Abstract
Background
MicroRNAs (miRNAs) are a novel class of gene regulators whose biogenesis involves hairpin structures called precursor miRNAs, or pre-miRNAs. A pre-miRNA is processed to make a miRNA:miRNA* duplex, which is then separated to generate a mature miRNA and a miRNA*. The mature miRNAs play key regulatory roles during embryonic development as well as other cellular processes. They are also implicated in control of viral infection as well as innate immunity. Direct experimental evidence for mosquito miRNAs has been recently reported in anopheline mosquitoes based on small-scale cloning efforts.
Results
We obtained approximately 130, 000 small RNA sequences from the yellow fever mosquito, Aedes aegypti, by 454 sequencing of samples that were isolated from mixed-age embryos and midguts from sugar-fed and blood-fed females, respectively. We also performed bioinformatics analysis on the Ae. aegypti genome assembly to identify evidence for additional miRNAs. The combination of these approaches uncovered 98 different pre-miRNAs in Ae. aegypti which could produce 86 distinct miRNAs. Thirteen miRNAs, including eight novel miRNAs identified in this study, are currently only found in mosquitoes. We also identified five potential revisions to previously annotated miRNAs at the miRNA termini, two cases of highly abundant miRNA* sequences, 14 miRNA clusters, and 17 cases where more than one pre-miRNA hairpin produces the same or highly similar mature miRNAs. A number of miRNAs showed higher levels in midgut from blood-fed female than that from sugar-fed female, which was confirmed by northern blots on two of these miRNAs. Northern blots also revealed several miRNAs that showed stage-specific expression. Detailed expression analysis of eight of the 13 mosquito-specific miRNAs in four divergent mosquito genera identified cases of clearly conserved expression patterns and obvious differences. Four of the 13 miRNAs are specific to certain lineage(s) within mosquitoes.
Conclusion
This study provides the first systematic analysis of miRNAs in Ae. aegypti and offers a substantially expanded list of miRNAs for all mosquitoes. New insights were gained on the evolution of conserved and lineage-specific miRNAs in mosquitoes. The expression profiles of a few miRNAs suggest stage-specific functions and functions related to embryonic development or blood feeding. A better understanding of the functions of these miRNAs will offer new insights in mosquito biology and may lead to novel approaches to combat mosquito-borne infectious diseases.
Background
MicroRNAs (miRNAs) are approximately 22 nucleotide long, non-coding RNAs that regulate the expression of cellular genes by binding to target mRNAs for cleavage or translational repression (reviewed in [1]). Thousands of miRNAs have been reported in animals and plants [2]. Many miRNAs are highly conserved across divergent species while others are specific to a particular evolutionary lineage (miRBase: http://microrna.sanger.ac.uk/). Lineage-specific miRNAs can arise from non-miRNA sequences, which has been observed in Drosophila, or by modifications of existing miRNAs [3,4]. MiRNA genes can occur in intergenic regions and within introns [5-8]. For some miRNAs, their biogenesis starts with transcription of miRNA genes, mostly by RNA polymerase II, which produces a primary miRNA (reviewed in [9]). A given primary miRNA can be either monocistronic, containing one mature miRNA, or polycistronic, containing multiple mature miRNAs. In Drosophila, the primary miRNA is processed by a Drosha-Pasha complex to yield small stem-loop structures that are approximately 70 nucleotides in length called pre-miRNAs [10]. Following export to the cytosol, the double-stranded pre-miRNA is recognized by Dicer-1, which complexes with Loqs and produces a ~22 bp duplex with 2 nt 3' overhangs [10]. The product is referred to as a miRNA:miRNA* duplex, which is further separated by a helicase and normally the miRNA* strand is rapidly degraded [5,9,11]. During "deep" sequencing studies, approximately 100 fold as many miRNAs as miRNA*s was observed for a given miRNA except in cases where both strands could generate functional miRNAs [7,12,13]. A new category of pre-miRNAs known as mirtrons are derived from introns and undergo Drosha-independent processing [14-16]. Some researchers suggest that the presence of mirtrons indicates that RNA sources may be less relevant for determining which RNAs become miRNAs than their structural characteristics [7]. Mature miRNAs pair with target mRNAs with the assistance of Argonaute to achieve translational repression and/or mRNA degradation (reviewed in [17]).
MicroRNAs play key regulatory roles during embryonic development, stem cell division, cancer development, neurogenesis, heart development, haematopoietic cell differentiation, and cell death (reviewed in [1,18]). They are also implicated in control of viral infection in vertebrates and in one recent report miRNAs were linked to malaria infection in mosquitoes [19-21]. Similarly, miRNAs are also linked to innate immunity (reviewed in [22]). There are reports describing Anopheles gambiae miRNAs on the basis of computational prediction or similarity to known miRNAs from other species [23-26]. Direct experimental evidence for mosquito miRNAs has been recently reported only in anopheline mosquitoes using small-scale sequencing [21,27]. There is also a study on genes involved in small RNA pathways in mosquitoes [28].
Using high throughput sequencing and bioinformatics approaches, we performed the first systematic analysis of miRNAs in Ae. aegypti. In the current study, we substantially expanded the list of miRNAs for all mosquitoes and uncovered a number of novel and mosquito-specific miRNAs. Northern and small RNA sequencing revealed several miRNAs that may play important roles during embryonic development and during blood feeding. Our analysis also offered insights into the evolution of conserved and lineage-specific miRNAs in mosquitoes.
