Abstract
The revolution in genetics disclosed the types of malformations that occur when expression of a particular gene is lost. In the case of tooth dentin, mutations in the two genes encoding type I collagen cause osteogenesis imperfecta, a bone condition that often includes dentin malformations. Besides collagen, there are a number of non-collagenous proteins in dentin. Among the genes encoding the dentin non-collagenous proteins, only mutations in DSPP (dentin sialophosphoprotein) cause inherited dental malformations. DSPP mutations cause dentinogenesis imperfecta types II and III, and dentin dysplasia type II. DSPP is the most abundant non-collagenous protein in dentin. DSPP protein is necessary for proper dentin formation, and understanding its structure and function should yield important insights into how dentin forms and biomineralization is controlled. DSPP is expressed and secreted by odontoblasts, the cells that make tooth dentin and that also maintain cell processes extending into the mineralized tissue. Following its secretion, DSPP is cleaved into smaller pieces by multiple extracellular proteases. For the last five years I have devoted myself to characterizing DSPP-derived proteins. DSPP is cleaved by proteases into three main parts : dentin sialoprotein (DSP), dentin glycoprotein (DGP), and dentin phosphoprotein (DPP). We have learned that DSP is a proteoglycan that forms covalent dimers, DGP is a phosphorylated glycoprotein, and DPP is a highly phosphorylated intrinsically disordered protein that shows extensive length polymorphisms due to the genetic heterogeneity of its coding region.
Keywords: teeth, dentin sialophosphoprotein, dentin dysplasia, dentinogenesis imperfecta, dentin sialoprotein, dentin glycoprotein, dentin phosphoprotein, intrinsically disordered proteins
Introduction
Dentin comprises the body of a tooth and discovering the mechanisms governing its formation promises future improvements in the diagnosis and treatment of pathologies affecting dentin, which can arise from genetic or environmental factors, injury and disease. Dentin forms in a defined extracellular space by matrix-mediated biomineralization. Secreted proteins regulate and control the mineralization process.
Most of the organic material (~85%) in dentin is type I collagen. Collagen triple helices assemble into long fibrils (50 to 200 nm in diameter) that are noted for their tensile strength and for giving dentin its flexibility and resilience. Collagen fibers also serve as a scaffold for mineralization1). The non-collagenous proteins in dentin are dominated by DSPP-derived proteins, that is, proteins generated by proteolysis of dentin sialophosphoprotein (DSPP)2), a multidomain protein with hundreds of post-translational modifications3). DSPP is expressed from the DSPP gene (4q21). It is processed by proteases into three protein products : dentin sialoprotein (DSP), dentin glycoprotein (DGP), and dentin phosphoprotein (DPP).
The importance of collagen and DSPP-derived proteins in dentin biomineralization is proven by the tooth defects that occur in their absence. Inherited dentin malformations are classified into 5 groups : three types of dentinogenesis imperfecta (DGI) and two types of dentin dysplasia (DD). DGI-I comprises dentin malformations associated with osteogenesis imperfecta (OI). OI is caused primarily by mutations in the two genes encoding type I collagen : COL1A1 (17q21.31-q22) and COL1A2 (7q22.1). The bone phenotype predominates in OI, but selected collagen gene mutations produce no clinically evident bony defects, even though dentin defects are manifested4). DGI-II has the same dental phenotype (opalescent dentin, bulbous crowns, obliterated pulp chambers) as DGI-I, but lacks bone symptoms. DGI-II is caused by mutations in DSPP. DSPP mutations also cause DGI-III and DD- II5—12). No genes besides DSPP have been implicated in the etiologies of isolated (non syndromic) inherited defects of dentin13). Thus, recent genetic studies accentuate the importance of DSPP in both normal and pathological dentin biomineralization.
This review focuses upon dentin sialophosphoprotein (DSPP), a chimeric extracellular matrix protein that is actually three proteins, as DSPP is cleaved by proteases into three separate parts2). The order of the three components is DSP-DGP-DPP, where DSP is dentin sialoprotein, DGP is dentin glycoprotein, and DPP is dentin phosphoprotein (or phosphophoryn). DSPP is expressed in teeth, predominantly by odontoblasts14). DSPP is detected in bone, but at only 1/400th the level as dentin15). DSPP-derived proteins are most easily obtained from developing teeth. Historically, the rat animal model was primarily used to study DSPP. Our success in clarifying the structural properties of DSPP-derived proteins, however, stems from pioneering the porcine animal model, which has the obvious advantage of tooth size (Fig. 1). Here we review our path to understanding the structure and proteolytic processing of DSPP, treating each structural/functional domain separately, according to their order in the parent DSPP protein.
Fig. 1. Tooth size and stage of development.
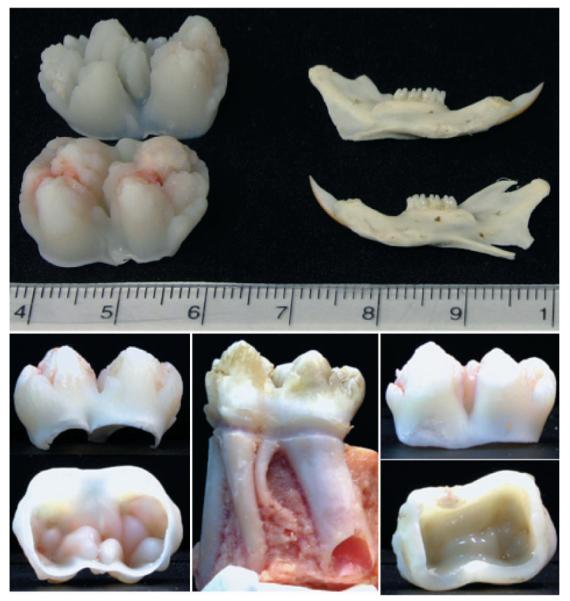
The developing molars used in our studies are just finishing crown formation, but have not yet started root development. They are about 2 cm in mesialdistal dimension, compared with an adult rat jaw, which is about 3 cm long. On the lower left are two views of the porcine 2nd molar from a 5 to 6 month old pig. Note the open pulp chamber allows easy removal of the pulp at the time of extraction, minimizing contamination by blood components. This contrasts with the erupted 1st molar (center and right). The crown was cut off with a rotary instrument and cleaned. Note the dentin in the 6 month 2nd molar (left) is at an early stage of development and the ceiling of the pulp chamber shows the forms of the cusps, while the 6 month 1st molar has a nearly smooth pulp ceiling. Despite being at an early stage of development, the dentin proteins extracted from the 2nd molars have already experienced extensive proteolysis.
