Abstract
KRAS is an oncogene that can be activated by mutations. Patients with non-small cell lung cancer who have KRAS mutations do not respond to tyrosine kinase inhibitors; therefore, accurate detection of KRAS mutations is important for deciding therapeutic strategies. Although sequencing-related techniques have been frequently used, they are usually too complex, have low sensitivity, and are time-consuming for routine screening in clinical situations. We evaluated peptide nucleic acid (PNA)-clamp smart amplification process version 2 (SmartAmp2) as a detection method for KRAS codon 12 mutations in patient specimens compared with traditional sequencing and polymerase chain reaction-related methods. Among 172 lung adenocarcinoma samples, direct sequencing, enzyme-enriched sequencing, and PNA-enriched sequencing showed that 16 (9.3%), 26 (15.7%), and 28 (16.3%) tumors, respectively, contained KRAS mutations in codon 12. Using PNA-clamp SmartAmp2, we could identify 31 (18.0%) tumors that had KRAS mutations in codon 12 within 60 minutes, three of which were undetected by polymerase chain reaction-related methods. On the other hand, we examined 30 nonmalignant peripheral lung tissue specimens and found no mutations in any of the samples using PNA-clamp SmartAmp2. In this study, we confirmed that PNA-clamp SmartAmp2 has high sensitivity and accuracy and is suitable for the clinical diagnosis of KRAS codon 12 mutations.
KRAS is an oncogene located on chromosome 12p12.1 that is activated by point mutations.1 When the point mutation increases the active GTP-bound form of KRAS, it activates transcription factors through binding to target proteins, including Raf and PI3K,2,3,4,5 which play an important role in lung cancer carcinogenesis.6 KRAS mutations occur in 33% of patients with non-small-cell lung cancer (NSCLC).7,8 Recent studies have shown that KRAS mutations are associated with poor prognosis in patients with resected NSCLC,9 lack of survival benefit from adjuvant chemotherapy,10 and resistance to epidermal growth factor receptor (EGFR) inhibitors such as erlotinib or gefitinib.9 Therefore, physicians who treat patients with NSCLC need to be able to determine whether their patients have such mutations to design optimal therapeutic strategies.
Detection of KRAS mutations is usually achieved by sequencing DNA amplified from tumor tissues. However, diagnostic sequencing techniques are too complex, time-consuming, and expensive for routine pretherapeutic screening. Adding to the complexity is the problem that clinical samples often contain a subpopulation of mutant cells mixed within an excess of normal tissue, sometimes causing the mutations to be missed by sequencing because of the limitations of the technology. Recently, several methods, such as polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP), single-strand conformation polymorphism (PCR-SSCP), mutant-allele-specific amplification, and mutant-enriched assays, have been reported for the detection of KRAS mutations. Some of them are more sensitive than simple sequencing, but all are still less desirable for routine clinical use because of their long turnaround times and complexity.
In previous reports, KRAS mutations have been estimated to occur in 3.0% to 15.0% of lung adenocarcinomas in Asian individuals.9,10,11,12,13 This variation in the prevalence of these mutations is thought to be partly an artifact resulting from the varying sensitivities and accuracies of different detection methods. The accurate diagnosis of KRAS mutations is clinically very important; therefore, reliable methods for the detection of KRAS codon 12 mutations should be established.
Recently, rapid, simple, and sensitive mutation detection assays, named smart amplification process version 2 (SmartAmp2) and peptide nucleic acid (PNA)-clamp SmartAmp2, have been developed.14 The latter method was applied for the detection of KRAS gene mutations in pancreatic cancer samples.15,16 The SmartAmp2 method is a unique genotyping technology that can detect a mutation within 30 minutes under isothermal conditions and in a single step.14 The basic principle of SmartAmp2 is that DNA amplification equals detection; no further analysis is required. A set of five specially designed primers that recognize a total of six distinct sequences on the target DNA enable the precise amplification of only mutant-specific sequences. The two primers are referred to as the folding primer and the turn-back primer and are engaged in amplifying the target through a self-priming mechanism. In addition, the PNA-clamp probe was designed for the wild-type allele sequence to allow amplification of all possible sequences dictated by the first two nucleotides in codon 12 of KRAS. PNA cannot serve as a primer for polymerization because it is not recognized by the polymerase as natural DNA, and it cannot be a substrate for the exonuclease activities of Taq polymerase. In addition, the melting temperature of a perfectly matched PNA-DNA duplex is higher than that of DNA-DNA of the same length, but a single mismatch destabilizes the PNA-DNA hybrids, causing a melting temperature shift of 10–18°C. The greater stability of the hybridized PNA probe inhibits the SmartAmp2 amplification; thus, amplification of the wild-type allele is suppressed. This effect is theoretically manifest at the initial elongation event from the turn-back primer on wild-type template or on a folding primer-synthesized strand (also early in the reaction).14
In this study, we compared the PNA-clamp SmartAmp2 assay with PCR-based direct sequencing, enzyme-enriched PCR-RFLP and sequencing, and PNA-enriched sequencing using clinical samples from 172 lung adenocarcinomas and demonstrated that the PNA-clamp SmartAmp2 assay was the most reliable method for the clinical diagnosis of KRAS codon 12 mutations.
