered and performed. Specimens were excluded if sufficient RNA was not available for testing. In addition, we tested 55 peripheral blood specimens from adult patients not known to have a MPN. These control specimens were anonymized samples submitted to the Stanford Molecular Pathology Laboratory for heritable disease testing. All specimens were collected in tubes with EDTA anticoagulant, and these samples were obtained in accordance with a Stanford institutional review board-approved protocol.
Quantitative Allele-Specific RT-PCR
RNA was extracted from peripheral blood buffy coat specimens by using the RNeasy Plus Mini Kit (Qiagen, Valencia, CA) according to the manufacturer's instructions, and quantified by using spectrophotometric measurements. The reverse transcription reaction was performed by using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA) following the manufacturer's instructions.
Primers and probes for JAK2 V617F and wild-type JAK2 transcripts (Table 1) were designed with the aid of Primer Express software (Applied Biosystems, version 3.0) by using a single forward primer and a single probe for both the wild-type and V617F-mutation reactions. The reverse primers were designed by using the amplification refractory mutation system principle,26 a strategy previously used by Jones et al.27 The 3′-ends of the reverse primers are complementary to the mutant or wild-type cDNA sequences, and an additional mismatch was incorporated to enhance allelic discrimination. All primers and probes were synthesized by Operon Biotechnologies, Inc (Huntsville, AL). Subsequent to the design and evaluation of the primers and most specimen testing, a single-nucleotide polymorphism was reported in the sequence corresponding to the 5′-end of the common forward primer QTJK2-FOR (5′-GA[A/T]AAAGCACACAGAAACTATTCAGAGTC-3′, rs17490221 is italic). The A/T genotype has only been reported in 1 out of ∼330 individuals analyzed. Given the low frequency of this single-nucleotide polymorphism and its presence in the 5′-end of the primer, we have not investigated this further.
Table 1.
Primer and Probe Sequences
| Primer | Sequence |
|---|---|
| QTJK2-FOR | 5′-GATAAAGCACACAGAAACTATTCAGAGTC-3′ |
| QTJK2-REV-MUT | 5′-AGAATATTCTCGTCTCCACAaAA-3′ |
| QTJK2-REV-WT | 5′-AGAATATTCTCGTCTCCACAaAC-3′ |
| QTJK2-PROBE | 5′-FAM-AGCTTGCTCATCATACTTGCTGCTTCAAAGAA-TAMRA-3′ |
The underlined A or C is complementary to the JAK2 V617F mutated base 1849G>T or the wild-type JAK2 1849G, respectively. The lowercase “a” is a mismatch incorporated in the reverse primers to enhance allelic discrimination. FAM, 6-carboxy-fluorescein; TAMRA, 6-carboxy-tetramethyl-rhodamine.
We used the Applied Biosystems Human GAPDH Endogenous Control Kit (VIC/TAMRA Probe) according to the manufacturer's instructions to quantify GAPDH. The JAK2 V617F, wild-type JAK2, and GAPDH primer and probe sets were confirmed to not amplify genomic DNA.
Plasmid standards for the JAK2 V617F and wild-type JAK2 assays were manufactured by DNA 2.0, Inc (Menlo Park, CA) to contain 236 bp of JAK2 cDNA sequence (JAK2 NM_004972.2:c.1691_1926) that includes the V617F codon in pJ201. The JAK2 V617F and wild-type JAK2 plasmid standards were linearized by BglI digestion (New England Biolabs, Ipswich, MA) and include seven 10-fold serial dilutions, ranging from 106 to 1 targets per well. The GAPDH cDNA standards were generated from Human HeLa S3 Cells Total RNA (Stratagene, La Jolla, CA) and include five fivefold serial dilutions corresponding to 250, 50, 10, 2, and 0.4 ng of input RNA per well.
Real-time quantitative PCR was performed in the ABI 7700 or ABI 7900HT (Applied Biosystems) with an initial 10-minute incubation at 95°C, followed by 40 cycles of 15 seconds at 95°C and 1 minute at 60°C. The three transcripts were examined in separate reactions. Each 96-well plate included standard curves as well as positive, negative, and no-template controls in duplicate; patient samples were run in triplicate. Samples from individuals not known to have a MPN were run in duplicate. Initially, these samples from individuals with no known MPN were run with the same amount of input RNA and cDNA as the patient samples. Since nonspecific amplification was observed in the mutant reaction in samples with high levels of wild-type JAK2 cDNA, all control samples were rerun with twice the normal cDNA input in the PCR step to better evaluate the likely spectrum of clinical samples. The JAK2 V617F and JAK2 wells included 1X TaqMan Universal PCR Master Mix (Applied Biosystems), 20 pmol QTJK2-FOR primer, 10 pmol QTJK2-PROBE probe, and 20 pmol of either QTJK2-REV-MUT for JAK2 V617F or QTJK2-REV-WT for wild-type JAK2, and 2 μl of cDNA with a final volume of 25 μl.