Dentin Sialoprotein (DSP)
The N-terminal domain of DSPP is dentin sialoprotein. DSP was discovered a quarter of a century ago as a prominent band on SDS-PAGE having an apparent molecular weight of 95-kDa and an N-terminal sequence of IPVPQLV16). Its molecular weight was measured by sedimentation equilibrium analysis to be only 52.57kDa, which included 29.6% carbohydrate17). The rat DSP and DPP cDNAs were the first to be cloned, but DNA sequencing errors led to the faulty conclusion that rat DSP translation terminates after 366 amino acids18), and that the rat DSP and DPP domains are expressed from a single bicistronic transcript19—21). Subsequently, the mouse DSPP cDNA showed the “bicistronic” concept was wrong, that DSP and DPP are expressed from a single open reading frame, and that DSP and DPP must be generated by proteolysis of a chimeric protein2). Since this finding, two cleavage sites, a major site after Tyr421 and a minor site after His406 have been proposed to define the C-terminus of rat DSP22). Unfortunately important unresolved inconsistencies arise by increasing the size of the rat DSP amino acid sequence from 366 to 421 amino acids. The rat DSP peptide backbone cannot possibly be that large, contain 29.6% carbohydrate, and also have a molecular weight as low as 52.57kDa. Subsequently, rat DSP products having a much higher apparent molecular weight (HMW-DSP) have been discovered. HMW-DSP reportedly has 10.3 phosphates per molecule and the original rat DSP 6.2, but these values too are suspect. DSP is highly heterogeneous due to variations in its post-translational modifications and its proteolytic processing (at multiple sites) from the secreted chimera. The average molecular weight of DSP fractions cannot be determined with precision, but molecular weight values are nonetheless are required to calculate phosphate levels on a per molecule basis. As inconsistencies in the published conclusions concerning rat DSP structure are still being resolved, we have directed our efforts to the study porcine DSP.
At the start, no genomic or cDNA sequences for porcine DSPP were available in the databases. We constructed and screened a unidirectional cDNA library derived from the pulp organ of developing pig teeth and isolated cDNA clones encoding DSP-only, as well as two DSPP clones that were probably affected by artifactual deletions involving the code for the highly redundant region of DPP in exon 523). The DSP-only transcript was completely unexpected, but is important because it helps us define the C-terminus of DSP. The DSP-only transcript was generated through the use of a polyadenylation signal in intron 4, thereby precluding transcription of the code for the rest of DSPP (encoded by exon 5). An in-frame translation termination signal (TGA) appears just four codons after the end of exon 4, so only three amino acids are added to the C-terminus of the DSP-only protein relative to the DSPP deduced amino acid sequence. The DSP-only open reading frame has 386 codons, with the first 15 encoding the signal peptide. (In this report we will number the amino acids in DSPP starting with Met1, so the numbers are inclusive of the signal peptide.) Another DSP-only transcript was previously reported for the rat24), but this cDNA is a likely cloning artifact as its generation cannot be explained by any known mechanism (the internal deletion does not obey the GT…AG rule for splicing). We expressed recombinant porcine DSP in bacteria as a GST fusion product and raised polyclonal antibodies that allowed us to isolate DSP from developing pig teeth25). Using this antibody we are able to track DSP and DSP-derived cleavage products throughout our purification procedures25—27).
In vivo, the first proteolytic cleavage of DSPP releases DPP from the C-terminus. Subsequent cleavages release DGP or extended DGP from the C-terminus of DSP-DGP. The C-terminus of pig DSP varies depending upon the cleavage site used. The main cleavage site is on the C-terminal side of Arg391, which was confirmed by demonstrating that Ser392 is at the N-terminus of DGP25,26). The genetic and proteolytic cleavage data are consistent and show that DSP is the N-terminal domain of DSPP and DSP is encoded by exons 2 through 4 of the DSPP. Exon 5 encodes both DGP and DPP. Based upon the characterization of DSPP cleavage products isolated from developing pig teeth, DSP extends from Ile16 to Arg391. Porcine DSP has 376 amino acids. Despite this clear definition of the boundaries of DSP, our research shows that the proteolytic processing of DSPP does not simply generate three cleavage products (DSP, DGP, and DPP) that accumulate to serve separate functions. Instead these domains are only somewhat stable intermediates in what appears to be the extracellular degradation of DSPP.
Knowing the primary structure of porcine DSP was only a start. It was clear from the beginning that post-translational modifications are an important part of DSP structure. The deduced amino acid sequence of porcine dentin sialoprotein (Fig. 2) contains 8 putative N-glycosylation sites23). There are several predicted O-glycosylation sites as well as four SerGly (SG) pairs, which could potentially be targeted by xylosyl-transferase to introduce glycan attachments28). We use efficient extraction and purification methods to isolate DSPP-derived proteins in quantity from developing pig teeth. Unerupted late crown formation stage molars are extracted from 5-month-old pigs as they are slaughtered at the Michigan State Meat Laboratory. The soft tissues and enamel are removed, and the dentin is pulverized using a jaw crusher. Dentin powder (40 g) is sequentially extracted with guanidine (G), acetic acid (A) and acetic acid/NaCl (AN). DGP and low molecular weight (LMW) DSP cleavage products are in the A extract, while DSP, DSP-DGP, and DPP are in the AN extract. These extracts are subsequently fractionated by size exclusion chromatography and reversed phase HPLC.