Materials and Methods
Cell lines, Patient Samples, and DNA Extraction
Tumor samples containing KRAS mutations obtained from 172 consecutive patients with lung adenocarcinoma who were surgically treated at the Gunma University Hospital, Japan, between July 2002 and July 2008 were used in this study. Thirty nonmalignant specimens from peripheral lung tissue surrounding the tumors were used to examine the specificity of the assays. Institutional approval and informed consents from all patients were obtained in writing. All tumor tissues were diagnosed as lung cancer by hematoxylin eosin staining. After surgical removal, all tumor samples were immediately frozen and stored at −80°C.
To assist in the comparison of detection methods, DNA harboring KRAS mutations was extracted from the lung cancer cell line A549 (American Type Culture Collection), which is homozygous for the KRAS codon 12 allele (G34A); DNA containing the wild-type KRAS codon 12 allele was prepared from EBC-1 (American Type Culture Collection). DNA samples were extracted from tumor tissues and cell lines using the QIAamp DNA Mini Kit (QIAGEN GmbH, Hilden, Germany) and serially diluted to a concentration representing 20 ng/μL. For the evaluation of sensitivity and specificity, DNA from the A549 cells was diluted with the DNA from the EBC-1 cells to give mutation/wild-type ratios of 50%, 10%, 5%, 1%, and 0.1%. The DNA templates extracted from the tumor samples and cell lines were stored at −20°C.
PNA-Clamp SmartAmp2
We performed PNA-clamp SmartAmp2 using the KRAS mutation detection kit (K.K. DNAFORM, Kanagawa, Japan). According to manufacturer's instructions, PNA-clamp SmartAmp2 reactions were assembled on ice and incubated at 60°C for 60 minutes. The Mx3000P system (Stratagene, CA) was used to maintain isothermal conditions and monitor the transition in fluorescence intensity of the intercalating SYBR Green I (Invitrogen, CA) during the reactions. Results of the PNA-clamp SmartAmp2 assay were evaluated according to the criterion of amplification versus non-amplification within 60 minutes. Each DNA sample was analyzed in triplicate.
PCR-Based Direct Sequencing
The PCR reactions were performed in a final volume of 25 μl containing 1X PCR Gold buffer, 1.5 mmol/L of MgCl2, 200 μmol/L dNTPs, 500 nmol/L each primer (KRAS-F and KRAS-R; Table 1), 1 unit of TaqDNA Gold polymerase (Applied Biosystems, CA), and 20 ng of genomic DNA. Thermal cycling conditions included preincubation at 94°C for 5 minutes, followed by 35 cycles at 94°C for 15 seconds, 60°C for 30 seconds, 72°C for 1 minute, and a final extension at 72°C for 5 minutes. The PCR products were then purified using the QIAquick PCR purification kit (QIAGEN GmbH). DNA sequencing was performed with the ABI PRISM 3100 DNA analyzer (Applied Biosystems, CA) using the ABI PRISM BigDye Terminator version 3.1 (Applied Biosystems) with the KRAS-F primer.
Table 1.
Primer and PNA-Clamp Probe Sequences
| Name | Sequences |
|---|---|
| KRAS-F | 5′-TGAAGTACAGTTCATTACGATACACG-3′ |
| KRAS-R | 5′-GGAAAGTAAAGTTCCCATATTAATGGT-3′ |
| KRASES-F | 5′-ACTGAATATAAACTTGTGGTAGTTGGCCCT-3′ |
| KRASES-R | 5′-TCAAAGAATGCTCCTGCACC-3′ |
| KRASSA-F | 5′-ATAAACTTGTGGTAGTTGGAGC-3′ |
| KRASSA-R | 5′-TGGCGTAGGCATGATTCTGAATTAGCTGTAT-3′ |
| PNA clamp probe | N′-CCTACGCCACCAGCTCC-C′ |
The mutant and mismatched sequences are shown Italicized and underlined.
