Abstract
Rapid, semiautomated, and fully automated multiplex real-time RT-PCR assays were developed and validated for the detection of influenza (Flu) A, Flu B, and respiratory syncytial virus (RSV) from nasopharyngeal specimens. The assays can detect human H1N1, H3N2, and swine-origin (S-OIV) H1N1 Flu A viruses and were effectively used to distinguish Flu A infections (of all subtypes) from Flu B and RSV infections during the current S-OIV outbreak in Milwaukee, WI. The analytical limits of detection were 10−2 to 101 TCID50/ml depending on the platform and analyte and showed only one minor cross-reaction among 23 common respiratory pathogens (intermittent cross-reaction to adenovirus at >107 TCID50/ml). A total of 100 clinical samples were tested by tissue culture, both automated assays, and the US Food and Drug Administration-approved ProFlu+ assay. Both the semiautomated and fully automated assays exhibited greater overall (Flu A, Flu B, and RSV combined) clinical sensitivities (93 and 96%, respectively) and individual Flu A sensitivities (100%) than the Food and Drug Administration-approved test (89% overall sensitivity and 93% Flu A sensitivity). All assays were 99% specific. During the S-OIV outbreak in Milwaukee, WI, the fully automated assay was used to test 1232 samples in 2 weeks. Flu A was detected in 134 clinical samples (126 H1N1 S-OIV, 5 H1N1 [human], and 1 untyped) with 100% positive agreement compared with other “in-house” validated molecular assays, with only 2 false-positive results. Such accurate testing using automated high-throughput molecule systems should allow clinicians and public health officials to react quickly and effectively during viral outbreaks.
Tissue culture had been the gold standard for respiratory virus diagnosis until approximately the mid-1990s when large multiplex RT-PCR assays first became available clinically and commercially.1,2,3,4,5,6,7 RT-PCR was quickly shown to be significantly more sensitive and highly specific for the detection of influenza (Flu) and respiratory syncytial viruses (RSV) (and most other respiratory viruses as well). Since that time, many laboratories (including the Midwest Respiratory Virus Program) have worked to develop automated platforms that would encompass nucleic acid extraction, RT-PCR amplification, and detection 8,9,10,11. The goals have been to decrease technician time to only a few minutes, to make the assay time only 2 to 3 hours (or faster), to decrease cost, and to keep the footprint of the equipment as small as possible. In early 2008, two multiplex RT-PCR assays received US Food and Drug Administration (FDA) clearance for the detection of Flu A, Flu B, and RSV A/B. xTAG RVP (Luminex Molecular Diagnostics, Toronto, ON, Canada) is capable of detecting 9 viruses and a combined 11 subtypes for 3 of these viruses.12 This is an open format assay, which increases the chance for contamination, and requires approximately 8 hours for completion and the use of a Luminex flow cytometer. The second FDA-approved multiplex RT-PCR assay is the ProFlu+ Assay (Prodesse, Waukesha, WI), which is capable of detecting Flu A, Flu B, and RSV simultaneously. The assay takes approximately 3 hours, excluding extraction time, and as approved, is semiautomated: using automated extraction via the NucliSENS easyMAG System (bioMérieux, Durham, NC) or the MagNA Pure LC Instrument (Roche, Indianapolis, IN) followed by manual transfer for real-time thermocycling on a Smart Cycler II (Cepheid, Sunnyvale, CA). In addition to these multiplex tests, the FDA, in September 2008, approved a series of singleplex RT-PCR assays developed by the Centers for Disease Control and Prevention that are capable of typing Flu viruses as A or B and further subtyping Flu A viruses as H1, H3, or H5 subtypes. However, the reagents are only readily available to members of the Centers for Disease Control and Prevention's Laboratory Response Network.13
The dearth of FDA-approved Flu A, Flu B, and RSV assays has led a number of laboratories to develop their own “in-house” singleplex or multiplex semiautomated assays using commercial automated extractors, real-time thermocyclers, and research use-only reagents. Our laboratory has recently developed semiautomated and fully automated multiplex RT-PCR assays capable of detecting Flu A, Flu B, and RSV A/B with a noncompetitive RNA internal control (MS2 RNA phage, ZeptoMetrix Corp., Buffalo, NY). The semiautomated assay uses the NucliSENS easyMAG System for automated nucleic acid extraction and a rapid microfluidics-based real-time thermocycler (Raider, HandyLab Inc., Ann Arbor, MI) for amplification and detection. The fully automated assay is run on the Jaguar (HandyLab, Inc.), which performs automated extraction followed by amplification and detection on a similar microfluidics-based real-time thermocycler. These platforms were chosen because they have an open format, which allows laboratories to run their own assays while maintaining high throughput with minimal hands-on technician time.
The assays were designed with Pleiades probes, which use modified bases for selecting specific melt temperatures and a minor groove binder (Epoch Biosciences, Inc., Bothell, WA).14 Although this chemistry is similar to that of previously described Epoch analyte specific reagents,15 those reagents required modification to perform optimally on the easyMAG/Raider platform. After optimization in the semiautomated system, the assay was adapted to the completely automated Jaguar system. On completion of analytical development, both assays were compared with tissue culture, an in-house RT-PCR-enzyme hybridization assay, and the FDA-approved ProFlu+ assay to determine specificity and sensitivity before routine use in our clinical laboratory.