Fig. 2. Porcine DSP post-translational modifications (PTMs).
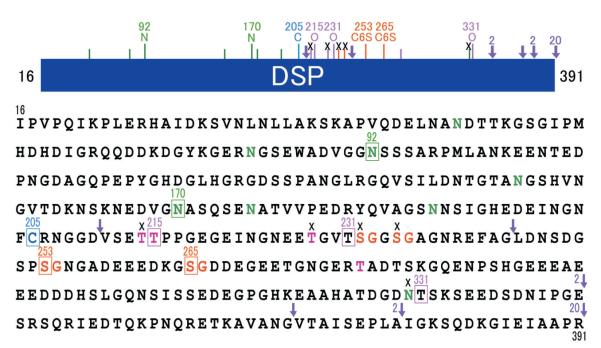
Porcine DSP extends from the DSPP amino terminus at Ile16 and extends to Arg391. The bar maps PTMs in the DSP region. Key : A short line marks a predicted glycosylation or phosphorylation site that has not been determined experimentally. An “x” indicates a predicted site that is not modified. Arrowheads mark known cleavage sites. The numbers 2 and 20 beside an arrowhead indicates that MMP-2 or MMP-20 make that cleavage, respectively. In the primary amino acid sequence of porcine DSP there are eight predicted N-linked glycosylation(N at positions 52, 82, 92, 150, 170, 176, 191 and 330) (green), four potential O-linked glycosylations (T at positions 214, 215, 228, and 279) (pink), and four potential glycan attachment sites (S at positions 232, 235, 253, and 265) (orange). We have demonstrated at the protein level that two of the potential N-linked sites (N92 and N170) (green squares) are glycosylated, but one (N330) is not (black cross). Two of the four predicted O-linked glycosylation sites (T214 and T228) are not glycosylated (black crosses), but one predicted site (T215) and two unpredicted sites (T231 and T331) are O-glycosylated (pink squares) ; and two of the four potential glycan attachment sites (S253 and S265) (orange squares) are used, but other two (S232 and S235) are not.
The magnitude of DSP glycosylation is evident from its apparent molecular mass on SDS-PAGE. DSP extracted from porcine teeth migrates as a smear extending from 100 to 280-kDa on Western blots. In contrast, the apparent molecular mass of recombinant DSP expressed in bacteria is 65-kDa and the predicted molecular mass of the unmodified DSP is 39-kDa. To gain information about sugar attachments on the purified porcine DSP glycoprotein, we conducted a series of deglycosylation digestions. First, N-linked oligosaccharide chains were removed by glycopeptidase A digestion ; second, O-linked oligosaccharide chains were removed by O-glycosidase digestion ; third, glycan attachments were removed by protease-free chondroitinase ABC digestion. Carbohydrates were released from DSP in each step, so DSP has N- and O-linked glycosylations as well as glycan attachments25). We also determined that the glycan attachments are comprised exclusively of chondroitin 6-sulfate. This was accomplished by digesting N- and O-deglycosylated DSP with four glycosaminoglycanases (protease-free chondroitinase ABC, heparitinase I, keratanase, and keratanase II) and then assaying the products on Western blots using a panel of monoclonal antibodies that recognize specific glycans. The results indicated that porcine DSP is a proteoglycan with a large and variable amount of chondroitin 6-sulfate.
The more we worked with DSP the more familiar it became. On SDS-PAGE and Western blots, DSP always appeared as two smear bands extending from 280-kDa to 100-kDa, so we decided to compare its migration on SDS-PAGE under oxidizing and reducing conditions to check for disulfide bridging. Under reducing conditions (with β-mercaptoethanol) the higher molecular weight smear disappears, as it is comprised of DSP dimers covalently linked by a disulfide bridge. Porcine DSP has only one cysteine residue per molecule, so the disulfide connection must join two DSP proteins at Cys205.
Our next objective was to determine the positions of the glycosylations on DSP. We used several strategies based upon the fact that amino acids carrying a post-translational modification show a “blank cycle” when sequenced by Edman degradation. We digested DSP with lysylendopeptidase or pronase and also isolated smaller DSP-derived cleavage products from dentin extracts. The DSP-derived peptides were assayed for the presence of glycosylations using the phenol sulfuric acid method, and characterized by N-terminal sequencing. The results show that DSP has two glycan attachment sites, at Ser253 and Ser265. All other potential glycan sites have been excluded (Fig. 2).
Dentin Glycoprotein (DGP)
We discovered the middle portion of DSPP as a prominent stains-all positive protein in porcine dentin extracts, and designated it dentin glycoprotein, or DGP26). DGP is a glycoprotein that migrates at 19-kDa on SDS-PAGE, and is reduced to 16-kDa following deglycosylation with glycopeptidase A. DGP is the 81 amino acid segment of DSPP (Ser392 to Gly472) between the DSP and DPP domains. DGP is abundant and easy to isolate from pig dentin, allowing us to characterize its entire amino acid sequence by protein methods. DGP has four phosphorylated serine residues (Ser453, Ser455, Ser457, and Ser462) and one glycosylated asparagine (Asn397). Any of five different complex biantenniary structures, both sialidated and unsialidated, can be attached to Asn397 (Fig. 3). Dentin glycoprotein has a novel amino acid sequence and only shares significant amino acid identity with homologous regions of other DSPP sequences. Pig DGP shares 81% amino acid identity with human DGP, and all five of the modified amino acids are conserved. Although the main DGP protein has 81 amino acids, recently we identified several larger versions of the protein (“extended DGPs”) that are generated by cleavage of DSP-DGP at alternative sites. The N-terminal sequence of a 22-kDa band has 105 amino acids (Val368-Gly472) and is 24 amino acids longer than DGP27).