Enzyme-Enriched PCR-RFLP and Sequencing
The enzyme-enriched PCR-RFLP is a two-step PCR procedure with intermittent restriction digestion to selectively eliminate wild-type alleles, thus enriching the mutated alleles. The first-step PCR reaction was performed in a final volume of 10 μl containing 1X PCR Gold buffer, 1.5 mmol/L MgCl2, 200 μmol/L dNTPs, 500 nmol/L each primer (KRASES-F and KRASES-R),17 0.4 units of TaqDNA Gold polymerase, and 14 ng of genomic DNA. The underlined base in Figure 1 represents the introduced mismatches used to generate MvaI recognition sites for wild-type KRAS sequence. Thermal cycling conditions included preincubation at 95°C for 10 minutes, followed by 20 cycles at 95°C for 15 seconds, 55°C for 30 seconds, 72°C for 30 seconds, and a final extension at 72°C for 5 minutes. After the first-step PCR, the product was digested with restriction enzyme MvaI (Takara, Kyoto, Japan). An aliquot of 5 μl of the first-step PCR product was incubated for 3 hours at 37°C in a total volume of 20 μl containing 1X K buffer and 10 units of MvaI to digest the amplified wild-type products. Then, the second-step PCR reaction was performed in a total volume of 25 μl containing 1X PCR Gold buffer, 1.5 mmol/L MgCl2, 250 μmol/L dNTPs, 500 nmol/L each primer (KRASES-F and KRASES-R), 1 unit of TaqDNA Gold polymerase, and 1 μl of the intermediate digestion products. Thermal cycling conditions included preincubation at 95°C for 10 minutes, followed by 30 cycles at 95°C for 15 seconds, 55°C for 30 seconds, 72°C for 30 seconds, and a final extension at 72°C for 5 minutes. An aliquot of 8 μl of the second-step PCR product was digested with the restriction enzyme MvaI at 37°C for 6 hours in a final volume of 10 μl containing 10 units of MvaI and 1X K buffer. The products were separated by electrophoresis on 3% agarose gel, stained with ethidium bromide, and photographed under ultraviolet light. KRAS mutant products that were not digested by MvaI were extracted from the 164-bp band using a QIAEX II gel extraction kit (QIAGEN GmbH) and sequenced under the aforementioned conditions.
Figure 1.
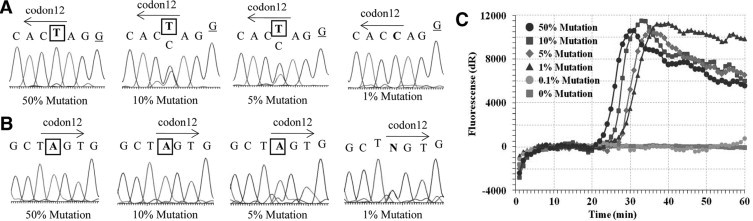
Evaluation of the sensitivity of each detection method for KRAS codon 12 mutations. A: sensitivity of enzyme-enriched sequencing (anti-sense strand). B: sensitivity of PNA-enriched sequencing (sense strand). C: sensitivity of PNA-clamp SmartAmp2. The amplification curve shows the presence of the KRAS mutation. The mutant and mismatched sequences are shown with boxed and underlined text, respectively.
PNA-Enriched Sequencing
PNA-enriched PCR was performed in a total volume of 25 μl containing 1X PCR Gold buffer, 1.5 mmol/L MgCl2, 200 μmol/L dNTPs, 500 nmol/L each primer (KRAS-F and KRAS-R), 1 μmol/L PNA clamp probe (Table 1),16 1 unit of TaqDNA Gold polymerase, and 20 ng of genomic DNA. Thermal cycling conditions included preincubation at 94°C for 5 minutes, followed by 40 cycles of 94°C for 15 seconds, 60°C for 30 seconds, 72°C for 1 minute, and a final extension at 72°C for 5 minutes. The PNA-enriched PCR products were purified and sequenced under the aforementioned conditions.
PNA-Clamp SmartAmp2-Based Sequencing
We performed PNA-clamp SmartAmp2-based sequencing in the 31 samples in which mutations were detected by the PNA-clamp SmartAmp2 assays. The PNA-clamp SmartAmp2 products were diluted 1000-fold with distilled water, and 1 μl of the diluted products were processed for PCR. The PCR reactions were performed in a total volume of 20 μl containing 1X PCR Gold buffer, 1.3 mmol/L MgCl2, 200 μmol/L dNTPs, 500 nmol/L each primer (KRASSA-F and KRASSA-R), 1 unit of TaqDNA Gold polymerase, and 1 μl of diluted DNA. Thermal cycling conditions included preincubation at 94°C for 5 minutes, followed by 35 cycles at 94°C for 30 seconds, 52°C for 30 seconds, 72°C for 30 seconds, and a final extension at 72°C for 7 minutes. The PCR products were purified and sequenced under the aforementioned conditions.
Results
Comparison of Sensitivities of KRAS Mutation Detection
The results of enzyme-enriched sequencing, PNA-enriched sequencing, and the PNA-clamp SmartAmp2 method for the detection of KRAS mutant alleles diluted with wild-type DNA are shown in Figure 1. The sample containing 1% mutant DNA could not be differentiated from the background noise by PNA-enriched sequencing (Figure 1A), and that containing 0.5% mutant DNA was not amplified by PCR with the PNA clamp probe. The detection limits of enzyme-enriched sequencing and PNA-enriched sequencing were 5% mutant DNA (Figure 1, A and B, respectively), whereas we could detect a sample containing 1% of mutant DNA clearly by PNA-clamp SmartAmp2 (Figure 1C). The mutant and mismatched sequences are shown with boxed and underlined text, respectively.