Beginning in March 2009, infections caused by a novel influenza virus [H1N1 swine origin influenza virus (S-OIV)] began to be recognized around the world.16,17 On April 28, 2009, our laboratory detected and confirmed (April 29) the first patient with the novel Flu A S-OIV in the state of Wisconsin. The lack of knowledge concerning this new strain as well as public and medical concern over an apparently unprotected population led to many public health decisions that increased the number of patients and physicians seeking specific viral diagnostic information in a rapid time frame. We describe the use of this rapid, fully automated Flu A, Flu B, and RSV assay/system in testing large numbers of patient specimens over a very short time period during the ongoing S-OIV outbreak. This outbreak demonstrates the importance of having rapid, reliable, sensitive, and specific assays that allow clinicians and public health officials to react quickly and effectively during viral outbreaks.
Materials and Methods
Primer/Probe Design and Coverage
Primers and probes were designed to highly conserved regions of the Flu A and Flu B matrix genes (M) and the RSV polymerase gene (L) and the bacteriophage MS2 A protein gene (internal control) (Table 1). The probes for Flu A, Flu B, and RSV were labeled with the fluorescent dye FAM, and the MS2 probe was labeled with AP-593 (Epoch Biosciences, Inc.). Primer, probe, and assay design was performed in our laboratory. Many primer/probe designs were obtained using the Influenza Primer Design Resource and are described in further detail in Bose et al18; coverage based on all Flu A, Flu B, and RSV GenBank entries (from before November 2008) is presented in Table 1.18,19
Table 1.
In Silico Coverage of Primers and Probes for the Flu A, Flu B, and RSV Assays
| Virus | Primer name | Primer and probe sequences | Final conc. (μM) | Target | Total sequences | Hits | Gaps | Coverage (%) |
|---|---|---|---|---|---|---|---|---|
| RSV | RSV-L3 | 5′-AATAAATCATAAGTCA*GTAGTA*GACCATGT-3′ | 1.00 | L gene | 16 | 16 | 0 | 100.0 |
| RSV-E4 | 5′-AATAAATCATAATAAGCT*GGTA*TTGA*TGCA-3′ | 0.25 | L gene | 16 | 16 | 0 | 100.0 | |
| RSV-FAM12 | 5′-MGB-FAM-TTGAT*GCA*GG*GA*ATTCA*CA-EDQ-3′ | 0.10 | L gene | 16 | 16 | 0 | 100.0 | |
| Total In Silico coverage for RSV primers and probes | 16 | 16 | 0 | 100.0 | ||||
| Flu A | INFA-L8 | 5′-AATAAATCATAAGTCAGA*GGTGACAGGAT-3′ | 2.00 | Matrix | 8015 | 7978 | 27 | 99.9 |
| INFA-E7 | 5′-AATAAATCATAACTCA*TGGA*ATGGCTAAAG-3′ | 0.50 | Matrix | 8015 | 7876 | 34 | 98.7 | |
| INFA-E8 | 5′-AATAAATCATAACTCA*TGGA*GTGGCTAAAG-3′ | 0.50 | Matrix | 8015 | 7600 | 34 | 95.2 | |
| INFA-FAM27 | 5′-MGB-FAM-AGACAA*GACCA*ATCCT-EDQ-3′ | 0.20 | Matrix | 8015 | 7972 | 30 | 99.8 | |
| Total In Silico coverage for Flu A primers and probes | 8015 | 7893 | 36 | 98.9 | ||||
| Flu B | DIF430-L13 | 5′-AATAAATCATAATGT*CGCT*GT*TTGGA-3′ | 1.00 | Matrix | 364 | 280 | 82 | 99.3 |
| DIF430-E9 | 5′-AATAAATCATAATGA*AAGCA*GGTA*GGCA-3′ | 2.00 | Matrix | 364 | 291 | 73 | 100.0 | |
| DIF430-FAM31 | 5′-MGB-FAM-G*TTTGGA*GA*CACAATT-EDQ-3′ | 0.20 | Matrix | 364 | 282 | 82 | 100.0 | |
| Total In Silico coverage for Flu B primers and probes | 364 | 280 | 82 | 99.3 | ||||
| MS2 Phage | MS2-L23 | 5′-AATAAATCATAAGGTCGGTA*CTAACA*TCAAG-3′ | 0.10 | A gene | Not applicable | |||
| MS2-E7 | 5′-AATAAATCATAAGCA*CGTTGT*CTGGAAGTT-3′ | 0.30 | A gene | |||||
| MS2-AP593–6 | 5′-MGB-AP593-CGTATCCA*GCTGCA*AA*CT-EDQ-3′ | 0.10 | A gene |
MGB, proprietary minor groove binder; FAM, proprietary fluorophore similar to 6-carboxyfluorescein (Epoch Biosciences, Inc.); EDQ, Eclipse Dark Quencher (Glen Research Corp., Sterling, VA); AP593, proprietary fluorophore similar to CalRed.
Asterisks indicate proprietary “Superbases” (Epoch Biosciences, Inc.).