Fig. 3. Porcine DGP post-translational modifications (PTMs).
Porcine DGP is the middle part of DSPP and extends from Ser392 and extends to Gly472. The bar maps PTMs in the DGP region, which has been entirely sequenced by protein methods. Although predicted, Asn394 is not glycosylated, but Asn 397 is glycosylated (green square) and four serines (Ser453, Ser455, Ser457 and Ser462) are phosphorylated (red squares). Biantennary glycan structures released from Asn397 were fluorescently labeled and characterized, and the five determined structures are shown.
The DGP domain of DSPP has not yet been reported from other species, possibly creating the impression that DGP is not conserved or important. While the general existence of DGP in mammalian dentin is not demonstrated, there are no reports of efforts to search for DGP in other organisms. The early landmark rat cloning studies suggested that no domain between DSP and DPP should exist, as DNA sequencing errors caused incorrect assignment of the coding region for DGP to be the 3′ non-coding region for DSP18) and 5′ non-coding region and leader sequence for DPP19). Until a survey is conducted of dentin proteins in other mammals, the question of the general importance of DGP in the process of odontogenesis cannot be answered. The alignment of human, porcine, and rat DSP-DGP amino acid sequences shows high similarity among these homologues (Fig. 4).
Fig. 4. Alignment of porcine, human, and rat DSP-DGP.
The pig(#NP_998942)23), human(#NP_055023)48), and rat(#AJ403971)49) DSP-DGP sequences (DPP domain not included). The human and pig sequences show 58.5% (269/460) identity ; the human and rat sequences show only 41.1% (189/460) identity. Because the pig primary amino acid sequence is so much more identical to the human sequence than is the rat, the pig may be a more relevant animal model for understanding the functions of human DSPP-derived proteins.
Proteolysis of DSPP
DSPP has never been isolated (or even detected with certainty) from dentin extracts. Very rapidly following its synthesis, DSPP is cleaved by an unidentified protease after Gly472, into DSP-DGP (Ile16 to Gly472) and DPP (Asp473 to C-terminus). We have successfully isolated these two cleavage products from pig dentin27). Even in molars at an early stage of development (before the onset of root formation), a large number of other DSPP pieces are observed in the extracts. There is intact DSP (Ile16 to Arg391), various N-terminal pieces of DSP, a “DSP proteoglycan core” comprised of short DSP peptides starting downstream of the disulfide bridge connection at Cys205 and containing the two glycan attachments on Ser253 and Ser265 but not continuing much further, extended DGPs (Ser345-Gly472 and Ile377-Gly472) and DGP (Ser392-Gly472). The DPP domain appears to be degraded almost at random, leaving a long, homogeneous smear of degradation products containing no discernible bands on SDS-PAGE.
Zymogram analyses of porcine dentin extracts identified three major proteases in dentin : gelatinase (MMP-2), enamelysin (MMP-20), and kallikrein 4 (KLK4). MMP-9 is reportedly found in rat29) and human30) dentin, but we could not detect it in porcine dentin extracts. Instead, we observed that DPP does not stain with Coomassie Brilliant Blue (CBB) and leaves a clear band on zymograms that might mistakenly be interpreted as a protease migrating under 100-kDa, which approximates the mobility of MMP-9, a glycosylated matrix metalloproteinase. In addition, blood proteins such as MMP-9 can easily contaminate dentin30). Bleeding often accompanies the extraction of unerupted teeth and the dental pulp must be removed immediately following extraction to avoid internal bleeding, which is nearly impossible if the roots have already formed. MMP-2, MMP-20, and KLK4 supply forming dentin with an impressive catalog of enzyme activities. The seepage of KLK4 from mantle dentin into the deepest enamel layer reportedly increases the degree of mineralization of the enamel covering the dentino-enamel junction (DEJ)31).
We isolated MMP-2 and MMP-20 from dentin and incubated the native proteases with DSP-DGP or DPP27). These enzymes showed no activity against DPP. MMP-20 cleaved DSP-DGP to generate DSP and DGP (S392-G472), and also cleaved DSP at multiple sites to generate the DSP proteoglycan core. MMP-2 generated both extended DGP forms. Clearly these two proteases play an important part of the process/degradation of DSPP.
In general we were surprised to see the extent of DSPP degradation so early in tooth development. Our findings suggest that DSPP, like enamel proteins, might be consumed in the process of biomineralization and may not play a structural role in erupted, functional teeth. Reports from the literature do not paint a consistent picture. DPP could not be detected in human dentin except in the immature dentin at the growing root tip32), while bovine and rabbit dentin sampled at three developmental stages showed progressively increasing amounts of DPP33). The DPP domain is known to be inherently unstable and is degraded in vitro by heating34)
The prevailing concept of dentinogenesis that envisions DSPP being cleaved into two stable products (DSP and DPP) that persist as structural proteins after initiating and regulating dentin mineralization is not at all consistent with our findings in pig. DPP, for instance, is thought to bind collagen and initiate oriented crystal growth, essentially burying DPP within the mineralized collagen. Wouldn’t this model predict that DPP would be protected from further proteolysis and persist in the matrix? Clearly more information is needed about the endurance DSPP-derived proteins past the formative stages and their continued presence to serve as structural elements in mature dentin.
Dentin Phosphoprotein (DPP)
DPP is the most acidic protein known, having an isoelectric point near 1.1 and stains very strongly with stains-all35), but not CBB. The cleavage site that generates DPP from DSPP is conserved, so that DPP from various mammals has the same N-terminal sequence : AspAspProAsn (DDPN)36,37). Rat DPP may be unique in that it reportedly exists in highly (> 400 res/1000) or moderately (~250 residues/1000) phosphorylated forms, although such variability comes from early studies and requires confirmation, and has not been observed in DPP characterized from other mammals. The length of DPP certainly varies among species. Rat, bovine, and porcine DPPs have an apparent molecular mass just under 100-kDa36,38), while human DPP migrates at about 140-kDa32). Based upon length variations among the human DPP genomic sequences in the databases, human DPP is believed to display length polymorphisms13), although size variations were not noted in the one study characterizing the human DPP protein32).