Detection of KRAS Codon 12 Mutations in Clinical Samples
We examined KRAS codon 12 mutations in 172 lung adenocarcinoma samples, in which we compared PCR-based direct sequencing, enzyme-enriched PCR-RFLP, enzyme-enriched sequencing, PNA-enriched sequencing, and PNA-clamp SmartAmp2. The results from two clinical samples analyzed with each method are shown in Figures 2 and 3. In case 6, we detected mutations by enzyme-enriched PCR-RFLP and sequencing (Figure 2D and 2E), PNA-enriched sequencing (Figure 2C), and PNA-clamp SmartAmp2 (Figure 2A), but not by PCR-based direct sequencing (Figure 2B). In case 8, we detected mutations by only PNA-clamp SmartAmp2 (Figure 3A), but not by any other methods (Figure 3B3C3D3E). This sample was verified by PNA-clamp SmartAmp2-based sequencing and confirmed to be harboring KRAS codon 12 mutations. Using PNA-enriched sequencing, the DNA template was not amplified by PCR with the PNA clamp probe (Figure 3C).
Figure 2.
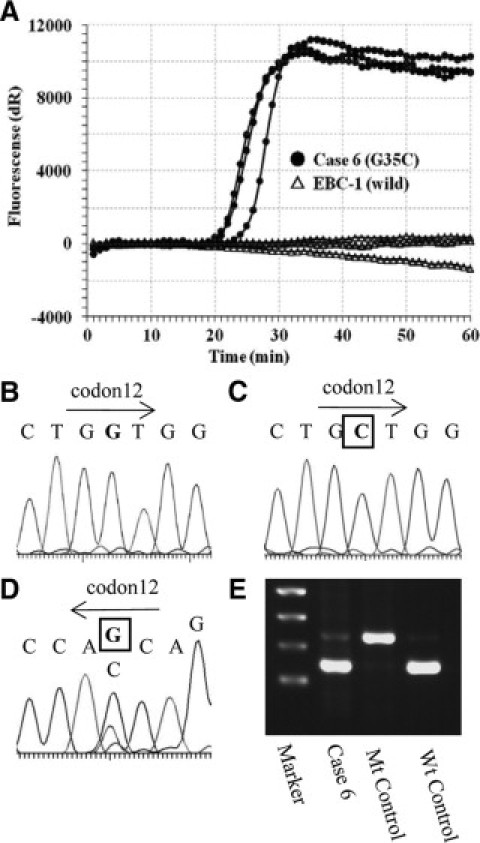
Results using different methods of detection for KRAS mutations (case 6). The results of PNA-clamp SmartAmp2 (A), PCR-based direct sequencing (sense strand sequence) (B), PNA-enriched sequencing (sense strand sequence) (C), enzyme-enriched sequencing (anti-sense strand sequence) (D), and enzyme-enriched PCR-RFLP (E) are shown. The sample contains KRAS mutation (G35C). The mutant sequences are shown with boxed text.
Figure 3.
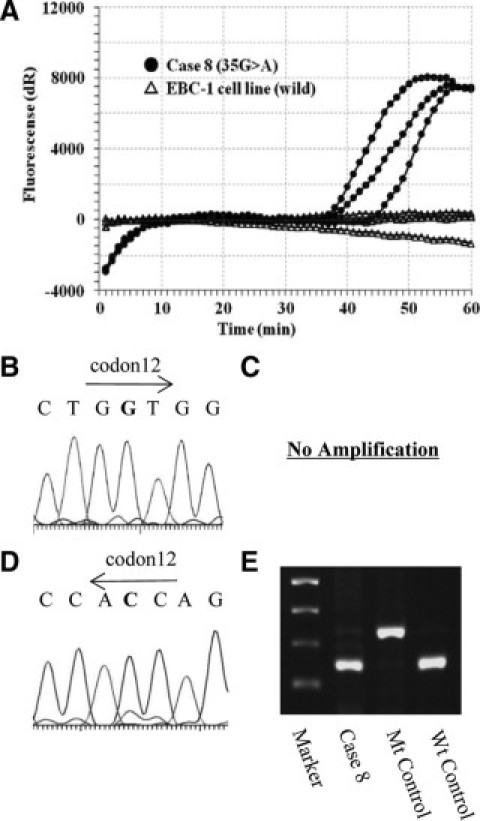
Results using different methods of detection for KRAS mutation (case 8). The results of PNA-clamp SmartAmp2 (A), PCR-based direct sequencing (sense strand sequence) (B), PNA-enriched sequencing (sense strand sequence) (C), enzyme-enriched sequencing (anti-sense strand sequence) (D), and enzyme-enriched PCR-RFLP (E) are shown. The sample contains KRAS mutation (G35A).