The mean threshold cycles for JAK2 V617F, wild-type JAK2, and GAPDH were calculated for each sample by using the ABI software package, and the appropriate standard curve was used to determine the number of JAK2 V617F or wild-type JAK2 copies or nanograms of RNA equivalents for GAPDH. An example of the amplification plots with associated standard curve for the JAK2 V617F standards is shown in Figure 1, A and B. To ensure that RNA of sufficient quality and quantity is present for accurate quantification, we require at least 0.4 ng of RNA equivalents for GAPDH be detected to report a result for the assay. Two patient specimens failed to meet this requirement and were excluded from subsequent analysis. The following three ratios were calculated for each specimen: the number of JAK2 V617F copies per the number of wild-type JAK2 copies; the number of JAK2 V617F copies per microgram of RNA equivalents based on the GAPDH measurement; and the number of wild-type JAK2 copies per microgram of RNA equivalents based on the GAPDH measurement. We report specimens as not detected if they are determined to have a JAK2 V617F/wild-type JAK2 ratio ≤0.0005. This working cutoff was derived by analyzing the nonspecific amplification detected for reactions containing plasmids with 105 wild-type JAK2 copies for nine separate experiments on different days. Using these data, we calculated the average and SD of the JAK2 V617F/wild-type JAK2 ratio and set the cutoff at least three SDs above the average. Low level positive samples with a JAK2 V617F/wild-type JAK2 ratio between 0.04 and 0.0005 were retested by using an independent reverse transcription reaction to confirm that the result was reproducible. Specimens in which the melting-curve analysis was negative but the JAK2 V617F/wild-type JAK2 ratio was >0.0005 were also retested by using an independent reverse transcription reaction.
Figure 1.
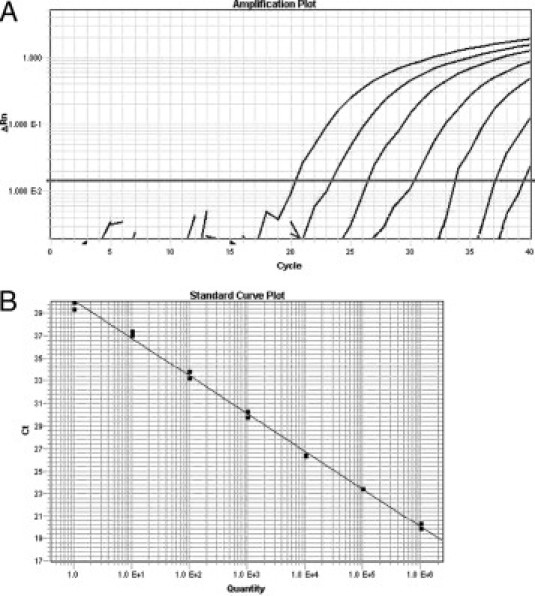
Examples of amplification and standard curve plots for JAK2 V617F quantification. A: Amplification plots of JAK2 V617F standards have the following number of copies per well (left to right): 106; 105; 104; 103; 102; 10; and 1. B: Standard curve with slope = −3.35; y-intercept = 40.1; and correlation coefficient = 0.997.
Sensitivity was determined by serial 10-fold dilutions of the HEL cell line (Coriell Cell Repository, Camden, NJ), which contains only JAK2 V617F mutant alleles, with the Stanford lymphoma cell line OCI-Ly8, which does not contain the JAK2 V617F mutation. The serial 10-fold dilutions spanned nine logs (100% to 0.000001%).
Reproducibility was measured by using three different approaches. Within-run reproducibility was measured by performing replicate analysis of a single specimen seven times on the same 96-well plate; we also reverse transcribed the same specimen in four different reactions and analyzed the reactions on the same 96-well plate. The between-run reproducibility was assessed by examining seven samples on three consecutive days. These seven samples had JAK2 V617F/wild-type JAK2 ratios that ranged from undetectable to 7.4 and included high, medium, and low transcript ratios.