Sample Preparation and Thermocycling Parameters
Samples run on the semiautomated NucliSENS easyMAG System/Raider thermocycler (easyMAG/Raider) were prepared by combining 400 μl of clinical sample (or surrogate samples prepared for analytical testing, as described later) with 10 μl of MS2 bacteriophage at a concentration of 1 × 106 plaque-forming units/ml. Samples were then combined with 1 ml of easyMAG lysis buffer, and RNA was extracted following the manufacturer's off-board lysis procedure with the generic 1.0 protocol. RNA was eluted in 25 μl of elution buffer. Then 3.4 μl of RNA was combined with 4.6 μl of RT-PCR mix containing 2× Platinum Tfi mix, SuperScript III (Invitrogen, Carlsbad, CA), and primers/probes for all targets including the internal control and loaded into a cartridge, which was placed in a Raider thermocycler. On the Jaguar platform, 475 μl of clinical sample (or surrogate samples prepared for analytical testing, as described later) was combined with 10 μl (4.75 × 105 plaque-forming units/ml) of MS2 bacteriophage and loaded into the Jaguar sample tubes. RNA was extracted and eluted in 10 μl of elution buffer and combined with 13.5 μl of RT-PCR mix (as described above). Then 8 μl of this mixture was transferred to one of two internal high-speed thermocyclers. All steps after sample addition proceeded in a fully automated format with sample transfer and cartridge loading occurring via a robotic arm. The final primer/probe concentrations in each platform were the same, as was the final amount of MS2/rxn (approximately 700 copies). The thermocycling parameters used in both systems were 20 minutes at 50°C (room temperature), 2 minutes at 95°C (denature), and 45 cycles of 15 seconds at 95°C, 30 seconds at 56°C, and 15 seconds at 76°C, followed by a subsequent melt analysis at 45–85°C at a rate of 0.1°C/second. This PCR protocol takes approximately 1.5 hours to run.
Result Analysis
Results were determined using a “workbook” (computational algorithm) in Microsoft Excel (Microsoft Corp., Redmond, WA) designed to convert the raw data produced by the Raider or the Jaguar to the final output format. Data were analyzed, and the tests were scored based on the amplification curves and melt profiles of the sample. Samples were considered positive if the FAM Ct value was ≤40, the amplification curve shape was appropriate, and the FAM melt profiles yielded a melting temperature (Tm) ±2°C of those expected for RSV (74°C), Flu B (68°C), or Flu A (63°C) (Figure 1A). Samples were considered indeterminate if the FAM Ct value was >40.0 with an appropriate FAM Tm and amplification curve shape; indeterminate samples were repeated once. Samples were considered negative if the internal control (MS2) was positive (Tm of 73°C in the AP-593 channel) (Figure 1B) and the FAM Ct value was >40.0 with an incorrect or nonexistent Tm or an abnormal amplification curve.
Figure 1.
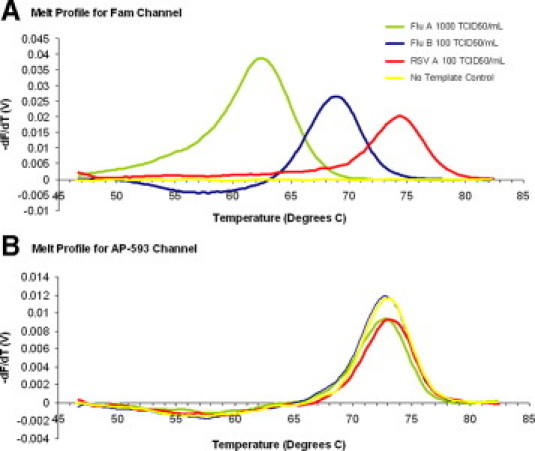
A: Melt profile of Flu A (green), Flu B (blue), and RSV (red) samples in the FAM channel. B: Melt profile of MS2 in the AP-593 channel.
Analytical Sensitivity (Limits of Detection)
Recent human virus strains of H1N1 [Flu A/WI/629-9/2008 (H1N1)], H3N2 [A/WI/629-2/2008 (H3N2)], Flu B (B/WI/629-1/2008), RSV A (RSV-A/WI/629-9-1/2008), and RSV B (RSV-B/WV/14617/1985) were quantitated by TCID50 and RNA copy number and used to determine the limits of detection (LOD) for each target.20,21,22,23,24,25,26 Serial 10-fold dilutions from 104 to 10−2 TCID50/ml were made in M4 viral transport medium (Remel, Lenexa, KS) and tested in both systems.
Specificity Testing against Common Respiratory Pathogens
Twenty-three common respiratory pathogens and commensal organisms were spiked at high concentrations (>104 TCID50 plaque-forming units, colony-forming units/ml) into M4 viral transport medium. The following organisms were obtained from the American Type Culture Collection (ATCC) (Manassas, VA) and were tested in both systems: adenovirus type 3 (ATCC VR-3), rhinovirus 1B (ATCC VR-481), coronavirus OC43 (ATCC VR-1558), herpes simplex virus type 1 (ATCC VR-1544), human parainfluenza viruses 1 (ATCC VR-94), 2 (ATCC VR-92), 3 (ATCC VR-93), and 4 (ATCC VR-1378), cytomegalovirus (ATCC VR-977), Escherichia coli (ATCC 11229), Staphylococcus epidermidis (ATCC 29641), Neisseria sicca (ATCC 9913), Streptococcus pyogenes (ATCC 19615), Pseudomonas aeruginosa (ATCC 17643), Haemophilus influenzae (ATCC 10211), Enterococcus faecalis (ATCC 12399), Streptococcus sanguinis (ATCC 10556), Streptococcus mitis (ATCC 49456), Eikenella corrodens (ATCC 51724), and Staphylococcus aureus (ATCC# 29247). Moraxella catarrhalis and Streptococcus agalactiae strains were kind donations from the Children's Hospital of Wisconsin (Milwaukee, WI). Human metapneumovirus was a kind gift from Dr. John Williams (Vanderbilt University, Nashville, TN). In addition, Flu A (A/WI/629-9/2008 (H1N1), Flu B (B/WI/629-1/2008), and RSV A (RSV-A/WI/629-9-1/2008) were tested at the following concentrations: 103, 102, and 102 TCID50/ml, respectively.