Among the 172 tumor samples used in this study, 31 (18.0%) samples were found to have KRAS codon 12 mutations by PNA-clamp SmartAmp2, whereas 16 (9.3%), 26 (15.7%), and 28 (16.3%) tumors were found to have KRAS codon 12 point mutations by PCR-based direct sequencing, enzyme-enriched sequencing, and PNA-enriched sequencing, respectively. The 31 (18.0%) tumors that had KRAS codon 12 mutations found by PNA-clamp SmartAmp2 included three tumors that were undetected by the other methods. A summary of the 31 KRAS mutations detected in this study is presented in Table 2. These mutations in the 31 tumors were verified by PNA-clamp SmartAmp2-based sequencing, and all tumors were confirmed to be harboring KRAS codon 12 mutations. The other 141 samples, in which KRAS codon 12 mutations were not detected by the PNA-clamp SmartAmp2, were shown to have the KRAS wild-type allele by all other methods. We examined 30 peripheral lung tissue specimens as nonmalignant samples and found no mutations in any of the samples using PNA-clamp SmartAmp2.
Table 2.
Comparison of Mutation Detection Methods in Clinical Samples
| Case | SmartAmp2 | Direct seq | Enzyme-enriched | PNA-enriched | AA change |
|---|---|---|---|---|---|
| 1 | Mut | Wt | Wt | G35T | G12V |
| 2 | Mut | Wt | G34T | G34T | G12C |
| 3 | Mut | Wt | G35A | G35A | G12D |
| 4 | Mut | G35T | G35T | G35T | G12V |
| 5 | Mut | Wt | G35A | G35A | G12D |
| 6 | Mut | Wt | G35C | G35C | G12A |
| 7 | Mut | G35T | G35T | G35T | G12V |
| 8 | Mut | Wt | Wt | Wt | G12D |
| 9 | Mut | G34T | G34T | G34T | G12C |
| 10 | Mut | Wt | Wt | Wt | G12C |
| 11 | Mut | G34T, G35T | Wt | G34T, G35T | G12F |
| 12 | Mut | G35T | G35T | G35T | G12V |
| 13 | Mut | G35T | G35T | G35T | G12V |
| 14 | Mut | G35C | G35C | G35C | G12A |
| 15 | Mut | G35C | G35C | G35C | G12A |
| 16 | Mut | G35A | G35A | G35A | G12D |
| 17 | Mut | G35A | G35A | G35A | G12D |
| 18 | Mut | Wt | G35A | G35A | G12D |
| 19 | Mut | G35A | G35A | G35A | G12D |
| 20 | Mut | G35T | G35T | G35T | G12V |
| 21 | Mut | Wt | G34T | G34T | G12C |
| 22 | Mut | Wt | G34T | G34T | G12C |
| 23 | Mut | Wt | G35A | G35A | G12D |
| 24 | Mut | G35A | G35A | G35A | G12D |
| 25 | Mut | G35A | G35A | G35A | G12D |
| 26 | Mut | Wt | Wt | Wt | G12C |
| 27 | Mut | Wt | G35A | G35A | G12D |
| 28 | Mut | G35A | G35A | G35A | G12D |
| 29 | Mut | G35A | G35A | G35A | G12D |
| 30 | Mut | Wt | G35A | G35A | G12D |
| 31 | Mut | Wt | G34T | G34T | G12C |
| Mut rate (n = 172) | 18.0% | 9.3% | 15.1% | 16.3% |
Direct seq, PCR-based direct sequencing; enzyme-enriched, enzyme-enriched sequencing; PNA-enriched, PNA-enriched sequencing; AA change, amino acid change; Mut, mutant allele; Wt, wild-type allele.
In this study, the most common mutation was the G35A transitional mutation (14/31; 45.2%). The G34T, G35T, G35C, and double point mutations (G34T and G35T) were identified in 22.6%, 19.4%, 9.7%, and 3.2% of the specimens, respectively. In addition to detecting all of these mutations, PNA-clamp SmartAmp2 provided reproducible amplification curves when each sample was examined in triplicate (Figures 2A and 3A). Furthermore, performing the PNA-clamp SmartAmp2 assay required only 60 minutes, whereas PNA-enriched sequencing and enzyme-enriched sequencing required over 9 hours and over 18 hours for the detection of KRAS codon 12 mutations, respectively.
Discussion
KRAS mutations, generally in codons 12 and 13 in exon 2 and codons 59 and 61 in exon 3, have been found in many tumor types, and they result in constitutive activation of the ras protein.18,19 The majority of the mutations (approximately 80 to 90%), especially those in NSCLC, are guanine to thymine transversions in codon 12, which result in constitutive activation of the KRAS protein.19,20
The tyrosine kinase inhibitors (TKIs) are expected to be effective EGFR-targeted agents for NSCLC. However, the effects of TKIs differ greatly among patients, and the presence of mutations in KRAS is considered one of the determinants of these differences in efficacy. Consequently, many methods have been developed and adapted for the clinical examination of KRAS codon 12 mutations. Some of these methods are more accurate and sensitive than traditional methods, but they are still lengthy and require several steps. Although these characteristics do not appear to be large drawbacks in specialized laboratories, they are less desirable for routine clinical diagnosis.