Melting-Curve Analysis
Detection of the JAK2 V617F mutation by melting-curve analysis was performed as previously described.28 In brief, this assay detects the JAK2 V617F mutation by PCR and probe-dissociation analysis by using the LightCycler instrument (Roche, Indianapolis, IN), and the sensitivity of the assay was reported to be 5% by using cell line dilutions.
Results
Although most published data indicated that JAK2 V617F is not found in healthy individuals,13 other groups using highly sensitive assays have reported detecting low levels of the JAK2 V617F mutation in healthy individuals.29,30,31 This may represent low level nonspecific amplification of the wild-type allele, or some healthy controls may in fact harbor low levels of the mutation. Consequently, we initially examined the specificity of our assay by using plasmids containing either wild-type JAK2 or JAK2 V617F to determine whether the wild-type plasmid would generate any amplification signal when assayed by using the V617F-mutation reaction. We consistently observed low level nonspecific amplification in the mutant reactions when ≥105 wild-type JAK2 copies were present in the reactions (Figure 2). Using plasmid standards, 100,000 wild-type JAK2 copies results in nonspecific amplification in the mutant reaction that is quantified at approximately 10 copies of JAK2 V617F. Conversely, we observed similar nonspecific amplification in the wild-type reaction when only JAK2 V617F copies were present. However, this low level nonspecific amplification of the V617F template in the wild-type reaction is predicted not to appreciably affect the JAK2 V617F/wild-type JAK2 cDNA ratio since the highest JAK2 V617F/wild-type JAK2 observed is 11.8.
Figure 2.
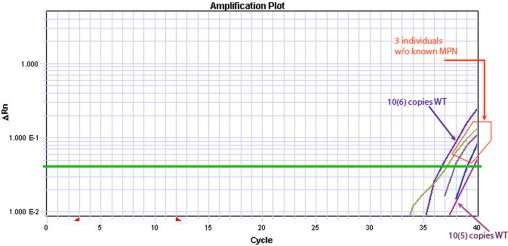
Low-level nonspecific amplification is observed in the mutant reaction when only wild-type JAK2 is present. We observed reproducible logarithmic amplification in the mutant reaction containing only known quantities of plasmids with the wild-type JAK2 cDNA sequence. The leftmost and rightmost amplification curves represent 106 and 105 wild-type JAK2 copies (WT), respectively. Similar low levels of mutant signal are detected in some specimens in our control population, as is shown for specimens from three individuals with no known MPN.
To further assess the potential for nonspecific amplification in clinical specimens, we determined the JAK2 V617F/wild-type JAK2 cDNA ratio for 55 specimens from patients with no known MPN. Initially, we identified three specimens with low level amplification in the JAK2 V617F reaction. To better evaluate the potential spectrum of clinical specimens and to confirm that we established a conservative positive/not detected cutoff value, we re-ran all 55 specimens with twice the input cDNA and found 29 of the specimens exhibited low levels of amplification in the JAK2 V617F reaction. Examples of these amplification curves are shown in Figure 2, and the JAK2 V617F/wild-type JAK2 ratios from the patients with no known MPN are depicted in Figure 3. Given the above findings and the low incidence of PV, ET, and PMF,32,33 we suspect that most, if not all, of these cases are false-positive results caused by nonspecific amplification. Such nonspecific amplification of the wild-type JAK2 allele has been previously described by Hammond et al.31 The authors used a similar mutation-specific primer design and hydrolysis probe chemistry to quantify the average number of JAK2 V617F DNA copies per cell. Furthermore, because many of the quantitative and highly sensitive JAK2 V617F assays use the same mutation-specific primer design described by Jones et al,27 we suggest that cases with very low levels of JAK2 V617F be interpreted with care. As was discussed by Hammond et al,31 this nonspecific amplification may be caused by low level mispriming of the mutation-specific primer or by the presence of low levels of 3′-truncated mutation-specific primers. Regardless of the mechanism, we were unable to eliminate this nonspecific amplification by limiting primer freeze-thaw cycles or by primer purification. Consequently, we used the nonspecific amplification observed for plasmid standards containing a known number of wild-type JAK2 copies to establish a working cutoff value of 0.0005 for use in our laboratory (Figure 3). We report results with ratios ≤0.0005 as “not detected” to minimize the possibility of generating a false-positive result. The clinical significance of patients with low JAK2 V617F/wild-type JAK2 transcript ratios is unclear at this time and will ultimately require longitudinal follow-up of these patients. Such work is underway.