Results and discussion
Discovery of 98 distinct pre-miRNA sequences in Ae. aegypti
As shown in Table 1, we have uncovered 98 different pre-miRNAs in Ae. aegypti which could produce 86 distinct miRNAs. Some of the 98 pre-miRNA sequences produce identical miRNAs and miRNA*s. Also included in Table 1 are 20 distinct miRNA* sequences that were uncovered by small RNA sequencing. Eighty-nine of the Ae. aegypti miRNA and miRNA* sequences showed a perfect match to small RNA sequences from at least one of the three samples (Table 1). There are clear variations of sequence counts among different miRNA species in these samples. However, to gain quantitative insights in the relative abundance of these miRNAs, further investigations are needed using methods such as northern blot, primer extension, and direct sequencing of millions of small RNA reads [11,12,21]. Twenty-nine of the 98 pre-miRNAs do not have small RNA sequences in the embryo and midgut samples. However, these 29 pre-miRNAs all form hairpins and are conserved among Ae. aegypti, Cx. quinquefasciatus, and An. gambiae, the three mosquito species with sequenced genomes.
Table 1.
Sequence, location, and expression of miRNAs in Aedes aegypti.
| Name1, 2 | mirBase Name | Sequence3 | Contig4 | Start4 | End4 | Strand | Embryo5 | Gut_SF5 | Gut_BF5 |
|---|---|---|---|---|---|---|---|---|---|
| aae-miR-M1-1 | aae-miR-2943-1 | TTAAGTAGGCACTTGCAGGCAAA | CONTIG_15246 | 25749 | 25829 | + | 28 | 0 | 0 |
| aae-miR-M1-2 | aae-miR-2943-2 | TTAAGTAGGCACTTGCAGGCAAA | CONTIG_15246 | 25911 | 25981 | + | 28 | 0 | 0 |
| aae-miR-M2 | aae-miR-2942 | TATTCGAGACTTCACGAGTTAAT | CONTIG_11944 | 151669 | 151750 | + | 0 | 0 | 7 |
| aae-miR-M3 | aae-miR-2945 | TGACTAGAGGCAGACTCGTTTA | CONTIG_2929 | 123975 | 124056 | + | 33 | 0 | 6 |
| aae-miR-M3* | aae-miR-2945* | AGCGGGTCCGTTTCTAGTGTCATG | CONTIG_2929 | 123975 | 124056 | + | 3 | 0 | 0 |
| aae-miR-M4a | aae-miR-2944a | GAAGGAACTTCTGCTGTGATCTGA | CONTIG_18252 | 93868 | 93929 | + | 91 | 2 | 0 |
| aae-miR-M4a* | aae-miR-2944a* | TATCACAGTAGTTGTACTTTAA | CONTIG_18252 | 93868 | 93929 | + | 3 | 0 | 0 |
| aae-miR-M4b | aae-miR-2944b | GAAGGAACTCCCGGTGTGATATA | CONTIG_18252 | 93731 | 93792 | + | 62 | 0 | 0 |
| aae-miR-M4b* | aae-miR-2944b* | TATCACAGCAGTAGTTACCTGA | CONTIG_18252 | 93731 | 93792 | + | 24 | 0 | 0 |
| aae-miR-N1-1 | aae-miR-2941-2 | TAGTACGGCTAGAACTCCACGGA | CONTIG_16241 | 318005 | 318105 | - | 288 | 1 | 1 |
| aae-miR-N1-2 | aae-miR-2941-1 | TAGTACGGCTAGAACTCCACGGA | CONTIG_16241 | 317700 | 317799 | - | 288 | 1 | 1 |
| aae-miR-N2 | aae-miR-2946 | TAGTACGGAAAAGATATGGGGA | CONTIG_16241 | 318135 | 318222 | - | 5 | 0 | 0 |
| aae-miR-1174 | aae-miR-1174 | TCAGATCTACTTAATACCCAT | CONTIG_7232 | 17531 | 17665 | + | |||
| aae-miR-1175 | aae-miR-1175 | TGAGATTCTACTTCTCCGACTTAA | CONTIG_7232 | 17737 | 17816 | + | 0 | 0 | 1 |
| aae-miR-1175* | aae-miR-1175* | TAAGTGGAGTAGTGGTCTCATCGCT | CONTIG_7232 | 17737 | 17816 | + | 0 | 2 | 59 |
| aae-miR-1890 | aae-miR-1890 | TGAAATCTTTGATTAGGTCTGG | CONTIG_10435 | 60786 | 60931 | + | 1 | 0 | 0 |
| aae-miR-1891-1 | aae-miR-1891-1 | TGAGGAGTTAATTTGCGTGTTT | CONTIG_10244 | 3984 | 4069 | - | 2 | 0 | 0 |
| aae-miR-1891-2 | aae-miR-1891-2 | TGAGGAGTTAATTTGCGTGTTT | CONTIG_18287 | 4656 | 4741 | + | 2 | 0 | 0 |
| aae-miR-1889 | aae-miR-1889 | CACGTTACAGATTGGGGTTTCC | CONTIG_4323 | 23786 | 23921 | - | 0 | 0 | 3 |
| aae-bantam | aae-bantam | TGAGATCATTTTGAAAGCTGATT | CONTIG_3301 | 208288 | 208426 | - | 1 | 2 | 2 |