ProFlu+ Assay
ProFlu+ kits were purchased from the manufacturer and run according to the package insert. Clinical samples (200 μl) were extracted using the easyMAG and eluted in 55 μl as directed.
Clinical Sensitivity and Specificity in Frozen Nasopharyngeal Swab Samples
One hundred nasopharyngeal swab samples tested previously (and stored at −80°C in M4 viral transport medium) at the Children's Hospital of Wisconsin were determined to be positive for Flu A (n = 16), Flu B (n = 21), or RSV (n = 20) or negative (n = 43) by tissue culture. An aliquot of each sample was tested blindly in the easyMAG/Raider assay, the Jaguar assay, and the FDA-approved ProFlu+ assay. Tissue culture results were used as the true result for sensitivity and specificity calculations. However, when there was disagreement between tissue culture, easyMAG/Raider, Jaguar, or ProFlu+ assays, the RV-8 (described below) was used to resolve discordant results. Indeterminate results were excluded from sensitivity and specificity calculations.
RT-PCR-Enzyme Hybridization Assay (RV-8)
In addition to the real-time RT-PCR assays, clinical samples were tested with an in-house large multiplex RT-PCR-enzyme hybridization assay that can simultaneously detect and type nine respiratory viruses (RV-8) including Flu A, Flu B, and RSV A/B for discrepant analysis. This assay targets different genomic regions (not necessarily different genes, but at least different portions of the gene) from those for the multiplex real-time assays. The RV-8 was run using 3 μl of RNA extracted from each clinical sample (the same RNA samples used for easyMAG/Raider testing). RNA was run in a 20-μl reverse transcription reaction using MuLV reverse transcriptase (Applied Biosystems, Foster City, CA). Then 10 μl of the reverse transcription reaction was run in a 50-μl large multiplex PCR using Fast Taq Polymerase (Roche) and the following primers: Flu A forward 5′-CTTCTAACCGAGGTCGAAACGTA-3′, Flu A reverse 5′-Biotin-ACAAAGCGTCTACGCTGCAGTCC-3′, Flu A probe 5′-HRP-TCAGGCCCCCTCAAAGCCGAAATCGC-3′, Flu B forward 5′-ATGGCCATCGGATCCTCAACTCACTC-3′, Flu B reverse 5′-Biotin-TCATGTCAGCTATTATGGAGCTGTT-3′, Flu B probe 5′-HRP-TATCCCAATTTGGTCAAGAGCACCGATTATCACCAG-3′, RSV A forward 5′-GCTGTCATCCAGCAAATACAC-3′, RSV A reverse 5′-Biotin-GAGTATTTTTATAGTGTCTTCTCTTCCT-3′, RSV A probe 5′-HRP-CAGAAACACATCAATAAGTTATGTGGCATGTTA-3′, RSV B forward 5′-GCTGTCATCCAGCAAATACAC-3′, RSV B reverse 5′-Biotin-GAGTATTTTTATAGTGTCTTCTCTTCCT-3′, and RSV B probe 5′-HRP-CAAAAACACCTAAACAAACTATGTGGTATGCTA-3′. These primers detect the Flu A matrix gene, the Flu B Nsp1 and Nsp2 genes, the RSV-A nucleoprotein gene, and the RSV-B nucleoprotein gene. The other targets in the assay include the parainfluenza virus type 1 (PIV-1) nucleoprotein gene, the PIV-2 hemagglutinin gene, the PIV-3 hemagglutinin gene, the human metapneumovirus fusion protein, and the adenovirus penton gene (primer sequences and analytical data are presented for these targets because they have no bearing on the current study). After PCR, 5 μl of the reaction was probed with each of the probes according to well described protocols for RT-PCR-enzyme hybridization assay.1,10 Analytical sensitivity was determined by testing serial dilutions of quantitated stocks of Flu A/WI/629-9/2008, Flu B/WI/629-1/2008, RSV-A/WI/625-9-1/0708, and RSV-B/WV/14617/85 in M4 viral transport medium (Remel). Analytical specificity was tested using high concentrations (≥105 TCID50 or colony-forming units/ml for viruses and bacteria, respectively) of the same bacteria and viruses used for specificity testing of the easyMAG/Raider and Jaguar assays. Cytomegalovirus, S. pyogenes, E. faecalis, S. sanguinis, S. mitis, and E. corrodens were not tested in this assay.
Precision
Precision was determined by calculating the coefficient of variation of the Ct values reported for the positive controls on each run over a 4-month time period. The positive controls consisted of quantitated virus diluted in M4 at the following concentrations: 101 TCID50/ml (RSV-A/WI/625-9-1/0708), 100 TCID50/ml (RSV-B/WV/14617/1985, ATCC VR1400), and 102 TCID50/ml (Flu A/WI/629-9/2008 and Flu B/WI/629-1/2008). In addition, the mean and SD in Tm of each analyte were calculated by analyzing all data (specimens and controls) obtained from clinical testing during a 2-week period.
Performance in S-OIV Outbreak of April/May 2009
The Jaguar assay has been successfully used as a screening assay since November 2008. Beginning in the last week of April 2009, 1232 specimens were tested over the first 2 weeks of the S-OIV outbreak.27 Samples run in the Jaguar assay that tested positive for Flu A were subtyped and confirmed as Flu A (H1N1) S-OIV at the Midwest Respiratory Virus Program, Medical College of Wisconsin.18,28
Results
Analytical Sensitivity of easyMAG/Raider and Jaguar Assays
The limits of detection were 100 (100), 10−2 (10−2), 10−1 (101), 10−1 (10−2), and 10−1 (10−1) TCID50/ml on the Jaguar and the easyMAG/Raider (in parentheses) for Flu A (H1N1), Flu A (H3N2), Flu B, RSV A, and RSV B, respectively.