Figure 3.

JAK2 V617F/wild-type JAK2 transcript ratios for parallel-tested specimens and samples from individuals with no known MPN. Ratios equal to zero are not depicted; however, the following number of zero values were observed for each category: positive (0); borderline positive (0); negative (10); and no known MPN (29). Borderline positive refers to results in which the probe-dissociation pattern is consistent with the presence of the JAK2 V617F mutation at a low level (<10% of the assayed cells), but cannot be considered definitive. Based on the nonspecific amplification observed with plasmids containing wild-type JAK2, we report specimens as not detected if they are determined to have JAK2 V617F/wild-type JAK2 ratio ≤0.0005 (dotted line).
We used cell line dilutions to estimate the limit of detection of the assay. The JAK2 V617F/wild-type JAK2 transcript ratio was 0.0021 for the 0.1% dilution and 0.0003 for the 0.01% dilution. Using the 0.0005 cutoff ratio at which we report specimens as not detected, the limit of detection for this assay is estimated to be 0.1%. This is among the lowest values reported in the literature for DNA-based and especially cDNA-based assays.34,35,36,37 One potential concern about using the HEL cell line, which contains only JAK2 V617F mutant alleles, for the measurement of the limit of detection is that it has been shown to have undergone significant genomic amplification at the JAK2 locus.38 However, in our experiments, the HEL cell line does not display increased expression of JAK2 V617F RNA relative to specimens characterized as positive by our quantitative assay. The normalized JAK2 V617F copies detected in the HEL cell line (measured as copies per microgram of RNA equivalents [52,600 JAK2 V617F copies/μg GAPDH RNA]) was below the median number of JAK2 V617F copies in the specimens that were characterized as positive by our quantitative assay (266,000 JAK2 V617F copies/μg GAPDH RNA, Table 2). Consequently, these data indicate that we can detect a JAK2 V617F-positive clone that comprises approximately 0.1% of the cell population.
Table 2.
Median (Range) of Values for Clinical Specimens Classified by the Quantitative Assay as JAK2 V617F-Positive or -Not Detected
| JAK2 V617F, copies | Wild-type JAK2, copies | GAPDH, ng | JAK2 V617F/μg GAPDH | Wild-type JAK2/μg GAPDH | JAK2 V617F/ Wild-type JAK2 | |
|---|---|---|---|---|---|---|
| JAK2 V617F- | 14,300 | 31,400 | 44.8 | 266,000 | 879,000 | 4.20E–01 |
| positive | (47.7–83,800) | (1330–101,000) | (1.00–136) | (352–83,800,000) | (12,000–67,100,000) | (1.42E-03–1.18E+01) |
| JAK2 V617F-not | 1.90 | 54,700 | 32.2 | 42.1 | 1,420,000 | 4.33E–05 |
| detected | (0–16.2) | (7610–193,000) | (1.21–106) | (0–1230) | (452,000–8,450,000) | (0–1.87E-04) |
Within-run reproducibility was initially measured by performing replicate analysis of a single cDNA specimen, and the resulting CV was calculated to be 14.6%. We also reverse transcribed the same specimen in different reactions, and the within-run CV was again calculated to be 14.6%. The between-run reproducibility showed an average CV of 11.8% (range, 3.6 to 23.3).
Parallel testing of specimens submitted to our laboratory for clinical JAK2 V617F mutation testing by melting-curve analysis28 demonstrates that our new quantitative JAK2 V617F assay provides valuable qualitative data. All specimens that were characterized as positive or borderline positive by the melting-curve analysis were also characterized as positive by the quantitative assay (Table 3, Figure 3). Borderline positive refers to results in which the probe-dissociation pattern is consistent with the presence of the JAK2 V617F mutation at a low level (<10% of the assayed cells) but cannot be considered definitive. The quantitative JAK2 V617F assay allowed definitive characterization of these seven specimens as containing the JAK2 V617F mutation. In addition, the quantitative assay identified the JAK2 V617F mutation in three specimens where it was not detected by melting-curve analysis. We favor that these specimens contain low levels of the JAK2 V617F mutation that are below the limit of sensitivity of the melting-curve assay because the JAK2 V617F/wild-type JAK2 transcript ratios are low (Figure 3), the results are reproducible, and the results are consistent with the patients' clinical history. Each of the three specimens was confirmed by repeat testing from an independent reverse transcription reaction. One patient had a reported history of PV, and another was reported to have a history of ET. The third patient was reported to have anemia, and a follow-up specimen from 1 year later was interpreted as having a borderline positive result by melting-curve analysis. Collectively, these data suggest that the real-time quantitative PCR assay can identify clinically relevant low level JAK2 V617F-positive cases that are not detected by less sensitive assays. This will likely be most useful in confirming the clinical suspicion of a diagnosis of MPNs in patients with low JAK2 V617F allele burdens. Another potential application of quantitative JAK2 V617F testing is to evaluate response to JAK2 inhibitor and other therapies. Because we do not know the effect of JAK2 inhibitors and other therapies on wild-type JAK2 and JAK2 V617F transcript levels, we did not want to rely solely on the JAK2 V617F/wild-type JAK2 transcript ratios to evaluate treatments. For this reason, we also quantify and normalize JAK2 V617F and wild-type JAK2 to GAPDH. We are performing additional testing by using pre- and posttreatment specimens to evaluate the utility of this assay in monitoring response to therapy.
