Abstract
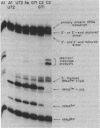
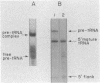
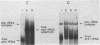
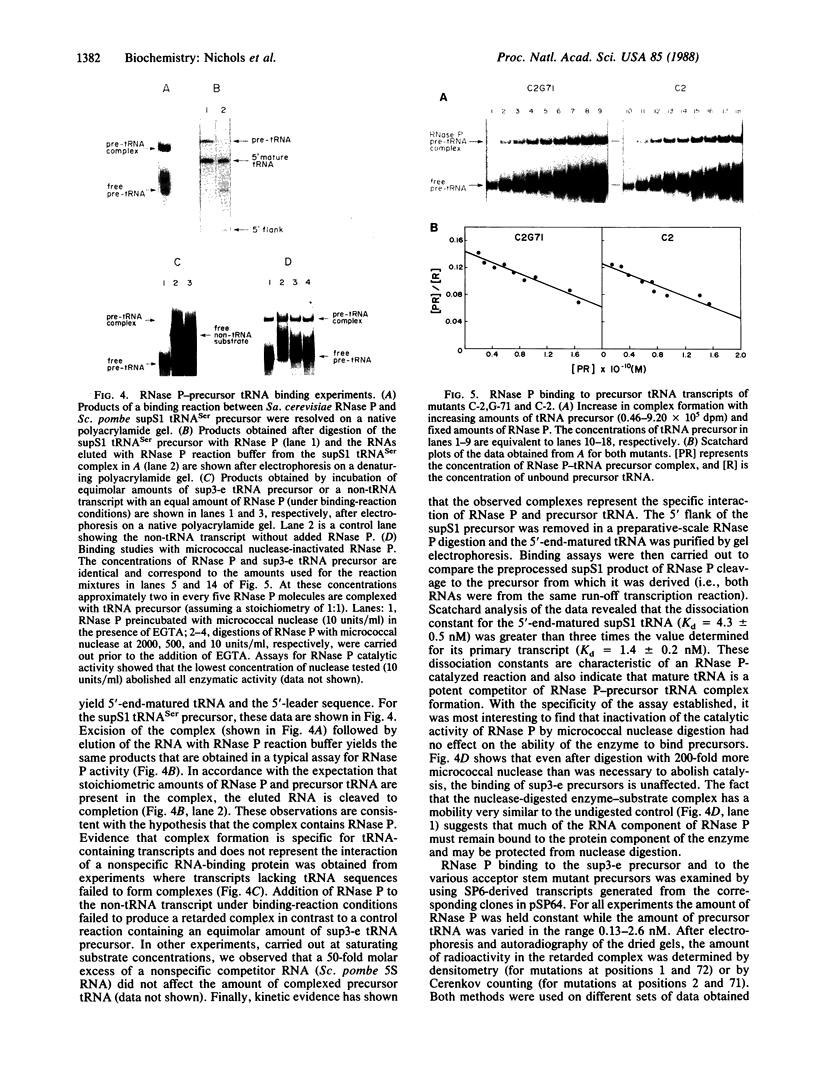
During tRNA biosynthesis the 5'-leader sequences in precursor tRNAs are removed by the ribonucleoprotein RNase P, an enzyme whose RNA moiety is required for activity. To clarify some aspects of the enzyme mechanism, we examined substrate binding and product formation with mutant precursor tRNAs. Mutations G-1----A or U-2----C in the Schizosaccharomyces pombe sup3-e tRNASer, which cause mispairing at or near the top of the acceptor stem, prevent the removal of the 5'-leader sequences by Saccharomyces cerevisiae RNase P. Equilibrium binding studies involving specific gel retardation of RNase P-precursor tRNA complexes showed that complexes with wild-type and A-1 and C-2 mutant precursor tRNAs had very similar dissociation constants (average Kd for sup3 = 1.5 +/- 0.2 nM). Thus, the 5'-terminal nucleotides of mature tRNA, on the 3' proximal side of the RNase P cleavage site, affect the enzyme's catalytic function but not substrate binding. The catalytic integrity of the RNA component of RNase P is not essential for binding of tRNA precursors, as demonstrated by gel retardation of micrococcal nuclease-inactivated enzyme. This suggests a possible role for the protein component of the enzyme in substrate binding. Upon restoration of base pairing to the acceptor stem in the A-1 or C-2 mutants, we found that, in addition to a requirement for pairing at these positions, conservation of the wild-type first and second nucleotides of the tRNA was necessary to obtain maximal cleavage by RNase P. This indicates a distinct sequence preference of this enzyme.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Castaño J. G., Ornberg R., Koster J. G., Tobian J. A., Zasloff M. Eukaryotic pre-tRNA 5' processing nuclease: copurification with a complex cylindrical particle. Cell. 1986 Aug 1;46(3):377–385. doi: 10.1016/0092-8674(86)90658-6. [DOI] [PubMed] [Google Scholar]
- Engelke D. R., Gegenheimer P., Abelson J. Nucleolytic processing of a tRNAArg-tRNAAsp dimeric precursor by a homologous component from Saccharomyces cerevisiae. J Biol Chem. 1985 Jan 25;260(2):1271–1279. [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber R. L., Altman S. In vitro processing of B. mori transfer RNA precursor molecules. Cell. 1979 Jun;17(2):389–397. doi: 10.1016/0092-8674(79)90165-x. [DOI] [PubMed] [Google Scholar]
- Genbauffe F. S., Chisholm G. E., Cooper T. G. Tau, sigma, and delta. A family of repeated elements in yeast. J Biol Chem. 1984 Aug 25;259(16):10518–10525. [PubMed] [Google Scholar]
- Gold H. A., Altman S. Reconstitution of RNAase P activity using inactive subunits from E. coli and HeLa cells. Cell. 1986 Jan 31;44(2):243–249. doi: 10.1016/0092-8674(86)90758-0. [DOI] [PubMed] [Google Scholar]