| aae-let-7 | aae-let-7 | TGAGGTAGTTGGTTGTATAGT | CONTIG_2929 | 321614 | 321684 | + | 0 | 0 | 5 |
| aae-miR-1 | aae-miR-1 | TGGAATGTAAAGAAGTATGGAG | CONTIG_25061 | 66256 | 66338 | + | 1 | 0 | 1 |
| aae-miR-10 | aae-miR-10 | ACCCTGTAGATCCGAATTTGTT | CONTIG_17639 | 8749 | 8833 | + | 1 | 0 | 0 |
| aae-miR-100 | aae-miR-100 | AACCCGTAGATCCGAACTTGTG | CONTIG_2929 | 307441 | 307568 | + | |||
| aae-miR-1000-1 | aae-miR-1000-1 | ATATTGTCCTGTCACAGCAGT | CONTIG_35246 | 172 | 265 | - | |||
| aae-miR-1000-2 | aae-miR-1000-2 | ATATTGTCCTGTCACAGCAGT | CONTIG_9774 | 12985 | 13078 | + | |||
| aae-miR-11 | aae-miR-11 | CATCACAGTCTGAGTTCTTGCTT | CONTIG_23911 | 46399 | 46488 | + | 1861 | 42 | 436 |
| aae-miR-11* | aae-miR-11* | CGAGAACTCCGGCTGTGACCTGTG | CONTIG_23911 | 46399 | 46488 | + | 4 | 0 | 2 |
| aae-miR-12 | aae-miR-12 | TGAGTATTACATCAGGTACTGGT | CONTIG_4323 | 23386 | 23503 | - | |||
| aae-miR-124 | aae-miR-124 | TAAGGCACGCGGTGAATGCCAAG | CONTIG_531 | 21343 | 21423 | - | 3 | 0 | 0 |
| aae-miR-125 | aae-miR-125 | TCCCTGAGACCCTAACTTGTGAC | CONTIG_2929 | 321885 | 321975 | + | 0 | 0 | 3 |
| aae-miR-133 | aae-miR-133 | TTGGTCCCCTTCAACCAGCTGT | CONTIG_24522 | 50243 | 50347 | - | |||
| aae-miR-137-1 | aae-miR-137-1 | TTATTGCTTGAGAATACACGTA | CONTIG_7773 | 15414 | 15486 | - | |||
| aae-miR-137-2 | aae-miR-137-2 | TTATTGCTTGAGAATACACGTA | CONTIG_29705 | 92457 | 92529 | + | |||
| aae-miR-13b | aae-miR-13 | TATCACAGCCATTTTGACGAGTT | CONTIG_12793 | 17619 | 17710 | - | 168 | 2 | 42 |
| aae-miR-14 | aae-miR-14 | TCAGTCTTTTTCTCTCTCCTAT | CONTIG_12112 | 693 | 785 | - | 1 | 1 | 6 |
| aae-miR-184 | aae-miR-184 | TGGACGGAGAACTGATAAGGGC | CONTIG_19030 | 3452 | 3535 | - | 957 | 76 | 1307 |
| aae-miR-190 | aae-miR-190 | AGATATGTTTGATATTCTTGGTTG | CONTIG_10108 | 2092 | 2180 | - | 83 | 2 | 43 |
| aae-miR-193 | aae-miR-193 | TACTGGCCTACTAAGTCCCAAC | CONTIG_19568 | 51035 | 51158 | + | |||
| aae-miR-2a | aae-miR-2a | TATCACAGCCAGCTTTGATGAGCT | CONTIG_12793 | 16130 | 16216 | - | 1217 | 80 | 249 |
| aae-miR-210 | aae-miR-210 | TTGTGCGTGTGACAACGGCTAT | CONTIG_19443 | 85221 | 85292 | - | 3 | 12 | 12 |
| aae-miR-219 | aae-miR-219 | TGATTGTCCAAACGCAATTCTTG | CONTIG_15776 | 56993 | 57078 | + | |||
| aae-miR-2b | aae-miR-2b | TATCACAGCCAGCTTTGAAGAGCG | CONTIG_12793 | 17471 | 17562 | - | 758 | 74 | 147 |
| aae-miR-2c | aae-miR-2c | TATCACAGCCAGCTTTGATGAGC | CONTIG_12793 | 17971 | 18048 | - | 1218 | 80 | 249 |
| aae-miR-252 | aae-miR-252 | CTAAGTACTAGTGCCGCAGGAGA | CONTIG_3685 | 79002 | 79177 | - | 38 | 0 | 1 |
| aae-miR-252* | aae-miR-252* | CCTGCTGCCCAAGTGCTTATCGAA | CONTIG_3685 | 79002 | 79177 | - | 5 | 0 | 0 |
| aae-miR-263 | aae-miR-263a | AATGGCACTGGAAGAATTCACGGG | CONTIG_27430 | 4746 | 4821 | - | 53 | 1 | 1 |
| aae-miR-263* | aae-miR-263a* | CGTGTTCTGGCAGTGGCATCCC | CONTIG_27430 | 4746 | 4821 | - | 6 | 0 | 0 |
| aae-miR-263b | aae-miR-263b | CTTGGCACTGGGAGAATTCACAG | CONTIG_1908 | 33267 | 33357 | + | 6 | 0 | 0 |
| aae-miR-263b* | aae-miR-263b* | TGGATCTTTTCGTGCCATCGT | CONTIG_1908 | 33267 | 33357 | + | 1 | 0 | 0 |
| aae-miR-275 | aae-miR-275 | TCAGGTACCTGAAGTAGCGCGCG | CONTIG_1651 | 96625 | 96720 | + | 0 | 0 | 5 |
| aae-miR-275* | aae-miR-275* | CGCGCTAAGCAGGAACCGAGACT | CONTIG_1651 | 96625 | 96720 | + | 1 | 0 | 6 |
| aae-miR-276-1 | aae-miR-276-1 | TAGGAACTTCATACCGTGCTCT | CONTIG_417 | 10988 | 11177 | - | |||
| aae-miR-276-2 | aae-miR-276-2 | TAGGAACTTCATACCGTGCTCT | CONTIG_7617 | 38953 | 39087 | + | |||
| aae-miR-277 | aae-miR-277 | TAAATGCACTATCTGGTACGACA | CONTIG_12646 | 3556 | 3644 | + | |||