Currently we are preparing a manuscript that will demonstrate that the DPP coding region shows extensive length variation among the DSPP alleles in pigs, and that these length polymorphisms translate into significant size variations in the DPP protein expressed from different DSPP alleles. This is a highly unusual and exciting finding. In fact, we are unaware of any other protein that shows such length polymorphisms. Length polymorphisms in the coding region for dentin matrix protein 1 (DMP1) proved useful in defining phylogenetic relationships among closely related species of bat39), but DMP1 size heterogeneity at the protein level has not been reported.
DPP isolated from our dentin extracts shows up as multiple (usually 4) bands migrating between 96-kDa and 100-kDa on SDS-PAGE. We determined that each of these bands has the same N-terminus (DDPN). Dephosphorylation reduced the apparent size of the DPP bands on SDS-PAGE, but did not alter the pattern of multiple bands. Deglycosylation did not affect the mobility or the pattern of the bands. We amplified the DPP coding region (exon 5 of DSPP) and cloned and characterized the products. The DPP coding regions varied in length, and all of the length variations maintained the same downstream reading frame, but we could not rule out that the length variations were, at least in part, artifacts generated during the PCR amplification step. The length variations were not due to alternative RNA splicing, as genomic DNA was used as template. No length polymorphisms were observed outside of the DPP redundant region, including the 3′ non-coding region.
The breakthrough came when we decided to isolate DPP protein from individual pigs, rather than follow our routine of combining teeth from 8 pigs for a large-scale isolation. DPP from individual pigs migrates as either one or two bands, depending upon the length of the DPP coding regions on the two DSPP alleles. Six DPP protein bands of different apparent sizes on SDS-PAGE were apparent in the dentin obtained from 22 pigs. Cloning and characterizing the DPP coding region from each pig showed that the DNA length polymorphisms were more extensive than even the protein data suggested. DPP bands of identical apparent size gave different sequences, and varied according to their pattern of deletions and insertions in the highly redundant DPP region. We do not expect that the DPP length polymorphisms are functionally significant, but rather, hypothesize that DPP can serve its functions despite significant variations in the length of its redundant region. Although we did not examine the structure of dentin in detail, there was no hint at a gross level of any defects or inherited dental malformations running through this population of pigs.
Is DSPP an Intrinsically Disordered Protein (IDP) ?
Many eukaryotic proteins are disordered under physiological conditions, and fold into ordered structures only on binding to their cellular targets40). Protein disorder may be important for understanding protein function41). Proteins, or regions of proteins, that do not appear to adopt a well-defined conformation (fold) under native conditions are increasingly being categorized as “intrinsically disordered”42). Binding to a partner (protein, crystals etc) may then induce folding of the IDP43). Whereas a globular protein exhibits a few well-ordered conformations, an IDP, in the absence of a suitable partner, exists in a multitude of conformations. The tendency of proteins to be intrinsically disordered can be predicted by analyses of their amino acid sequences. We used a Web-based computational software (PONDR) as a preliminary method to assess if DSPP might be an intrinsically disordered protein (Fig. 5)44).
Fig. 5. Predictions of protein disordered region for porcine DSPP, enamelin, ameloblastin, and albumin.
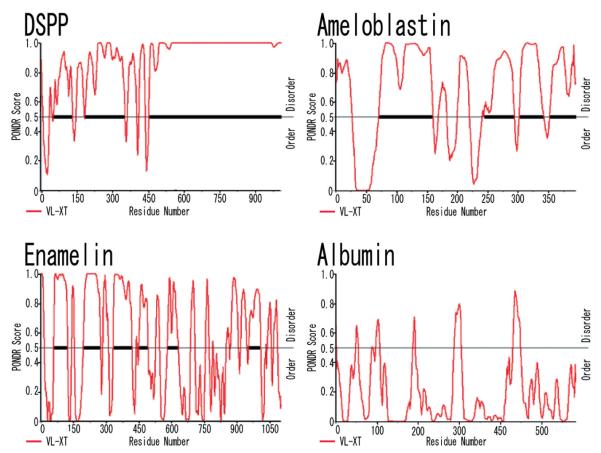
Sample output from the Predictor of Natural Disordered Regions (PON-DR®), showing predictions for porcine DPP. Access to PONDR® was provided by Molecular Kinetics (6201 La Pas Trail-Ste 160, Indianapolis, IN 46268 ; 317-280-8737 ; main@molecularkinetics.com). VL-XT is copyright© 1999 by the WSU Research Foundation, all rights reserved. PONDR® is copyright© 2004 by Molecular Kinetics, all rights reserved. From this plot it is seen that predictors agree on all DPP region(473—1008) as being disordered.
DSPP is not only predicted to be an intrinsically disordered protein, but the highly redundant region of DPP flat-lines at the highest possible score for a continuous region extending for over 350 amino acids. Porcine enamel extracellular matrix proteins, enamelin45) and ameloblastin46) also display multiple regions of intrinsic disorder, but albumin does not. DSP has two disordered segments, and both contain known glycosylation sites. The first segment is about 75 amino acids long, and extends from amino acids ~ 45-120. Asn92 is glycosylated and Asn82 is in the appropriate context for glycosylation but its modification status is not yet determined. The second DSP disordered region is larger (~160 aa), extending from ~190 to 350. This segment contains the inter-molecular disulfide bridge at Cys205, two O-linked glycosylations at Thr215 and Thr231, and the two glycan attachment sites at Ser253 and Ser265. The third DSPP disordered region is the largest, and lies outside the DSP domain. It extends from amino acid 460 all the way to the DSPP C-terminus. This last segment includes a small part of the DGP C-terminal region and all of the DPP domain. Recently, NMR analysis showed that bovine DPP is a molecule of uniformly high mobility, which is consistent with it being an intrinsically disordered protein47).