To assess the usefulness of PNA-clamp SmartAmp2 for routine clinical diagnosis, we applied this technique for the detection of KRAS codon 12 mutations in tumor samples from 172 patients with lung adenocarcinoma and compared it to other conventional methods. In the evaluation of sensitivity using DNA from cell lines, the PNA-clamp SmartAmp2 detected as little as 1% of the mutant allele in the DNA samples with no nonspecific amplification, which often reduces the sensitivity of sequencing, electrophoresis, and other methods by elevating the background noise. For this reason, the sensitivity of this method was the best of all those tested in this study.
On the other hand, when we examined 30 nonmalignant peripheral lung tissue specimens, there were no false-positive results with the PNA-clamp SmartAmp2 assay. Similarly, the 141 patients who were identified as having the wild-type allele of KRAS codon 12 by PNA-clamp SmartAmp2 were also shown to have the wild-type allele by the other PCR-related methods. In this study, we performed the PNA-clamp SmartAmp2 assays in triplicate, and the three results were matched in each sample. Moreover, using PNA-clamp SmartAmp2, the risk of sample contamination, which is an unacceptable but ever-present concern using PCR-related methods, is also reduced because PNA-clamp SmartAmp2 is performed in a closed tube. In contrast, many PCR-related methods are performed in two or three steps and require opening the tubes containing PCR products to perform the second- or third-step reactions. The PNA-clamp SmartAmp2 has high sensitivity, accuracy, and reproducibility; thus, we are confident that the PNA-clamp SmartAmp2 assay will be widely used as a reliable method for the clinical diagnosis of KRAS codon 12 mutations.
In evaluating the 172 clinical samples, 16 (9.3%) and 26 (15.7%) tumors were identified as having KRAS codon 12 mutations by direct sequencing and enzyme-enriched sequencing, and these mutation rates were consistent with those reported in the past for KRAS codon 12 mutations in Asian patients with NSCLC.9,10,11,12,13 On the other hand, we demonstrated that 31 (18%) of 172 patients with lung adenocarcinoma had KRAS codon 12 mutations by SmartAmp2, including three patients whose mutant alleles had not been detected by the other methods. These results differ from those reported in a previous study where there was perfect concordance between PNA-clamp SmartAmp 2 and PCR-based direct sequencing of clinical samples from patients with pancreatic cancer.16 To determine the basis for this difference, we evaluated the slides from the tumors with mutant KRAS DNA that were detected by PNA-clamp SmartAmp2 but not by PCR-related methods and observed that they had a lower percentage of tumor cells or more marked fibrosis than the tumors that were detected by both SmartAmp2 and PCR sequencing. The results of the enzyme-enriched sequencing of these samples were similar to the results obtained when mutations were present at frequencies of 5% to 10% (Figure 1A), suggesting that only a small fraction of the cells harbored the KRAS codon 12 mutations. Therefore, it is likely that the different proportions of tumors cells in the samples could account for the different results in our study compared with the study using pancreatic tumors. The PNA-clamp SmartAmp2 assay was shown to have the highest sensitivity using DNA from the cell lines, in which we could detect small amounts of mutant DNA that were not detected by the other methods. Thus, the higher sensitivity of the PNA-clamp SmartAmp2 assay may make it more suitable for detecting even the presence of small amounts of DNA containing KRAS codon 12 mutations in these samples with fibrosis in clinical use.
This method could find widespread clinical application in determining therapeutic strategies for patients with NSCLC because of its high sensitivity, specificity, and ease of use. Furthermore the results we showed implied that a highly sensitive method, such as the PNA-clamp SmartAmp2, may alter some therapeutic strategies. For example, it is known that patients whose tumors have the wild-type KRAS allele have possibility to respond to TKIs, and patients with the wild-type KRAS allele and a mutated EGFR are most likely to respond to TKIs. In this study, we identified 156 patients who appeared to have the wild-type KRAS allele and that TKIs might be effective for these patients by direct sequencing; however, 15 of these patients were shown to actually have KRAS codon 12 mutations by PNA-clamp SmartAmp2. According to this result, TKIs might be ineffective for these 15 patients.
While many highly sensitive methods, such as PNA-clamp SmartAmp2, have been developed and adapted to detect very low levels of oncogenes, the association between the effects of agents such as the TKIs and the presence of extremely small levels of the mutant alleles is not precisely known. In this study, we demonstrated that we could detect the presence of DNA containing KRAS codon 12 mutations at a level of 1% of the wild-type allele by PNA-clamp SmartAmp2; however, it is not clear whether TKIs would be effective if the mutant KRAS codon 12 allele were present at a level of 1% in individual patients. Additional research is needed to clarify this point. To assess how low levels of mutant alleles affect the efficacy of TKIs and to determine the minimum level of mutant allele that is clinically significant, a highly sensitive detection method is essential, and PNA-clamp SmartAmp2 should be an effective tool for such studies.
Recently, the presence of KRAS mutations in plasma, serum, and urine has become a focus of interest.21,22 The PNA-clamp SmartAmp2 assay may also contribute to the development of mutant DNA detection methods in these specimens, which contain extremely small amounts of mutant cells.