| aae-miR-278 | aae-miR-278 | TCGGTGGGACTTTCGTCCGTTT | CONTIG_1172 | 33037 | 33128 | + | 6 | 0 | 2 |
| aae-miR-279 | aae-miR-279 | TGACTAGATCCACACTCATTAA | CONTIG_17556 | 116934 | 117006 | - | 26 | 0 | 2 |
| aae-miR-281 | aae-miR-281 | CTGTCATGGAATTGCTCTCTTTA | CONTIG_27100 | 34036 | 34204 | + | 1 | 16 | 50 |
| aae-miR-281* | aae-miR-281* | AAAGAGAGCTATCCGTCGACAGTA | CONTIG_27100 | 34036 | 34204 | + | 164 | 5686 | 4806 |
| aae-miR-282-1 | aae-miR-282-1 | AATCTAGCCTCTCCTAGGCTTTGTCTG | CONTIG_15805 | 19414 | 19548 | + | |||
| aae-miR-282-1* | aae-miR-282-1* | ACATAGCCTGACAGAGGTTAGG | CONTIG_15805 | 19414 | 19548 | + | 2 | 0 | 0 |
| aae-miR-282-2 | aae-miR-282-2 | AATCTAGCCTCTCCTAGGCTTTGTCTG | CONTIG_15982 | 5479 | 5613 | - | |||
| aae-miR-282-2* | aae-miR-282-2* | ACATAGCCTGACAGAGGTTAGG | CONTIG_15982 | 5479 | 5613 | - | 2 | 0 | 0 |
| aae-miR-283 | aae-miR-283 | CAATATCAGCTGGTAATTCTGGG | CONTIG_4323 | 32334 | 32426 | - | 6 | 1 | 131 |
| aae-miR-285 | aae-miR-285 | TAGCACCATTCGAAATCAGT | CONTIG_1634 | 212427 | 212492 | - | 0 | 1 | 0 |
| aae-miR-286a-1 | aae-miR-286b-1 | TGACTAGACCGAACACTCGTATCC | CONTIG_21241 | 18407 | 18503 | + | 8 | 0 | 0 |
| aae-miR-286a-2 | aae-miR-286b-2 | TGACTAGACCGAACACTCGTATCC | CONTIG_8291 | 17656 | 17752 | + | 8 | 0 | 0 |
| aae-miR-286b | aae-miR-286a | TGACTAGACCGAACACTCGCGTCCT | CONTIG_18252 | 93413 | 93509 | + | 10 | 0 | 0 |
| aae-miR-305 | aae-miR-305 | ATTGTACTTCATCAGGTGCTCTGG | CONTIG_1651 | 105677 | 105766 | + | 0 | 0 | 2 |
| aae-miR-305* | aae-miR-305* | CGGCACATGTTGGAGTACACTTAA | CONTIG_1651 | 105677 | 105766 | + | 0 | 0 | 7 |
| aae-miR-306 | aae-miR-306 | TCAGGTACTGAGTGACTCTCAG | CONTIG_24640 | 55004 | 55128 | + | 119 | 0 | 27 |
| aae-miR-307 | aae-miR-307 | TCACAACCTCCTTGAGTGAGCGA | CONTIG_1157 | 147943 | 148043 | - | 1 | 0 | 0 |
| aae-miR-308 | aae-miR-308 | AATCACAGGAGTATACTGTGAG | CONTIG_6324 | 6462 | 6528 | + | |||
| aae-miR-308* | aae-miR-308* | CGCGGTATATTCTTGTGGCTTGA | CONTIG_6324 | 6462 | 6528 | + | 2 | 0 | 0 |
| aae-miR-31 | aae-miR-31 | TGGCAAGATGTTGGCATAGCTGAAA | CONTIG_21960 | 98153 | 98301 | - | 1 | 0 | 3 |
| aae-miR-315 | aae-miR-315 | TTTTGATTGTTGCTCAGAAAGCC | CONTIG_21501 | 12498 | 12562 | + | 27 | 1 | 0 |
| aae-miR-315* | aae-miR-315* | CTTTCGAGCAGTAATCAAAGTc | CONTIG_21501 | 12498 | 12562 | + | 5 | 0 | 0 |
| aae-miR-316 | aae-miR-316 | TGTCTTTTTCCGCTTACTGCCG | CONTIG_13460 | 10738 | 10828 | - | 0 | 0 | 1 |
| aae-miR-317-1 | aae-miR-317-1 | TGAACACAGCTGGTGGTATCTCAGT | CONTIG_12640 | 88437 | 88524 | + | 36 | 38 | 737 |
| aae-miR-317-2 | aae-miR-317-2 | TGAACACAGCTGGTGGTATCTCAGT | CONTIG_8451 | 9550 | 9637 | - | 36 | 38 | 737 |
| aae-miR-3a-1 | aae-miR-309a-1 | TCACTGGGCAAAGTTTGTCGCA | CONTIG_21241 | 18967 | 19043 | + | 6 | 0 | 0 |
| aae-miR-3a-2 | aae-miR-309a-2 | TCACTGGGCAAAGTTTGTCGCA | CONTIG_8291 | 18216 | 18292 | + | 6 | 0 | 0 |
| aae-miR-3b | aaw-miR-309b | TCACTGGGCATAGTTTGTCGCA | CONTIG_18252 | 94047 | 94117 | + | 3 | 0 | 0 |
| aae-miR-3b* | aae-miR-309b* | CGTCAAACTCCGTTCAGTTGGTG | CONTIG_18252 | 94047 | 94117 | + | 1 | 0 | 0 |
| aae-miR-33 | aae-miR-33 | GTGCATTGTAGTTGCATTGCA | CONTIG_18815 | 108787 | 108866 | + | |||
| aae-miR-34 | aae-miR-34 | TGGCAGTGTGGTTAGCTGGTTGTG | CONTIG_12646 | 4278 | 4400 | + | 44 | 55 | 639 |
| aae-miR-375 | aae-miR-375 | TTTGTTCGTTTGGCTCGAGTTA | CONTIG_14081 | 238834 | 238944 | - | 1 | 0 | 0 |
| aae-miR-7 | aae-miR-7 | TGGAAGACTAGTGATTTTGTTGTT | CONTIG_31115 | 46028 | 46112 | + | 14 | 0 | 0 |