Specificity Testing against Common Respiratory Pathogens in easyMAG/Raider and Jaguar Assays
Of the 23 nonspecific organisms tested, only adenovirus type 3 gave a positive result (Flu A) on the easyMAG/Raider at a concentration of 107 TCID50/ml. No dilutions below 107 TCID50/ml showed any sign of cross-reactivity. H. influenzae gave an indeterminate result on the easyMAG/Raider system at high concentrations. Cross- reactivity was not seen in the Jaguar assay.
Analytical Sensitivity and Specificity of the RV-8 Assay
The RV-8 assay is still undergoing some slight modifications in an effort to incorporate an internal control and automate it; however, in the version used in this experiment the limits of detection are 101, 101, 102, and 102 TCID50/ml for Flu A, Flu B, RSV A, and RSV B, respectively. In addition, no cross-reaction was seen when nonspecific organisms were probed with the Flu A, Flu B, RSV A, or RSV B probes.
Precision Testing
The monthly coefficient of variation of the Ct for each sample run on the Jaguar platform was less than 5% each month with the exception of RSV B during December (Table 2).
Table 2.
Precision of Flu A, Flu B, and RSV Jaguar Assay
| December |
January |
February |
March |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Virus | Conc. TCID50/ml | N | CV (%) | N | CV (%) | N | CV (%) | N | CV (%) |
| Mean precision of Ct values for positive controls | |||||||||
| RSV A | 101 | 7 | 3.97 | 26 | 3.75 | 22 | 4.87 | 16 | 2.38 |
| RSV B | 100 | 26 | 7.97 | 26 | 3.04 | 31 | 5.00 | 10 | 4.12 |
| Flu A | 102 | 6 | 2.74 | 21 | 1.94 | 15 | 1.59 | ||
| Flu B | 102 | 5 | 3.03 | 22 | 4.86 | 10 | 1.59 | ||
| Precision of Tm over 2 weeks of clinical testing | No. controls | No. clinical samples | Mean Tm | SD of Tm |
|---|---|---|---|---|
| RSV A/B | 17 | 6 | 73.8 | 0.53 |
| Flu A | 8 | 1 | 62.8 | 0.90 |
| Flu B | 6 | 5 | 67.7 | 0.79 |
| MS2 | 50 | 50 | 72.8 | 0.43 |
CV, coefficient of variation.
Clinical Sensitivity and Specificity
The semiautomated and fully automated Flu A, Flu B, and RSV assays described here exhibit greater sensitivity, positive predictive value, and negative predictive value than the FDA-approved ProFlu+ assay (though the results are not statistically significant). All assays have the same specificity (Table 3).
Table 3.
Performance of the easyMAG/Raider, Jaguar, ProFlu+, and RV-8 Assays Compared with Tissue Culture
| ProFlu+ (No. tested = 98; 6 indeterminates) |
easyMAG/Raider (No. tested = 100; 3 indeterminates) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Virus | Sens. (CI) | Spec. (CI) | PPV | NPV | Virus | Sens. (CI) | Spec. (CI) | PPV | NPV |
| Flu A (N = 14) | 93 (66–100) | 100 (95–100) | 100 | 99 | Flu A (N = 16) | 100 (79–100) | 100 (96–100) | 100 | 100 |
| Flu B (N = 20) | 80 (56–94) | 100 (95–100) | 100 | 95 | Flu B (N = 18) | 89 (67–99)* | 100 (95–100) | 100 | 98 |
| RSV (N = 19) | 95 (73–100) | 95 (82–100) | 82 | 99 | RSV (N = 20) | 100 (82–100) | 99 (93–100) | 87 | 100 |
| Total | 89 (77–96) | 99 (97–100) | 92 | 97 | Total | 93 (82–98) | 99 (98–100) | 95 | 99 |
| Jaguar (No. tested = 100; 7 indeterminates) |
RV-8 (No. tested = 85; 0 indeterminates) |
||||||
|---|---|---|---|---|---|---|---|
| Virus | Sens. (CI) | Spec. (CI) | PPV | NPV | Virus | Sens. (CI) | Spec. (CI) |
| Flu A (N = 15) | 100 (74–100) | 100 (96–100) | 100 | 99 | Flu A (N = 13) | 100 (75–100) | 100 (95–100) |
| Flu B (N = 20) | 90 (70–99)† | 100 (95–100) | 100 | 97 | Flu B (N = 20) | 100 (83–100) | 100 (95–100) |
| RSV (N = 20) | 100 (83–100) | 98 (91–100) | 87 | 100 | RSV (N = 17) | 94 (71–100) | 99 (92–100) |
| Total | 96 (87–99) | 99 (97–99) | 94 | 99 | Total | 98 (89–100) | 99.5 (97–100) |
Tissue culture results were used to calculate sensitivity and specificity. The RV-8 was used to resolve discordant results between the easyMAG/Raider, Jaguar, or ProFlu+ assays; however, this result was only noted for discussion and was not factored into the sensitivity and specificity calculations. Indeterminate results were excluded from sensitivity and specificity results.