Porcine DPP averages a little over 500 amino acids in length and contains about 250 phosphates per molecule. The finding that DPP can support extensive changes in its length without a loss of function adds one more novel feature to a protein that is already the most acidic ever discovered.
Future Directions
By focusing on the structure of DSPP, its post-translational modifications, and processing by proteases, we are gaining factual information that will allow us to pose realistic mechanistic hypotheses concerning its functional roles in dentin biomineralization. We continue to map the post-translational modifications of DSPP, but would also like to clarify if DSPP persists in mature dentin as a structural component, or if it is degraded as part of the maturation process. We are also mapping the locations of DSPP derived proteins within developing dentin to determine if any parts specifically concentrate in peritubular or intertubular dentin. Finally, we are characterizing the association of DSPP derived proteins with collagen, hyaluronic acid, and other matrix constituents. We hope to achieve a better understanding of how dentin forms, and enjoy the excitement of making new discoveries along the way.
Acknowledgements
I thank the National Institute of Craniofacial and Dental Research of the National Institutes of Health (NIDCR/NIH) and the many principle investigators who have allowed me to collaborate with them and supported my research with their grant funding, including Drs. Bartlett (DE016276), Margolis (DE016376), Clarkson (DE015599), Hu (DE011301), and Simmer (DE015846).
References
- 1).Nanci A. In: Dentin-Pulp Complex, in Ten Cate’s Oral Histology Development, Structure, and Function. Nanci A, editor. St. Louis, MO, USA; Mosby: 2003. pp. 192–239. [Google Scholar]
- 2).MacDougall M, Simmons D, Luan X, Nydegger J, Feng J, Gu TT. Dentin phosphoprotein and dentin sialoprotein are cleavage products expressed from a single transcript coded by a gene on human chromosome 4. Dentin phosphoprotein DNA sequence determination. J. Biol. Chem. 1997;272:835–842. doi: 10.1074/jbc.272.2.835. [DOI] [PubMed] [Google Scholar]
- 3).Qin C, Baba O, Butler WT. Post-translational modifications of sibling proteins and their roles in osteogenesis and dentinogenesis. Crit. Rev. Oral Biol. Med. 2004;15:126–136. doi: 10.1177/154411130401500302. [DOI] [PubMed] [Google Scholar]
- 4).Pallos D, Hart PS, Cortelli JR, Vian S, Wright JT, Korkko J, Brunoni D, Hart TC. Novel COL1A1 mutation (G559C) [correction of G599C] associated with mild osteogenesis imperfecta and dentinogenesis imperfecta. Arch. Oral Biol. 2001;46:459–470. doi: 10.1016/s0003-9969(00)00130-8. [DOI] [PubMed] [Google Scholar]
- 5).Zhang X, Zhao J, Li C, Gao S, Qiu C, Liu P, Wu G, Qiang B, Lo WH, Shen Y. DSPP mutation in dentinogenesis imperfecta Shields type II. Nat. Genet. 2001;27:151–152. doi: 10.1038/84765. [DOI] [PubMed] [Google Scholar]
- 6).Xiao S, Yu C, Chou X, Yuan W, Wang Y, Bu L, Fu G, Qian M, Yang J, Shi Y, Hu L, Han B, Wang Z, Huang W, Liu J, Chen Z, Zhao G, Kong X. Dentinogenesis imperfecta 1 with or without progressive hearing loss is associated with distinct mutations in DSPP. Nat. Genet. 2001;27:201–204. doi: 10.1038/84848. [DOI] [PubMed] [Google Scholar]
- 7).Rajpar MH, Koch MJ, Davies RM, Mellody KT, Kielty CM, Dixon MJ. Mutation of the signal peptide region of the bicistronic gene DSPP affects translocation to the endoplasmic reticulum and results in defective dentine biomineralization. Hum. Mol. Genet. 2002;11:2559–2565. doi: 10.1093/hmg/11.21.2559. [DOI] [PubMed] [Google Scholar]
- 8).Malmgren B, Lindskog S, Elgadi A, Norgren S. Clinical, histopathologic, and genetic investigation in two large families with dentinogenesis imperfecta type II. Hum. Genet. 2004;114:491–498. doi: 10.1007/s00439-004-1084-z. [DOI] [PubMed] [Google Scholar]
- 9).Kim JW, Nam SH, Jang KT, Lee SH, Kim CC, Hahn SH, Hu JC, Simmer JP. A novel splice acceptor mutation in the DSPP gene causing dentinogenesis imperfecta type II. Hum. Genet. 2004;115:248–254. doi: 10.1007/s00439-004-1143-5. [DOI] [PubMed] [Google Scholar]
- 10).Kim JW, Hu JC, Lee JI, Moon SK, Kim YJ, Jang KT, Lee SH, Kim CC, Hahn SH, Simmer JP. Mutational hot spot in the DSPP gene causing dentinogenesis imperfecta type II. Hum. Genet. 2005;116:186–191. doi: 10.1007/s00439-004-1223-6. [DOI] [PubMed] [Google Scholar]
- 11).Dong J, Gu T, Jeffords L, MacDougall M. Dentin phosphoprotein compound mutation in dentin sialophosphoprotein causes dentinogenesis imperfecta type III. Am. J. Med. Genet. 2005;132:305–309. doi: 10.1002/ajmg.a.30460. [DOI] [PubMed] [Google Scholar]