| aae-miR-71 | aae-miR-71 | AGAAAGACATGGGTAGTGAGATA | CONTIG_12793 | 18215 | 18396 | - | 39 | 2 | 7 |
| aae-miR-71* | aae-miR-71* | TCTCACTACCTTGTCTTTCATG | CONTIG_12793 | 18215 | 18396 | - | 6 | 0 | 0 |
| aae-miR-79 | aae-miR-79 | ATAAAGCTAGATTACCAAAGCAT | CONTIG_24640 | 55215 | 55284 | + | 2 | 0 | 0 |
| aae-miR-8 | aae-miR-8 | TAATACTGTCAGGTAAAGATGTC | CONTIG_16942 | 34594 | 34660 | + | 32 | 1 | 38 |
| aae-miR-8* | aae-miR-8* | CATCTTACCGGGCAGCATTAGA | CONTIG_16942 | 34594 | 34660 | + | 7 | 2 | 4 |
| aae-miR-87 | aae-miR-87 | GGTGAGCAAATTTTCAGGTGT | CONTIG_15587 | 30157 | 30251 | + | |||
| aae-miR-927 | aae-miR-927 | TTTAGAATTCCTACGCTTTACC | CONTIG_1795 | 170460 | 170534 | + | |||
| aae-miR-929-1 | aae-miR-929-1 | ATTGACTCTAGTAGGGAGTCC | CONTIG_12461 | 76647 | 76734 | - | |||
| aae-miR-929-2 | aae-miR-929-2 | ATTGACTCTAGTAGGGAGTCC | CONTIG_34315 | 3294 | 3381 | + | |||
| aae-miR-92a | aae-miR-92a | TATTGCACTTGTCCCGGCCTAT | CONTIG_6821 | 67755 | 67832 | + | 74 | 0 | 5 |
| aae-miR-92a* | aae-miR-92a* | CGGTACGGACAGGGGCAACATT | CONTIG_6821 | 67755 | 67832 | + | 6 | 0 | 0 |
| aae-miR-92b | aae-miR-92b | AATTGCACTTGTCCCGGCCTGC | CONTIG_6824 | 4049 | 4131 | + | 74 | 0 | 5 |
| aae-miR-92b* | aae-miR-92b* | AGGTCGTGACTTGTGCCCGTTTG | CONTIG_6824 | 4049 | 4131 | + | 12 | 0 | 0 |
| aae-miR-932 | aae-miR-932 | TCAATTCCGTAGTGCATTGCAG | CONTIG_28378 | 72232 | 72319 | - | |||
| aae-miR-957 | aae-miR-957 | TGAAACCGTCCAAAACTGAGGC | CONTIG_591 | 147619 | 147787 | + | |||
| aae-miR-965 | aae-miR-965 | TAAGCGTATAGCTTTTCCC | CONTIG_3410 | 87378 | 87524 | + | |||
| aae-miR-970 | aae-miR-970 | TCATAAGACACACGCGGCTAT | CONTIG_11324 | 29252 | 29338 | + | 1 | 0 | 3 |
| aae-miR-981 | aae-miR-981 | TTCGTTGTCGACGAAACCTGCA | CONTIG_7302 | 184768 | 184853 | - | |||
| aae-miR-988 | aae-miR-988 | CCCCTTGTTGCAAACCTCACGC | CONTIG_17684 | 77966 | 78086 | - | 3 | 0 | 1 |
| aae-miR-988* | aae-miR-988* | GTGTGCTTTGTGACAATGAGA | CONTIG_17684 | 77966 | 78086 | - | 2 | 0 | 2 |
| aae-miR-989 | aae-miR-989 | TGTGATGTGACGTAGTGGTAC | CONTIG_6774 | 51048 | 51137 | - | 2 | 33 | 3 |
| aae-miR-993 | aae-miR-993 | GAAGCTCGTTTCTATAGAGGTATCT | CONTIG_28305 | 82579 | 82660 | + | |||
| aae-miR-996 | aae-miR-996 | TGACTAGATTACATGCTCGTCT | CONTIG_17556 | 112174 | 112267 | - | |||
| aae-miR-998 | aae-miR-998 | TAGCACCATGAGATTCAGCTC | CONTIG_23911 | 46676 | 46764 | + | 164 | 6 | 42 |
| aae-miR-999 | aae-miR-999 | TGTTAACTGTAAGACTGTGTCT | CONTIG_6027 | 163128 | 163269 | + | |||
| aae-miR-9a-1 | aae-miR-9a-1 | TCTTTGGTTATCTAGCTGTATGA | CONTIG_19539 | 116871 | 116951 | + | 16 | 1 | 2 |
| aae-miR-9a-2 | aae-miR-9a-2 | TCTTTGGTTATCTAGCTGTATGA | CONTIG_19541 | 4061 | 4141 | + | 16 | 1 | 2 |
| aae-miR-9b | aae-miR-9b | TCTTTGGTGATTTTAGCTGTATGC | CONTIG_24640 | 55513 | 55604 | + | 134 | 0 | 69 |
| aae-miR-9c | aae-miR-9c | TCTAAAGCTTTAGTACCAGAGGTC | CONTIG_24640 | 28160 | 28257 | + | 5 | 0 | 0 |
| aae-miR-iab-4-1 | aae-miR-iab-4-1 | CGGTATACCTTCAGTATACGTAAC | CONTIG_17255 | 44653 | 44726 | - | |||
| aae-miR-iab-4-2 | aae-miR-iab-4-2 | CGGTATACCTTCAGTATACGTAAC | CONTIG_23219 | 23581 | 23654 | + |
Notes:
1. The first block (miR-M1-1 through miR-N2) are novel miRNAs that are discovered in this study. We have not detected homology to any known miRNAs or genomic sequences outside of mosquito species. The second block (miR-1174 through miR-1889) contains miRNAs that are homologous to previously reported "mosquito-specific" miRNAs. The third block, comprised of the remaining miRNAs, contains miRNAs that have homologues outside of mosquitoes.