Sens., sensitivity; CI, confidence interval; Spec., specificity; PPV, positive predictive value; NPV, negative predictive value.
Of 2 RSV/FluB dual positives, one was correctly identified as RSV/FluB; the other was identified as only RSV.
Of 2 RSV/FluB dual positives, both were identified only as RSV.
Performance in S-OIV Outbreak in April/May 2009
Of the 1232 samples screened in the Children's Hospital of Wisconsin laboratory over a 10-day period, we detected 134 Flu A-positive samples (Table 4).18,28 The Jaguar assay demonstrated 100% positive agreement compared with two separate in-house validated molecular assays directed to different genetic targets but had two false-positive results (99.81% negative agreement).18,28 Twenty-two negative samples were tested by all three molecular assays with complete agreement. All Flu A-positive samples were sent to the Midwest Respiratory Virus Program clinical laboratory for further subtyping. Subtyping results indicated 126 H1N1 S-OIV strains, 5 human H1N1 strains, and 1 Flu A unsubtyped (Table 4). All clinical samples were tested and confirmed at the Midwest Respiratory Virus Program. Twenty-three of 23 of the clinical samples were confirmed as influenza A (H1N1) S-OIV by the Wisconsin State Laboratory of Hygiene using the Centers for Disease Control and Prevention Laboratory Response Network influenza RT-PCR typing and subtyping assays.
Table 4.
Samples Tested in the Flu A, Flu B, and RSV Jaguar Assay During an Outbreak of S-OIV in Milwaukee, WI, During April and May of 2009
| Result | |
|---|---|
| Jaguar (N = 1232 samples tested) | |
| Flu A | 134 |
| Flu B | 35 |
| RSV | 12 |
| Negative | 1015 |
| Flu A subtyping (N = 134)18, 28 | |
| H1N1 (S-OIV) | 126 |
| H1N1 (human) | 5 |
| H3N2 | 0 |
| Untyped | 1 |
| False-positive | 2 |
Discussion
Analytically the semiautomated and fully automated assays described in this report are highly sensitive for Flu A, Flu B, and RSV A/B. All analytes can be detected in both platforms at concentrations of less then 10 TCID50/ml, which is comparable with detection seen with the ProFlu+ assay (LOD range from 102 to 10−1 TCID50/ml). Although the LOD for most targets are equivalent on both easyMAG/Raider and Jaguar systems, two exceptions are that the LOD of Flu B is approximately 2 log units better on the Jaguar then on the easyMAG/Raider platform and the LOD of RSV-A was 1 log unit better on the easyMAG/Raider platform. Both assay formats are also very specific. Testing of 23 nonspecific organisms in both formats revealed that only adenovirus type 3 showed intermittent cross-reactivity in the easyMAG/Raider format at a very high concentration of 107 TCID50/ml. In addition, comparison of samples tested by tissue culture with those tested with the easyMAG/Raider, Jaguar, and ProFlu+ assays showed that all were 99% specific. The only discrepancies observed were that RSV was detected in a specimen that was negative by tissue culture. RSV was also detected in a Flu B-positive tissue culture sample, and its presence was confirmed during discrepant analysis as a dual Flu B and RSV infection. Clinical testing demonstrated that the easyMAG/Raider and Jaguar assays exhibited greater sensitivity than the ProFlu+ assay for all three analytes (although these differences are not statistically significant). Overall sensitivities of the easyMAG/Raider and Jaguar formats were both greater than that observed for the ProFlu+ assay.
Testing with these primers and probes was performed on a large number of virus isolates consisting of H1 to H15 subtypes of Flu A, Yamagata and Victoria lineages of Flu B, and A and B subtypes of RSV. In addition, at least 15 samples of each virus were collected in Milwaukee, WI, in 2008 and tested with the assay (these samples were used to calculate clinical sensitivity and specificity). Collectively, these viruses were from various hosts (for Flu A viruses), collection years (dating back to 1977 for Flu A, to 2004 for Flu B, and to 1985 for RSV), and geographic locations and showed that Tm values ranged from 62.4 to 64.1, 68.1 to 69.8, and 73.0 to 74.7 for Flu A, Flu B, and RSV, respectively. These data are not shown because although each target was tested with the same analyte-specific primers/probes at the proper concentrations, the compositions of the nonspecific primers/probes were not necessarily the same as those in the completed assay. However, during the course of our assay optimization we observed that changing any of the oligonucleotides, with the exception of the analyte-specific probe, yielded very little change in Tm for a given virus. For this reason we are confident that the Tm ranges reported above for Flu A, Flu B, and RSV viruses are highly representative of their true ranges using the finalized version of the assay described in this article.
Both assay formats have rapid sample turnaround times with limited technician time. The Jaguar or easyMAG/Raider platforms require approximately 20 or 40 minutes of technician time and can return results in 3.5 or 2.5 hours, respectively, for 24 samples. Automated multiplex assays can decrease the numbers of laboratory tests ordered per patient, the technician time used per sample, the cost per virus detected, and the overall chance for contamination.29,30,31 In addition, the Jaguar assay proved efficient in rapidly and accurately screening a large number of Flu A, Flu B, and RSV samples during the current outbreak of a novel Flu A (H1N1) virus S-OIV in Milwaukee, WI. Flu A samples were reflexed into several subtyping assays that also confirmed the Flu A typing result (including 23 Flu A-positive samples, which were confirmed as S-OIV by the Centers for Disease Control and Prevention's subtyping assay at the Wisconsin State Laboratory of Hygiene). These subtyping results indicated that this Flu A, Flu B, and RSV assay could successfully detect the matrix gene of all three circulating strains of Flu A (H1N1 human, H3N2 human, and H1N1 S-OIV). This outbreak demonstrates the importance of having rapid, reliable, sensitive, and specific assays that allow clinicians and public health officials to react quickly and effectively during viral outbreaks.