In the CRYSTAL trial, it was shown that the EGFR inhibitor cetuximab is effective in only those patients with colorectal cancer who have the wild-type KRAS allele.23,25 These reports show that the diagnosis of KRAS mutations is essential for not only lung cancer therapies, but also colorectal cancer therapies. In colorectal cancer, KRAS mutations were detected in codon 13 as well as codon 12. If the PNA-clamp SmartAmp2 is adapted for the detection of KRAS codon 13 mutations, it could also be a suitable tool for deciding therapeutic strategies in colorectal cancer.
In conclusion, we have shown that PNA-clamp SmartAmp2 is highly sensitive, accurate, and reproducible for the clinical diagnosis of KRAS codon 12 mutations. Using this method, we detected small amounts of mutant DNA that were not detected by traditional methods and identified patients for whom this assay provided clinical information that was unavailable from other tests and that may contribute to therapeutic decisions. The sensitivity of this assay highlights the need to clarify the association between the effect of TKIs and low levels of mutant KRAS alleles, and points to important new areas of study.
Acknowledgements
We thank Ms. Aiko Matsumoto for secretarial assistance and Ms. Tomoko Senbongi for technical assistance.
Footnotes
Supported by grant-in-aid for scientific research KAKENHI 19390359 from the Japan Society for the Promotion of Science.
References
- 1.Barbacid M. Ras genes. Annu Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- 2.Kikuchi A, Demo SD, Ye ZH, Chen YW, Williams LT. ralGDS family members interact with the effector loop of ras p21. Mol Cell Biol. 1994;14:7483–7491. doi: 10.1128/mcb.14.11.7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodriguez-Viciana P, Warne PH, Dhand R, Vanhaesebroeck B, Gout I, Fry MJ, Waterfield MD, Downward J. Phosphatidylinositol-3-OH kinase as a direct target of Ras. Nature. 1994;370:527–532. doi: 10.1038/370527a0. [DOI] [PubMed] [Google Scholar]
- 4.Shields JM, Pruitt K, McFall A, Shaub A, Der CJ. Understanding Ras: ‘it ain't over ‘til it's over’. Trends Cell Biol. 2000;10:147–154. doi: 10.1016/s0962-8924(00)01740-2. [DOI] [PubMed] [Google Scholar]
- 5.Vojtek AB, Cooper JA. Rho family members: activators of MAP kinase cascades. Cell. 1995;82:527–529. doi: 10.1016/0092-8674(95)90023-3. [DOI] [PubMed] [Google Scholar]
- 6.Fisher GH, Wellen SL, Klimstra D, Lenczowski JM, Tichelaar JW, Lizak MJ, Whitsett JA, Koretsky A, Varmus HE. Induction and apoptotic regression of lung adenocarcinomas by regulation of a K-Ras transgene in the presence and absence of tumor suppressor genes. Genes Dev. 2001;15:2349–3262. doi: 10.1101/gad.947701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nelson HH, Christiani DC, Mark EJ, Wiencke JK, Wain JC, Kelsey KT. Implications and prognostic value of K-ras mutation for early-stage lung cancer in women. J Natl Cancer Inst. 1999;91:2032–2038. doi: 10.1093/jnci/91.23.2032. [DOI] [PubMed] [Google Scholar]
- 8.Gandara DR, Lara PN, Lau DH, Mack P, Gumerlock PH. Molecular-clinical correlative studies in non-small cell lung cancer: application of a three-tiered approach. Lung Cancer. 2001;34:S75–S80. doi: 10.1016/s0169-5002(01)00368-3. [DOI] [PubMed] [Google Scholar]
- 9.Pao W, Wang TY, Riely GJ, Miller VA, Pan Q, Ladanyi M, Zakowski MF, Heelan RT, Kris MG, Varmus HE. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005;2:57–61. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shepherd FA, Rosell R. Weighing tumor biology in treatment decisions for patients with non-small cell lung cancer. J Thorac Oncol. 2008;3:198–199. doi: 10.1097/01.JTO.0000269737.05962.a0. [DOI] [PubMed] [Google Scholar]
- 11.Kosaka T, Yatabe Y, Endoh H, Kuwano H, Takahashi T, Mitsudomi T. Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer Res. 2004;64:8919–8923. doi: 10.1158/0008-5472.CAN-04-2818. [DOI] [PubMed] [Google Scholar]
- 12.Toyooka S, Tsukuda K, Ouchida M, Tanino M, Inaki Y, Kobayashi K, Yano M, Soh J, Kobatake T, Shimizu N, Shimizu K. Detection of codon 61 point mutations of the K-ras gene in lung and colorectal cancers by enriched PCR. Oncol Rep. 2003;10:1455–1459. doi: 10.3892/or.10.5.1455. [DOI] [PubMed] [Google Scholar]