2. Naming in this column is temporary. Mirbase assigned names are shown in column 2, which was received after the acceptance of the manuscript. "-1", and "-2" suffixes refer to different hairpins that produce the same mature miRNA. "a", and "b" suffixes refer to different hairpins that produce similar but not identical miRNAs.
3. Underlined sequences are cases where there are extra bases at the 5' or 3' ends and they are the majority in at least six small RNA sequences. Italicized sequences are cases where such sequences are detected but they are either not the majority or there are less than six 454 sequence hits. As poly-As are added to the small RNAs, adenine(s) at the 3' end cannot be confirmed by this sequencing approach.
4. The contig, start, and end positions refer to the locations of the pre-miRNA hairpins.
5. The last three columns are the number of sequence hits in small RNA libraries obtained by 454 sequencing. The total small RNA reads are 55, 000, 33, 000, and 42, 000 in embryos, sugar-fed midguts (Gut_SF), and blood-fed midguts (Gut_BF), respectively. The total number of hits for all Ae. aegypti miRNAs are 8369, 6260, and 9922 in embryos, sugar-fed midguts (Gut_SF), and blood-fed midguts (Gut_BF), respectively. There are a few cases where we did not distinguish the hits from nearly identical miRNAs.
Possible revisions at the ends of known miRNAs and cases of abundant miRNA* sequences
As shown in Table 1, there are a few cases where the Ae. aegypti miRNA sequences, as indicated by direct sequencing, start or end with one or a few extra nucleotides compared to the known miRNAs reported from D. melanogaster or anopheline mosquitoes (miRBase). To minimize the contribution of sequencing error, we only consider cases where there are at least six such sequences in the 454 database and these differences are the majority. These miRNAs include aae-miR-2a, aae-miR-210, aae-miR-263b, aae-miR-281, and aae-miR-283. Because internal sequence variations between miRNAs from different species could simply result from species differences, we did not include in the above list the aae-miRNAs that only had internal sequence variations compared to known miRNAs. On the other hand, shifts at the 5' or 3' ends could either suggest a difference between species or imprecise annotation at the miRNA termini. Thus the above 5 aae-miRNAs provide leads to further studies to investigate whether these previously reported miRNA sequences need to be revised.
In vast majority of the cases, mature miRNAs are much more abundant than miRNA*. However, miR-281* and miR-1175* are at least a few dozen fold more abundant than their miRNA sequences. In both cases, the miRNA and the miRNA* sequences are 100% identical among Ae. aegypti, Cx. quinquiefasciatus, and An. gambiae. It is therefore possible that miR-281* and miR-1175* are functional. There are a few other cases in which the miRNA* is more abundant than the miRNA sequences (Table 1). However, the numbers of hits are low in these cases and it is difficult to assess how significant the differences may be.
miRNA gene clusters and duplications: evolutionary implications
There are 14 clusters of miRNAs that are defined as more than one miRNA hairpin within 10 kb [29]. Twelve of these clusters have members that are separated by less than one kb. All these clusters can be identified in Table 1 and Additional file 1 by tracking and sorting the contigs and start and end positions of the pre-miRNAs. Two previously identified clusters are worth noting. The first is the cluster that includes miR-9b, miR-79, and miR-306. We previously identified this cluster in both An. gambiae and D. melanogaster and we thought miR-306 was missing in the Ae. aegypti cluster [27]. However, small RNA sequencing and closer analysis of the Ae. aegypti assembly suggest that miR-306 is indeed present in Ae. aegypti and the relative positions of the three miRNAs in the cluster are conserved among all three species. The aae-miR-306 shows 2 mismatches in the 22 bp overlap compared to aga-miR-306 and dme-miR-306.
The second cluster includes miR-12 and miR-283, which flank either miR-304 in D. melanogaster or miR-1889 in An. gambiae [27]. MiR-1889 has similarity to the reverse-complementary sequence of miR-304 but the difference is significant enough for miRbase to assign a unique name for it. Given that the D. melanogaster miR-304 and the An. gambiae miR-1889 are flanked by the same miRNAs and that this miRNA cluster is found in an intron of orthologous genes in the two species, miR-304 and miR-1889 may have a common origin. Through small RNA sequencing and a closer analysis of the genome assembly, we identified miR-1889 in Ae. aegypti (Table 1 and Additional file 1), which was previously thought to be missing in the Ae. aegypti cluster [27]. The newly identified aae-miR-1889 shows 3 mismatches in the 21 bp overlap with aga-miR-1889. The identification of aae-miR-1889 further supports the strand orientation of the mosquito miR-1889. It is tempting to suggest that one of the two miRNA hairpins, miR-304 or miR-1889, was inverted during evolution. It is also possible that fruit flies and mosquitoes utilize different strands of the hairpin as mature miRNA. Wecurrently do not have evidence to support either of these hypotheses.
There are 17 cases where one of the pre-miRNAs is duplicated and thus more than one pre-miRNA hairpin produces the same or highly similar mature miRNAs. These pre-miRNAs are shown either with a suffix of "-1" and "-2" for hairpins that produce identical miRNAs or with a suffix of "a" or "b" for hairpins that produce highly similar miRNAs (Table 1). These miRNAs are a rich source for future comparative analysis to uncover the evolutionary patterns of miRNA duplication and the process of creating novel miRNAs in mosquitoes. It remains to be determined whether the rather common miRNA duplication observed in Ae. aegypti reflect the importance of duplication for the generation of new miRNAs in mosquitoes. In this regard, it is interesting to note that while duplication is a common mechanism to generate new miRNAs in plants (e.g., [30]), duplication was thought to be not important in Drosophila [3].