Acknowledgements
We acknowledge Jacob Metallo for his help with primer and probe design and coverage calculations. We also acknowledge Jessica Trost and Rose Chen for their help in growing and quantitating virus strains used in this study.
Footnotes
Supported by the Midwest Respiratory Virus Program Clinical Laboratory.
References
- 1.Fan J, Henrickson KJ, Savatski LL. Rapid simultaneous diagnosis of infections with respiratory syncytial viruses A and B, influenza viruses A and B, and human parainfluenza virus types 1, 2, and 3 by multiplex quantitative reverse transcription-polymerase chain reaction-enzyme hybridization assay (Hexaplex) Clin Infect Dis. 1998;26:1397–1402. doi: 10.1086/516357. [DOI] [PubMed] [Google Scholar]
- 2.Henrickson KJ. Advances in the laboratory diagnosis of viral respiratory disease. Pediatr Infect Dis J. 2004;23(Suppl.):S6–S10. doi: 10.1097/01.inf.0000108187.63151.ea. [DOI] [PubMed] [Google Scholar]
- 3.Kehl S, Henrickson KJ. Comparison of three diagnostic methods: hexaplex, tissue culture, and rapid EIA for the detection of respiratory viruses in children. J Clin Microbiol. 2001;39:1696–1701. doi: 10.1128/JCM.39.5.1696-1701.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koenig M, Kosha S, Hickman M, Heath D, Riddell S, Aldous S. Detection of influenza virus from throat and pharyngeal swabs with a nested duplex light cycler RT-PCR. Diag Microbiol Inf Dis. 2003;46:35–37. doi: 10.1016/s0732-8893(02)00552-7. [DOI] [PubMed] [Google Scholar]
- 5.Liolios L, Jenney A, Spelman D, Kotsimbos T, Catton M, Wesselingh S. Comparison of a multiplex reverse transcription-PCR-enzyme hybridization assay with conventional viral culture and immunofluorescence techniques for the detection of seven viral respiratory pathogens. J Clin Microbiol. 2001;39:2779–2783. doi: 10.1128/JCM.39.8.2779-2783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mahony J, Chong S, Merante F, Yaghoubian S, Sinha T, Lisle S, Janeczko R. Development of a respiratory virus panel test for detection of twenty human respiratory viruses by use of multiplex PCR and a fluid microbead-based assay. J Clin Microbiol. 2007;45:2965–2970. doi: 10.1128/JCM.02436-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nolte FS, Marshall DJ, Rasberry C, Schievelbein S, Banks GG, Storch GA, Arens MQ, Buller RS, Prudent JR. MultiCode-PLx system for multiplexed detection of seventeen respiratory viruses. J Clin Microbiol. 2007;45:2779–2786. doi: 10.1128/JCM.00669-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henrickson KJ, Kraft A, Shaw J, Canter D. Comparison of electronic microarray (NGEN RVA) to enzyme hybridization assay (Hexaplex) for multiplex RT-PCR detection of common respiratory viruses in children. Clin Microbiol Newsl. 2007;29:113–119. doi: 10.1016/j.clinmicnews.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang Y, Tang H, Duffy S, Hong Y, Norman S, Ghosh M, He J, Bose ME, Henrickson KJ, Fan J, Kraft AJ, Weisburg WG, Mather EL. A multiplex assay for simultaneously typing and subtyping influenza viruses on an electronic microarray. J Clin Microbiol. 2009;47:390–396. doi: 10.1128/JCM.01807-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar S, Wang L, Fan J, Kraft AJ, Bose ME, Tiwari S, Dyke M, Haigis R, Luo T, Ghosh M, Tang H, Haghnia M, Mather EL, Weisburg WG, Henrickson KJ. Detection of 11 common viral and bacterial pathogens causing community-acquired pneumonia or sepsis in asymptomatic patients using a multiplex reverse transcription PCR assay with manual (enzyme hybridization) or automated detection (electronic microarray) J Clin Microbiol. 2008;46:3063–3072. doi: 10.1128/JCM.00625-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee WM, Grindle K, Pappas T, Marshall DJ, Moser MJ, Beaty EL, Shult PA, Prudent JR, Gern JE. High-throughput sensitive, and accurate multiplex PCR-microsphere flow cytometry system for large-scale comprehensive detection of respiratory viruses. J Clin Microbiol. 2007;45:2626–2634. doi: 10.1128/JCM.02501-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merante F, Yaghoubian S, Janeczko R. Principles of the xTAG™ respiratory viral panel assay (RVP assay) J Clin Virol. 2007;40(Suppl 1):S31–S35. doi: 10.1016/S1386-6532(07)70007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.FDA clears new CDC test to detect human influenza. J Environ Health. 2008;71:62. [PubMed] [Google Scholar]