Table 3.
Results from Parallel-Tested Laboratory Samples and Individuals without a Known MPN
| Quantitative assay results, n |
||
|---|---|---|
| Results | Positive | Not detected |
| Melting-curve results | ||
| Positive | 19 | 0 |
| Borderline* | 7 | 0 |
| Not detected | 3† | 24 |
| No known MPN | 0 | 55 |
Borderline (positive) refers to results in which the probe dissociation pattern is consistent with the presence of the JAK2 V617F mutation at a low level (<10% of the assayed cells), but cannot be considered definitive.
Two patients had a reported history of PV or ET. The third patient had a reported diagnosis of anemia, and a follow-up specimen from 1 year later was interpreted as having a borderline (positive) result by melting-curve analysis.
Discussion
We chose to develop an RNA-based assay for the detection and quantification of the JAK2 V617F mutation because published data indicated that it may have several advantages over the more standard DNA-based testing. First, analysis of paired plasma samples from MPN patients suggests that analysis of mRNA may provide higher sensitivity for detecting JAK2 V617F as compared with analysis of DNA.24 Second, an RNA-based assay allows for a more direct analysis of JAK2 V617F expression level, and also accommodates the examination of platelet RNA. Prior work suggested that analysis of platelets may be a more sensitive approach for detecting JAK2 V617F in BCR-ABL1-negative MPN patients as compared with analysis of granulocytes alone.25 We suggest that examination of RNA from buffy coats (which include platelets and granulocytes) may also offer increased sensitivity when compared with looking at a single cell population. Finally, unlike many of the more sensitive published assays, our assay does not require isolation of granulocytes, because this procedure is difficult to integrate into most laboratories' workflow. Further testing with additional clinical specimens will ultimately be required to determine whether these potential advantages outweigh the disadvantages of working with RNA, including RNA lability and the added work of a reverse-transcription step.
In summary, we developed an assay that uses allele-specific PCR and real-time detection with hydrolysis probes for the quantification of JAK2 V617F and wild-type JAK2 transcript levels by using plasmid standards. The reproducibility, sensitivity, and specificity are comparable with other published real-time methods for the detection of point mutations. This assay has a low limit of detection for JAK2 V617F transcripts, and it was specifically designed and evaluated for the detection of JAK2 V617F transcripts and subsequent minimal residual disease monitoring in the clinical molecular pathology laboratory.
Acknowledgements
We thank Sabrina Khan for assistance with specimen processing and Alex McMillan for useful discussions regarding statistical analysis.
Footnotes
Supported in part by the National Institutes of Health under grant 2PO1CA49605.