Novel miRNAs that are potentially specific to mosquitoes
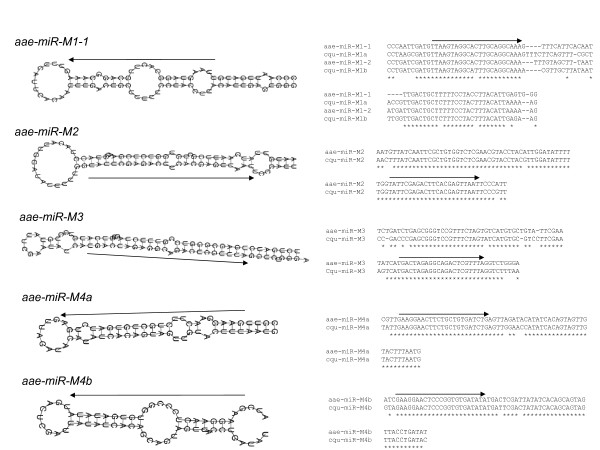
Nine of the 98 pre-miRNA hairpins are novel and currently have only been found in mosquitoes. These nine pre-miRNAs produce seven distinct mature miRNAs (miR-M1, -M2, -M3, -M4a and -M4b; miR-N1 and miR-N2). All seven mature miRNAs have multiple hits from small RNA sequencing, confirming their status as miRNAs. A few of these also have hits in the miRNA* strand. Shown in Figures 1 and 2 are the sequence alignments of the novel pre-miRNA sequences discovered in this study and the hairpins they form. Two physically linked pre-miRNA hairpins (miR-M1-1 and miR-M1-2) produce the same miR-M1 in Ae. aegypti. Two physically linked pre-miRNA hairpins produce similar but not identical miR-M4a and miR-M4b. MiR-M1, -M2, -M3, -M4a and -M4b are found in all three available mosquito genome assemblies.
Figure 1.
Alignments and stem-loop structures of five novel mosquito pre-miRNAs. See Table 1 for naming and sequence locations of these miRNAs. Left panels are the hairpin structures. Right panels are the sequence alignments between Ae. aegypti miRNAs (aae-miRNAs) and Cx. quinquefasciatus miRNAs (cqu-miRNAs). Arrows point to the mature miRNA sequences from 5' to 3'. In the case of miR-M1, there are two physically linked copies in both Ae. aegypti and Cx. quinquefasciatus, as shown in the alignment and in Table 1. The two copies in Ae. aegypti produce the same mature miRNAs and the hairpins are named -1 and -2. The two copies in Cx. quinquefasciatus are named -1a and -1b because their mature miRNAs differ by one nucleotide. Only the hairpin structure for aae-miR-M1-1 is shown. All five miRNAs shown in panel A have homologs in An. gambiae.
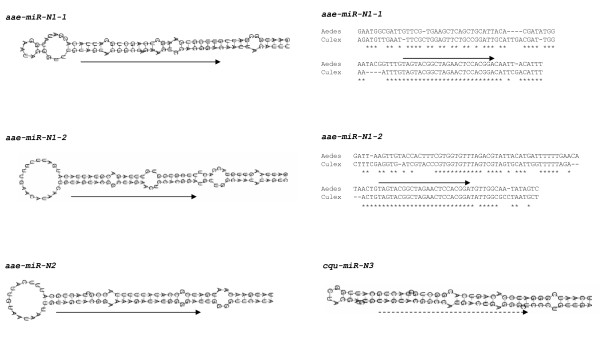
Figure 2.
Alignments and stem-loop structures of a novel miRNA cluster within the intron of a gene encoding a transcription factor. See Table 1 for naming and sequence locations of these miRNAs. There are two hairpins for the same miR-N1 (-1 and -2) in both Ae. aegypti and Cx. quinquefasciatus. Only the Ae. aegypti hairpin structures for the miR-N1 pre-miRNAs are shown. Aae-miR-N2 and cqu-miR-N3 are only found in Ae. aegypti and Cx. quinquefasciatus, respectively. Thus there are no alignments for these two miRNAs. Arrows point to the mature miRNA sequences from 5' to 3'. Dashed arrow for cqu-miR-N3 reflects the fact that we do not yet have the direct sequence for this miRNA. The mature cqu-miR-N3 sequence was predicted according to the conserved seed sequence shared with miR-N1 and miR-N2, which was confirmed by northern blots using anti cqu-miR-N3 as a probe (see Figure 7).
Two physically linked pre-miRNA hairpins (miR-N1-1 and miR-N1-2) produce the same miR-N1 and they are also in close proximity to the miR-N2 hairpin in Ae. aegypti. The three hairpins are in the first intron of a gene in Ae. aegypti (Vectorbase Gene ID AAEL009263) encoding a putative transcription factor with a basic leucine zipper domain. Sequence analysis suggests that miR-N1 is found in the orthologous gene in Cx. quinquefasciatus but not found in An. gambiae. MiR-N2 is only found in Ae. aegypti. MiR-N1 also exists in two hairpins in the intron of the homologous gene in Cx. quinquefasciatusand there is a third hairpin that has a predicted miRNA with a similar 5' sequence as miR-N1. We name this miRNA miR-N3 and it is only found in Cx. quinquefasciatus (Vectorbase Gene ID CPIJ000468). Cqu-miR-N3 is not listed in Table 1, which only shows miRNAs from Ae. aegypti.