- 14.Lukhtanov EA, Lokhov SG, Gorn VV, Podyminogin MA, Mahoney W. Novel DNA probes with low background and high hybridization-triggered fluorescence. Nucleic Acids Res. 2007;35:e30. doi: 10.1093/nar/gkl1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hymas WC, Hillyard DR. Evaluation of Nanogen MGB Alert® detection reagents in a multiplex real-time PCR for influenza virus types A and B and respiratory syncytial virus. J Virol Methods. 2009;156:124–128. doi: 10.1016/j.jviromet.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 16.CDC Update: Swine-origin influenza A (H1N1) virus—United States and other countries. MMWR CDC Surveill Summ. 2009;58:421. [PubMed] [Google Scholar]
- 17.CDC Outbreak of swine-origin influenza A (H1N1) virus infection—Mexico, March. MMWR.CDC Surveill Summ. 2009;58(Dispatch):1–3. [PubMed] [Google Scholar]
- 18.Bose ME, Beck ET, Ledeboer N, Kehl SC, Jurgens LA, Patitucci T, Witt L, LaGue E, Darga P, He J, Fan J, Kumar S, Henrickson KJ. Rapid semi-automated subtyping of influenza during the 2009 swine-origin influenza A H1N1 virus epidemic in Milwaukee, Wisconsin. J Clin Microbiol. 2009;47:2779–2786. doi: 10.1128/JCM.00999-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bose ME, Littrell JC, Patzer AD, Kraft AJ, Metallo JA, Fan J, Henrickson KJ. The Influenza Primer Design Resource—a new tool for translating influenza sequence data into effective diagnostics. Influenza Other Respi Viruses. 2008;2:23–31. doi: 10.1111/j.1750-2659.2007.00031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bianchi E, Liang XP, Ingallinella P, Finotto M, Chastain MA, Fan J, Fu TM, Manger M, Wen E, Shi L, Ionescu R, Emini EA, Cortese R, Ciliberto J, Shiver JW, Pessi A. A universal influenza B vaccine based on the maturational cleavage site of the hemagglutinin precursor. J Virol. 2005;79:7380–7388. doi: 10.1128/JVI.79.12.7380-7388.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan J, Liang XP, Horton MS, Perry HC, Citron MP, Heidecker GJ, Fu TM, Joyce J, Przysiecki CT, Keller PM, Garsky VM, Ionescu R, Shi L, Chastain MA, Condra JH, Davies MA, Liao J, Emini EA, Shiver JW. Preclinical study of influenza virus a M2 peptide conjugate vaccines in mice, ferrets and rhesus monkeys. Vaccine. 2004;22:2993–3003. doi: 10.1016/j.vaccine.2004.02.021. [DOI] [PubMed] [Google Scholar]
- 22.Hall CB, Douglas RG. Clinical useful method for the isolation of respiratory syncytial virus. J Infect Dis. 1975;131:1–5. doi: 10.1093/infdis/131.1.1. [DOI] [PubMed] [Google Scholar]
- 23.Hall CB, Douglas RG, Geiman JM. Respiratory syncytial virus infection in infants: quantitation and duration of shedding. J Pediatr. 1976;89:11–15. doi: 10.1016/s0022-3476(76)80918-3. [DOI] [PubMed] [Google Scholar]
- 24.Hsiung GD. Virus assay, neutralization test and antiviral assay. In: Hsiung GD, Fong CK, Landry ML, editors. Hsiung's diagnostic virology. Yale University Press; New Haven, CT: 1994. pp. 46–55. [Google Scholar]
- 25.Nguyen DC, Uyeki TM, Jadhao S, Maines T, Shaw M, Matsuoka Y, Smith C, Rowe T, Lu X, Hall H, Xu X, Balish A, Klimov A, Tumpey TM, Swayne DE, Huynh LPT, Nghiem HK, Nguyen HHT, Hoang LT, Cox NJ, Katz JM. Isolation and characterization of avian influenza viruses, including highly pathogenic H5N1, from poultry in live bird markets in Hanoi. Vietnam, in 2001. J Virol. 2005;79:4201–4212. doi: 10.1128/JVI.79.7.4201-4212.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reed LJ, Muench H. A simple method of estimating fifty percent endpoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 27.Kumar S, Chusid MJ, Willoughby RE, Havens PL, Kehl SC, Ledeboer NA, Li S, Henrickson KJ. Introduction of a novel swine-origin influenza A (H1N1) virus into Milwaukee. Wisconsin in 2009. Viruses. 2009;1:72–83. doi: 10.3390/v1010072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He J, Bose ME, Beck ET, Fan J, Tiwari S, Metallo J, Jurgens L, Kehl SC, Ledeboer N, Kumar S, Weisburg W, Henrickson KJ. Rapid multiplex RT-PCR typing of influenza A and B and subtyping of influenza A into H1, 2, 3, 5, 7, 9. N1 (human), N1 (animal), N2 and N7 including typing of novel swine-origin influenza A (H1N1) virus during the current 2009 outbreak in Milwaukee, WI. J Clin Microbiol. 2009;47:2772–2778. doi: 10.1128/JCM.00998-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barenfanger J, Drake C, Leon N, Mueller T, Troutt Y. Clinical and financial benefits of rapid detection of respiratory viruses: an outcomes study. J Clin Microbiol. 2000;38:2824–2828. doi: 10.1128/jcm.38.8.2824-2828.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henrickson KJ. Cost-effective use of rapid diagnostic techniques in the treatment and prevention of viral respiratory infections. Pediatr Ann. 2005;34:24–31. doi: 10.3928/0090-4481-20050101-08. [DOI] [PubMed] [Google Scholar]
- 31.Woo PCY, Chiu SS, Seto WH, Peiris M. Cost-effectiveness of rapid diagnosis of viral respiratory tract infections in pediatric patients. J Clin Microbiol. 1997;35:1579–1581. doi: 10.1128/jcm.35.6.1579-1581.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]