- Greer C. L., Peebles C. L., Gegenheimer P., Abelson J. Mechanism of action of a yeast RNA ligase in tRNA splicing. Cell. 1983 Feb;32(2):537–546. doi: 10.1016/0092-8674(83)90473-7. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983 Dec;35(3 Pt 2):849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- Hottinger H., Pearson D., Yamao F., Gamulin V., Cooley L., Cooper T., Söll D. Nonsense suppression in Schizosaccharomyces pombe: the S. pombe Sup3-e tRNASerUGA gene is active in S. cerevisiae. Mol Gen Genet. 1982;188(2):219–224. doi: 10.1007/BF00332678. [DOI] [PubMed] [Google Scholar]
- Krupp G., Cherayil B., Frendewey D., Nishikawa S., Söll D. Two RNA species co-purify with RNase P from the fission yeast Schizosaccharomyces pombe. EMBO J. 1986 Jul;5(7):1697–1703. doi: 10.1002/j.1460-2075.1986.tb04413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson D., Willis I., Hottinger H., Bell J., Kumar A., Leupold U., Söll D. Mutations preventing expression of sup3 tRNASer nonsense suppressors of Schizosaccharomyces pombe. Mol Cell Biol. 1985 Apr;5(4):808–815. doi: 10.1128/mcb.5.4.808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phizicky E. M., Schwartz R. C., Abelson J. Saccharomyces cerevisiae tRNA ligase. Purification of the protein and isolation of the structural gene. J Biol Chem. 1986 Feb 25;261(6):2978–2986. [PubMed] [Google Scholar]
- Reich C., Gardiner K. J., Olsen G. J., Pace B., Marsh T. L., Pace N. R. The RNA component of the Bacillus subtilis RNase P. Sequence, activity, and partial secondary structure. J Biol Chem. 1986 Jun 15;261(17):7888–7893. [PubMed] [Google Scholar]
- Smith J. D. Transcription and processing of transfer RNA precursors. Prog Nucleic Acid Res Mol Biol. 1976;16:25–73. doi: 10.1016/s0079-6603(08)60755-2. [DOI] [PubMed] [Google Scholar]
- Stewart T. S., Söll D., Sharp S. Point mutations in the 5' ICR and anticodon region of a Drosophila tRNAArg gene decrease in vitro transcription. Nucleic Acids Res. 1985 Jan 25;13(2):435–447. doi: 10.1093/nar/13.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis I., Frendewey D., Nichols M., Hottinger-Werlen A., Schaack J., Söll D. A single base change in the intron of a serine tRNA affects the rate of RNase P cleavage in vitro and suppressor activity in vivo in Saccharomyces cerevisiae. J Biol Chem. 1986 May 5;261(13):5878–5885. [PubMed] [Google Scholar]
- Willis I., Hottinger H., Pearson D., Chisholm V., Leupold U., Söll D. Mutations affecting excision of the intron from a eukaryotic dimeric tRNA precursor. EMBO J. 1984 Jul;3(7):1573–1580. doi: 10.1002/j.1460-2075.1984.tb02013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis I., Nichols M., Chisholm V., Söll D., Heyer W. D., Szankasi P., Amstutz H., Munz P., Kohli J. Functional complementation between mutations in a yeast suppressor tRNA gene reveals potential for evolution of tRNA sequences. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7860–7864. doi: 10.1073/pnas.83.20.7860. [DOI] [PMC free article] [PubMed] [Google Scholar]