References
- 1.James C, Ugo V, Le Couédic JP, Staerk J, Delhommeau F, Lacout C, Garçon L, Raslova H, Berger R, Bennaceur-Griscelli A, Villeval JL, Constantinescu SN, Casadevall N, Vainchenker W. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434:1144–1148. doi: 10.1038/nature03546. [DOI] [PubMed] [Google Scholar]
- 2.Levine RL, Wadleigh M, Cools J, Ebert BL, Wernig G, Huntly BJ, Boggon TJ, Wlodarska I, Clark JJ, Moore S, Adelsperger J, Koo S, Lee JC, Gabriel S, Mercher T, D'Andrea A, Fröhling S, Döhner K, Marynen P, Vandenberghe P, Mesa RA, Tefferi A, Griffin JD, Eck MJ, Sellers WR, Meyerson M, Golub TR, Lee SJ, Gilliland DG. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7:387–397. doi: 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 3.Baxter EJ, Scott LM, Campbell PJ, East C, Fourouclas N, Swanton S, Vassiliou GS, Bench AJ, Boyd EM, Curtin N, Scott MA, Erber WN, Green AR. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365:1054–1061. doi: 10.1016/S0140-6736(05)71142-9. [DOI] [PubMed] [Google Scholar]
- 4.Kralovics R, Passamonti F, Buser AS, Teo SS, Tiedt R, Passweg JR, Tichelli A, Cazzola M, Skoda RC. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med. 2005;352:1779–1790. doi: 10.1056/NEJMoa051113. [DOI] [PubMed] [Google Scholar]
- 5.Zhao R, Xing S, Li Z, Fu X, Li Q, Krantz SB, Zhao ZJ. Identification of an acquired JAK2 mutation in polycythemia vera. J Biol Chem. 2005;280:22788–22792. doi: 10.1074/jbc.C500138200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saharinen P, Takaluoma K, Silvennoinen O. Regulation of the Jak2 tyrosine kinase by its pseudokinase domain. Mol Cell Biol. 2000;20:3387–3395. doi: 10.1128/mcb.20.10.3387-3395.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morgan KJ, Gilliland DG. A role for JAK2 mutations in myeloproliferative diseases. Annu Rev Med. 2008;59:213–222. doi: 10.1146/annurev.med.59.061506.154159. [DOI] [PubMed] [Google Scholar]
- 8.Wang YL, Vandris K, Jones A, Cross NC, Christos P, Adriano F, Silver RT. JAK2 mutations are present in all cases of polycythemia vera. Leukemia. 2008;22:1289. doi: 10.1038/sj.leu.2405047. [DOI] [PubMed] [Google Scholar]
- 9.James C, Ugo V, Casadevall N, Constantinescu SN, Vainchenker W. A JAK2 mutation in myeloproliferative disorders: pathogenesis and therapeutic and scientific prospects. Trends Mol Med. 2005;11:546–554. doi: 10.1016/j.molmed.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 10.Steensma DP, Dewald GW, Lasho TL, Powell HL, McClure RF, Levine RL, Gilliland DG, Tefferi A. The JAK2 V617F activating tyrosine kinase mutation is an infrequent event in both “atypical” myeloproliferative disorders and myelodysplastic syndromes. Blood. 2005;106:1207–1209. doi: 10.1182/blood-2005-03-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jelinek J, Oki Y, Gharibyan V, Bueso-Ramos C, Prchal JT, Verstovsek S, Beran M, Estey E, Kantarjian HM, Issa JP. JAK2 mutation 1849G>T is rare in acute leukemias but can be found in CMML: Philadelphia chromosome-negative CML and megakaryocytic leukemia. Blood. 2005;106:3370–3373. doi: 10.1182/blood-2005-05-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levine RL, Loriaux M, Huntly BJ, Loh ML, Beran M, Stoffregen E, Berger R, Clark JJ, Willis SG, Nguyen KT, Flores NJ, Estey E, Gattermann N, Armstrong S, Look AT, Griffin JD, Bernard OA, Heinrich MC, Gilliland DG, Druker B, Deininger MW. The JAK2V617F activating mutation occurs in chronic myelomonocytic leukemia and acute myeloid leukemia, but not in acute lymphoblastic leukemia or chronic lymphocytic leukemia. Blood. 2005;106:3377–3379. doi: 10.1182/blood-2005-05-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelson ME, Steensma DP. JAK2 V617F in myeloid disorders: what do we know now, and where are we headed? Leuk Lymphoma. 2006;47:177–194. doi: 10.1080/10428190500301348. [DOI] [PubMed] [Google Scholar]