- 12).Holappa H, Nieminen P, Tolva L, Lukinmaa PL, Alaluusua S. Splicing site mutations in dentin sialophosphoprotein causing dentinogenesis imperfecta type II. Eur. J. Oral Sci. 2006;114:381–384. doi: 10.1111/j.1600-0722.2006.00391.x. [DOI] [PubMed] [Google Scholar]
- 13).Kim JW, Simmer JP. Hereditary dentin defects. J. Dent. Res. 2007;86:392–399. doi: 10.1177/154405910708600502. [DOI] [PubMed] [Google Scholar]
- 14).Begue-Kirn C, Krebsbach PH, Bartlett JD, Butler WT. Dentin sialoprotein, dentin phosphoprotein, enamelysin and ameloblastin : tooth-specific molecules that are distinctively expressed during murine dental differentiation. Eur. J. Oral Sci. 1998;106:963–970. doi: 10.1046/j.0909-8836.1998.eos106510.x. [DOI] [PubMed] [Google Scholar]
- 15).Qin C, Brunn JC, Cadena E, Ridall A, Tsujigiwa H, Nagatsuka H, Nagai N, Butler WT. The expression of dentin sialophosphoprotein gene in bone. J. Dent. Res. 2002;81:392–394. doi: 10.1177/154405910208100607. [DOI] [PubMed] [Google Scholar]
- 16).Butler WT, Bhown M, Dimuzio MT, Linde A. Nonocollagenous proteins of dentin. Isolation and partial characterization of rat dentin proteins and proteoglycans using a three-step preparative method. Coll. Relat. Res. 1981;1:187–199. doi: 10.1016/s0174-173x(81)80019-2. [DOI] [PubMed] [Google Scholar]
- 17).Butler WT. Dentin extracellular matrix and dentinogenesis. Oper. Dent. 1992;(Suppl 5):18–23. [PubMed] [Google Scholar]
- 18).Ritchie HH, Hou H, Veis A, Butler WT. Cloning and sequence determination of rat dentin sialoprotein, a novel dentin protein. J. Biol. Chem. 1994;269:3698–3702. [PubMed] [Google Scholar]
- 19).Ritchie HH, Wang LH. Sequence determination of an extremely acidic rat dentin phosphoprotein. J. Biol. Chem. 1996;271:21695–21698. doi: 10.1074/jbc.271.36.21695. [DOI] [PubMed] [Google Scholar]
- 20).Ritchie H, Wang LH. A mammalian bicistronic transcript encoding two dentin-specific proteins. Biochem. Biophys. Res. Commun. 1997;231:425–428. doi: 10.1006/bbrc.1997.6126. [DOI] [PubMed] [Google Scholar]
- 21).Ritchie HH, Ritchie DG, Wang LH. Six decades of dentinogenesis research. Historical and prospective views on phosphophoryn and dentin sialoprotein. Eur. J. Oral Sci. 1998;106(Suppl 1):211–220. doi: 10.1111/j.1600-0722.1998.tb02178.x. [DOI] [PubMed] [Google Scholar]
- 22).Qin C, Cook RG, Orkiszewski RS, Butler WT. Identification and characterization of the carboxyl-terminal region of rat dentin sialoprotein. J. Biol. Chem. 2001;276:904–909. doi: 10.1074/jbc.M006271200. [DOI] [PubMed] [Google Scholar]
- 23).Yamakoshi Y, Hu JC, Liu S, Zhang C, Oida S, Fukae M, Simmer JP. Characterization of porcine dentin sialoprotein (DSP) and dentin sialophosphoprotein (DSPP) cDNA clones. Eur. J. Oral Sci. 2003;111:60–67. doi: 10.1034/j.1600-0722.2003.00009.x. [DOI] [PubMed] [Google Scholar]
- 24).Ritchie HH, Li X. A novel rat dentin mRNA coding only for dentin sialoprotein. Eur. J. Oral Sci. 2001;109:342–347. doi: 10.1034/j.1600-0722.2001.00093.x. [DOI] [PubMed] [Google Scholar]
- 25).Yamakoshi Y, Hu JC, Fukae M, Iwata T, Kim JW, Zhang H, Simmer JP. Porcine dentin sialoprotein is a proteoglycan with glycosaminoglycan chains containing chondroitin 6-sulfate. J. Biol. Chem. 2005;280:1552–1560. doi: 10.1074/jbc.M409606200. [DOI] [PubMed] [Google Scholar]
- 26).Yamakoshi Y, Hu JC, Fukae M, Zhang H, Simmer JP. Dentin glycoprotein : the protein in the middle of the dentin sialophosphoprotein chimera. J. Biol. Chem. 2005;280:17472–17479. doi: 10.1074/jbc.M413220200. [DOI] [PubMed] [Google Scholar]
- 27).Yamakoshi Y, Hu JC, Iwata T, Kobayashi K, Fukae M, Simmer JP. Dentin sialophosphoprotein is processed by MMP-2 and MMP-20 in vitro and in vivo. J. Biol. Chem. 2006;281:38235–28243. doi: 10.1074/jbc.M607767200. [DOI] [PubMed] [Google Scholar]
- 28).Gotting C, Kuhn J, Kleesiek K. Human xylosyltransferases in health and disease. Cell Mol. Life Sci. 2007;64:1498–1517. doi: 10.1007/s00018-007-7069-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29).Goldberg M, Septier D, Bourd K, Hall R, George A, Goldberg H, Menashi S. Immunohistochemical localization of MMP-2, MMP-9, TIMP-1, and TIMP-2 in the forming rat incisor. Connect. Tissue Res. 2003;44:143–153. doi: 10.1080/03008200390223927. [DOI] [PubMed] [Google Scholar]
- 30).Mazzoni A, Mannello F, Tay FR, Tonti GA, Papa S, Mazzotti G, Di Lenarda R, Pashley DH, Breschi L. Zymographic analysis and characterization of MMP-2 and -9 forms in human sound dentin. J. Dent. Res. 2007;86:436–440. doi: 10.1177/154405910708600509. [DOI] [PubMed] [Google Scholar]