- 13.Huang CL, Taki T, Adachi M, Konishi T, Higashiyama M, Kinoshita M, Hadama T, Miyake M. Mutations of p53 and K-ras genes as prognostic factors for non-small cell lung cancer. Int J Oncol. 1998;12:553–563. doi: 10.3892/ijo.12.3.553. [DOI] [PubMed] [Google Scholar]
- 14.Mitani Y, Lezhava A, Kawai Y, Kikuchi T, Oguchi-Katayama A, Kogo Y, Itoh M, Miyagi T, Takakura H, Hoshi K, Kato C, Arakawa T, Shibata K, Fukui K, Masui R, Kuramitsu S, Kiyotani K, Chalk A, Tsunekawa K, Murakami M, Kamataki T, Oka T, Shimada H, Cizdziel PE, Hayashizaki Y. Rapid SNP diagnostics using asymmetric isothermal amplification and a new mismatch-suppression technology. Nat Methods. 2007;4:257–262. doi: 10.1038/nmeth1007. [DOI] [PubMed] [Google Scholar]
- 15.Hoshi K, Takakura H, Mitani Y, Tatsumi K, Momiyama N, Ichikawa Y, Togo S, Miyagi T, Kawai Y, Kogo Y, Kikuchi T, Kato C, Arakawa T, Uno S, Cizdziel PE, Lezhava A, Ogawa N, Hayashizaki Y, Shimada H. Rapid detection of epidermal growth factor receptor mutations in lung cancer by the SMart-amplification process. Clin Cancer Res. 2007;13:4974–4983. doi: 10.1158/1078-0432.CCR-07-0509. [DOI] [PubMed] [Google Scholar]
- 16.Tatsumi K, Mitani Y, Watanabe J, Takakura H, Hoshi K, Kawai Y, Kikuchi T, Kogo Y, Oguchi-Katayama A, Tomaru Y, Kanamori H, Baba M, Ishidao T, Usui K, Itoh M, Cizdziel PE, Lezhava A, Ueda M, Ichikawa Y, Endo I, Togo S, Shimada H, Hayashizaki Y. Rapid screening assay for KRAS mutations by the modified Smart amplification process. J Mol Diagn. 2008;10:520–526. doi: 10.2353/jmoldx.2008.080024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimura K, Nagasaka T, Hoshizima N, Sasamoto H, Notohara K, Takeda M, Kominami K, Iishii T, Tanaka N, Matsubara N. No duplicate KRAS mutation is identified on the same allele in gastric or colorectal cancer cells with multiple KRAS mutations. J Int Med Res. 2007;35:450–457. doi: 10.1177/147323000703500403. [DOI] [PubMed] [Google Scholar]
- 18.Bos JL. ras oncogenes in human cancer. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- 19.Keohavong P, DeMichele MA, Melacrinos AC, Landreneau RJ, Weyant RJ, Siegfried JM. Detection of K-ras mutations in lung carcinomas: relationship to prognosis. Clin Cancer Res. 1996;2:411–418. [PubMed] [Google Scholar]
- 20.Rodenhuis S, van de Wetering ML, Mooi WJ, Evers SG, van Zandwijk N, Bos JL. Mutational activation of the K-ras oncogene: a possible pathogenetic factor in adenocarcinoma of the lung. N Engl J Med. 1987;317:929–935. doi: 10.1056/NEJM198710083171504. [DOI] [PubMed] [Google Scholar]
- 21.Su YH, Wang M, Brenner DE, Norton PA, Block TM. Detection of mutated K-ras DNA in urine, plasma, and serum of patients with colorectal carcinoma or adenomatous polyps. Ann NY Acad Sci. 2008;1137:197–206. doi: 10.1196/annals.1448.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Di Fiore F, Charbonnier F, Lefebure B, Laurent M, Le Pessot F, Michel P, Frebourg T. Clinical interest of KRAS mutation detection in blood for anti-EGFR therapies in metastatic colorectal cancer. Br J Cancer. 2008;99:551–552. doi: 10.1038/sj.bjc.6604451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jean GW, Shah SR. Epidermal growth factor receptor monoclonal antibodies for the treatment of metastatic colorectal cancer. Pharmacotherapy. 2008;28:742–754. doi: 10.1592/phco.28.6.742. [DOI] [PubMed] [Google Scholar]
- 25.Khambata-Ford S, Garrett CR, Meropol NJ, Basik M, Harbison CT, Wu S, Wong TW, Huang X, Takimoto CH, Godwin AK, Tan BR, Krishnamurthi SS, Burris HA, 3rd, Poplin EA, Hidalgo M, Baselga J, Clark EA, Mauro DJ. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J Clin Oncol. 2007;25:3230–3237. doi: 10.1200/JCO.2006.10.5437. [DOI] [PubMed] [Google Scholar]
Uncited reference
- 24.Ramos FJ, Macarulla T, Capdevila J, Elez E, Tabernero J. Understanding the predictive role of K-ras for epidermal growth factor receptor-targeted therapies in colorectal cancer. Clin Colorectal Cancer. 2008;7:S52–S57. doi: 10.3816/CCC.2008.s.008. [DOI] [PubMed] [Google Scholar]