- 14.Steensma DP. JAK2 V617F in myeloid disorders: molecular diagnostic techniques and their clinical utility: a paper from the 2005 William Beaumont Hospital Symposium on Molecular Pathology. J Mol Diagn. 2006;8:397–411. doi: 10.2353/jmoldx.2006.060007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verstovsek S, Silver RT, Cross NC, Tefferi A. JAK2V617F mutational frequency in polycythemia vera: 100%, >90%, less? Leukemia. 2006;20:2067. doi: 10.1038/sj.leu.2404379. [DOI] [PubMed] [Google Scholar]
- 16.Girodon F, Schaeffer C, Cleyrat C, Mounier M, Lafont I, Santos FD, Vidal A, Maynadié M, Hermouet S. Frequent reduction or absence of detection of the JAK2-mutated clone in JAK2V617F-positive patients within the first years of hydroxyurea therapy. Haematologica. 2008;93:1723–1727. doi: 10.3324/haematol.13081. [DOI] [PubMed] [Google Scholar]
- 17.Antonioli E, Guglielmelli P, Poli G, Bogani C, Pancrazzi A, Longo G, Ponziani V, Tozzi L, Pieri L, Santini V, Bosi A, Vannucchi AM. Influence of JAK2V617F allele burden on phenotype in essential thrombocythemia. Haematologica. 2008;93:41–48. doi: 10.3324/haematol.11653. [DOI] [PubMed] [Google Scholar]
- 18.Pardanani A, Hood J, Lasho T, Levine RL, Martin MB, Noronha G, Finke C, Mak CC, Mesa R, Zhu H, Soll R, Gilliland DG, Tefferi A. TG101209, a small molecule JAK2-selective kinase inhibitor potently inhibits myeloproliferative disorder-associated JAK2V617F and MPLW515L/K mutations. Leukemia. 2007;21:1658–1668. doi: 10.1038/sj.leu.2404750. [DOI] [PubMed] [Google Scholar]
- 19.Wernig G, Kharas MG, Okabe R, Moore SA, Leeman DS, Cullen DE, Gozo M, McDowell EP, Levine RL, Doukas J, Mak CC, Noronha G, Martin M, Ko YD, Lee BH, Soll RM, Tefferi A, Hood JD, Gilliland DG. Efficacy of TG101348, a selective JAK2 inhibitor, in treatment of a murine model of JAK2V617F-induced polycythemia vera. Cancer Cell. 2008;13:311–320. doi: 10.1016/j.ccr.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 20.Lasho TL, Tefferi A, Hood JD, Verstovsek S, Gilliland DG, Pardanani A. TG101348, a JAK2-selective antagonist, inhibits primary hematopoietic cells derived from myeloproliferative disorder patients with JAK2V617F, MPLW515K or JAK2 exon 12 mutations as well as mutation negative patients. Leukemia. 2008;22:1790–1792. doi: 10.1038/leu.2008.56. [DOI] [PubMed] [Google Scholar]
- 21.Tiedt R, Hao-Shen H, Sobas MA, Looser R, Dirnhofer S, Schwaller J, Skoda RC. Ratio of mutant JAK2–V617F to wild-type Jak2 determines the MPD phenotypes in transgenic mice. Blood. 2008;111:3931–3940. doi: 10.1182/blood-2007-08-107748. [DOI] [PubMed] [Google Scholar]
- 22.Xing S, Wanting TH, Zhao W, Ma J, Wang S, Xu X, Li Q, Fu X, Xu M, Zhao ZJ. Transgenic expression of JAK2V617F causes myeloproliferative disorders in mice. Blood. 2008;111:5109–5117. doi: 10.1182/blood-2007-05-091579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vannucchi AM, Antonioli E, Guglielmelli P, Pardanani A, Tefferi A. Clinical correlates of JAK2V617F presence or allele burden in myeloproliferative neoplasms: a critical reappraisal. Leukemia. 2008;22:1299–1307. doi: 10.1038/leu.2008.113. [DOI] [PubMed] [Google Scholar]
- 24.Ma W, Kantarjian H, Zhang X, Yeh CH, Zhang ZJ, Verstovsek S, O'Brien S, Giles F, Albitar M. Plasma levels of JAK2 mRNA in patients with chronic myeloproliferative diseases with and without V617F mutation: implications for prognosis and disease biology. Int J Lab Hematol. 2009 doi: 10.1111/j.1751-553X.2009.01140.x. [DOI] [PubMed] [Google Scholar]
- 25.Toyama K, Karasawa M, Yamane A, Irisawa H, Yokohama A, Saitoh T, Handa H, Matsushima T, Sawamura M, Miyawaki S, Murakami H, Nojima Y, Tsukamoto N. JAK2-V617F mutation analysis of granulocytes and platelets from patients with chronic myeloproliferative disorders: advantage of studying platelets. Br J Haematol. 2007;139:64–69. doi: 10.1111/j.1365-2141.2007.06755.x. [DOI] [PubMed] [Google Scholar]