- 31).Fukae M, Tanabe T, Nagano T, Ando H, Yamakoshi Y, Yamada M, Simmer JP, Oida S. Odontoblasts enhance the maturation of enamel crystals by secreting EMSP1 at the enamel-dentin junction. J. Dent. Res. 2002;81:668–672. doi: 10.1177/154405910208101003. [DOI] [PubMed] [Google Scholar]
- 32).Chang SR, Chiego D, Jr., Clarkson BH. Characterization and identification of a human dentin phosphophoryn. Calcif. Tissue Int. 1996;59:149–153. doi: 10.1007/s002239900101. [DOI] [PubMed] [Google Scholar]
- 33).Fujisawa R, Kuboki Y. Increase of dentin phosphophoryn with dentin formation. Connect. Tissue Res. 1988;17:231–238. doi: 10.3109/03008208809017474. [DOI] [PubMed] [Google Scholar]
- 34).Ibaraki K, Shimokawa H, Sasaki S. An analysis of the biochemical and biosynthetic properties of dentin phosphoprotein. Matrix. 1991;11:115–124. doi: 10.1016/s0934-8832(11)80215-5. [DOI] [PubMed] [Google Scholar]
- 35).Jonsson M, Fredriksson S. Isoelectric focusing of the phosphoprotein of rat-incisor dentin in ampholine and acid pH gradients. Evidence for carrier ampholyte-protein complexes. J. Chromatogr. 1978;157:234–242. doi: 10.1016/s0021-9673(00)92338-0. [DOI] [PubMed] [Google Scholar]
- 36).Butler WT, Bhown M, DiMuzio MT, Cothran WC, Linde A. Multiple forms of rat dentin phosphoproteins. Arch. Biochem. Biophys. 1983;225:178–186. doi: 10.1016/0003-9861(83)90021-8. [DOI] [PubMed] [Google Scholar]
- 37).Huq NL, Cross KJ, Talbo GH, Riley PF, Loganathan A, Crossley MA, Perich JW, Reynolds EC. N-terminal sequence analysis of bovine dentin phosphophoryn after conversion of phosphoseryl to S-propylcysteinyl residues. J. Dent. Res. 2000;79:1914–1919. doi: 10.1177/00220345000790111701. [DOI] [PubMed] [Google Scholar]
- 38).Termine JD, Belcourt AB, Miyamoto MS, Conn KM. Properties of dissociatively extracted fetal tooth matrix proteins. II. Separation and purification of fetal bovine dentin phosphoprotein. J. Biol. Chem. 1980;255:9769–9772. [PubMed] [Google Scholar]
- 39).Van Den Bussche RA, Reeder SA, Hansen EW, Hoofer SR. Utility of the dentin matrix protein 1 (DMP1) gene for resolving mammalian intraordinal phylogenetic relationships. Mol. Phylogenet. Evol. 2003;26:89–101. doi: 10.1016/s1055-7903(02)00297-x. [DOI] [PubMed] [Google Scholar]
- 40).Linding R, Russell RB, Neduva V, Gibson TJ. GlobPlot : Exploring protein sequences for globularity and disorder. Nucleic Acids Res. 2003;31:3701–3708. doi: 10.1093/nar/gkg519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41).Verkhivker GM, Bouzida D, Gehlhaar DK, Rejto PA, Freer ST, Rose PW. Simulating disorder-order transitions in molecular recognition of unstructured proteins : where folding meets binding. Proc. Natl. Acad. Sci. USA. 2003;100:5148–5153. doi: 10.1073/pnas.0531373100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42).Hansen JC, Lu X, Ross ED, Woody RW. Intrinsic protein disorder, amino acid composition, and histone terminal domains. J. Biol. Chem. 2006;281:1853–1856. doi: 10.1074/jbc.R500022200. [DOI] [PubMed] [Google Scholar]
- 43).Receveur-Brechot V, Bourhis JM, Uversky VN, Canard B, Longhi S. Assessing protein disorder and induced folding. Proteins. 2006;62:24–45. doi: 10.1002/prot.20750. [DOI] [PubMed] [Google Scholar]
- 44).Li X, Romero P, Rani M, Dunker AK, Obradovic Z. Predicting protein disorder for N-, C-, and internal regions. Genome Inform. Ser. Workshop Genome Inform. 1999;10:30–40. [PubMed] [Google Scholar]
- 45).Hu C-C, Fukae M, Uchida T, Qian Q, Zhang CH, Ryu OH, Tanabe T, Yamakoshi Y, Murakami C, Dohi N, Shimizu M, Simmer JP. Cloning and characterization of porcine enamelin mRNAs. J. Dent. Res. 1997;76:1720–1729. doi: 10.1177/00220345970760110201. [DOI] [PubMed] [Google Scholar]
- 46).Hu CC, Fukae M, Uchida T, Qian Q, Zhang CH, Ryu OH, Tanabe T, Yamakoshi Y, Murakami C, Dohi N, Shimizu M, Simmer JP. Sheathlin : cloning, cDNA/polypeptide sequences, and immunolocalization of porcine enamel sheath proteins. J. Dent. Res. 1997;76:648–657. doi: 10.1177/00220345970760020501. [DOI] [PubMed] [Google Scholar]
- 47).Cross KJ, Huq NL, Reynolds EC. Protein dynamics of bovine dentin phosphophoryn. J. Pept. Res. 2005;66:59–67. doi: 10.1111/j.1399-3011.2005.00273.x. [DOI] [PubMed] [Google Scholar]
- 48).Gu K, Chang S, Ritchie HH, Clarkson BH, Rutherford RB. Molecular cloning of a human dentin sialophosphoprotein gene. Eur. J. Oral Sci. 2000;108:35–42. doi: 10.1034/j.1600-0722.2000.00765.x. [DOI] [PubMed] [Google Scholar]
- 49).Ritchie HH, Wang LH, Knudtson K. A novel rat 523 amino acid phosphophoryn : nucleotide sequence and genomic organization. Biochim. Biophys. Acta. 2001;1520:212–222. doi: 10.1016/s0167-4781(01)00274-3. [DOI] [PubMed] [Google Scholar]