- 26.Newton CR, Graham A, Heptinstall LE, Powell SJ, Summers C, Kalsheker N, Smith JC, Markham AF. Analysis of any point mutation in DNA: the amplification refractory mutation system (ARMS) Nucleic Acids Res. 1989;17:2503–2516. doi: 10.1093/nar/17.7.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones AV, Kreil S, Zoi K, Waghorn K, Curtis C, Zhang L, Score J, Seear R, Chase AJ, Grand FH, White H, Zoi C, Loukopoulos D, Terpos E, Vervessou EC, Schultheis B, Emig M, Ernst T, Lengfelder E, Hehlmann R, Hochhaus A, Oscier D, Silver RT, Reiter A, Cross NC. Widespread occurrence of the JAK2 V617F mutation in chronic myeloproliferative disorders. Blood. 2005;106:2162–2168. doi: 10.1182/blood-2005-03-1320. [DOI] [PubMed] [Google Scholar]
- 28.Lay M, Mariappan R, Gotlib J, Dietz L, Sebastian S, Schrijver I, Zehnder JL. Detection of the JAK2 V617F mutation by LightCycler PCR and probe dissociation analysis. J Mol Diagn. 2006;8:330–334. doi: 10.2353/jmoldx.2006.050130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sidon P, El Housni H, Dessars B, Heimann P. The JAK2V617F mutation is detectable at very low level in peripheral blood of healthy donors. Leukemia. 2006;20:1622. doi: 10.1038/sj.leu.2404292. [DOI] [PubMed] [Google Scholar]
- 30.Hammond E, Shaw K, Herrmann R. The JAK2 V617F mutation is detectable in granulocyte populations at greater than two copies per cell among individuals with myeloproliferative disorders. Leukemia. 2007;21:815–816. doi: 10.1038/sj.leu.2404567. [DOI] [PubMed] [Google Scholar]
- 31.Hammond E, Shaw K, Carnley B, P'ng S, James I, Herrmann R. Quantitative determination of JAK2 V617F by TaqMan: an absolute measure of averaged copies per cell that may be associated with the different types of myeloproliferative disorders. J Mol Diagn. 2007;9:242–248. doi: 10.2353/jmoldx.2007.060125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Modan B. An epidemiological study of polycythemia vera. Blood. 1965;26:657–667. [PubMed] [Google Scholar]
- 33.Mesa RA, Silverstein MN, Jacobsen SJ, Wollan PC, Tefferi A. Population-based incidence and survival figures in essential thrombocythemia and agnogenic myeloid metaplasia: an Olmsted County Study, 1976–1995. Am J Hematol. 1999;61:10–15. doi: 10.1002/(sici)1096-8652(199905)61:1<10::aid-ajh3>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 34.Greiner TC. Diagnostic assays for the JAK2 V617F mutation in chronic myeloproliferative disorders. Am J Clin Pathol. 2006;125:651–653. doi: 10.1309/NXXT-GRCX-D0TM-A3C2. [DOI] [PubMed] [Google Scholar]
- 35.Nussenzveig RH, Swierczek SI, Jelinek J, Gaikwad A, Liu E, Verstovsek S, Prchal JF, Prchal JT. Polycythemia vera is not initiated by JAK2V617F mutation. Exp Hematol. 2007;35:32–38. doi: 10.1016/j.exphem.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 36.Kröger N, Badbaran A, Holler E, Hahn J, Kobbe G, Bornhäuser M, Reiter A, Zabelina T, Zander AR, Fehse B. Monitoring of the JAK2–V617F mutation by highly sensitive quantitative real-time PCR after allogeneic stem cell transplantation in patients with myelofibrosis. Blood. 2007;109:1316–1321. doi: 10.1182/blood-2006-08-039909. [DOI] [PubMed] [Google Scholar]
- 37.Vannucchi AM, Pancrazzi A, Bogani C, Antonioli E, Guglielmelli P. A quantitative assay for JAK2(V617F) mutation in myeloproliferative disorders by ARMS-PCR and capillary electrophoresis. Leukemia. 2006;20:1055–1060. doi: 10.1038/sj.leu.2404209. [DOI] [PubMed] [Google Scholar]
- 38.Quentmeier H, MacLeod RA, Zaborski M, Drexler HG. JAK2 V617F tyrosine kinase mutation in cell lines derived from myeloproliferative disorders. Leukemia. 2006;20:471–476. doi: 10.1038/sj.leu.2404081. [DOI] [PubMed] [Google Scholar]