Thus, we have identified eight novel mosquito-specific miRNAs in this study. We define "mosquito-specific" miRNAs here as those that are currently only found in mosquitoes. BLAST searches using low stringent parameters (word size at seven, e-value cut-off at 10) failed to identify any reliable homologues from miRBase or non-redundant GenBank sequences. We also performed oligomap comparisons [31] of the "mosquito-specific" miRNAs to all miRBase sequences using default parameters and did not identify any match in any other organism. Oligomap [31] is designed for comparisons of short sequences such as miRNAs, allowing gaps and mismatches. Taken together, the evidence indicates that what we are reporting in this study are novel miRNAs. This study increased the number of novel miRNAs that are only found in mosquitoes from five [21,27] to 13. It is important to emphasize that some of these so-called "mosquito-specific" miRNAs may be discovered outside mosquitoes as future efforts of genome and small RNA sequencing expand to more and more organisms.
Expression patterns of conserved miRNAs
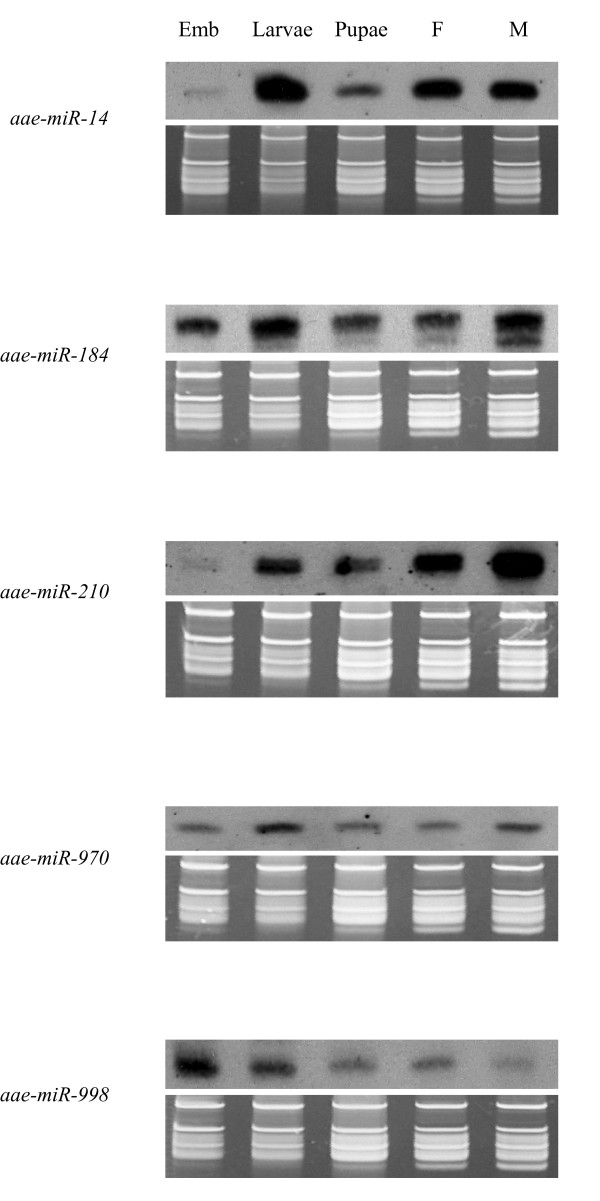
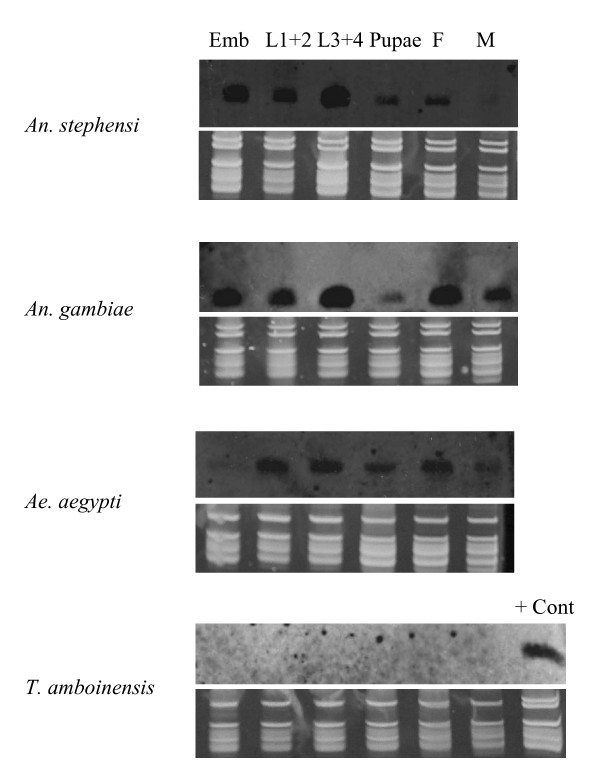
We chose nine conserved miRNAs and eight mosquito-specific miRNAs for further analysis using northern blot to confirm the small RNA sequencing results and to determine the expression patterns of these miRNAs in different developmental stages. Expression analysis of the eight mosquito-specific miRNAs is described in the context of a multi-species survey in a later section. The nine conserved miRNAs include let-7, miR-1, -133, -14, -184, -210, -9a, -970, and -998. All nine miRNAs showed signals at ~20 nt by northern during at least one of the developmental stages. Shown in Figure 3 are the expression patterns of five of the nine conserved miRNAs. The patterns of presence/absence of these miRNAs in embryo, larvae, pupa, and adult stages are similar to the patterns found in D. melanogaster [32] and An. stephensi [27].
Figure 3.
Expression patterns of Ae. aegypti homologs of previously known miRNAs. Only Ae. aegypti RNA samples were examined. The top panels are northern results and the bottom panels are RNA gel images for verification of small ribosomal RNA and tRNA integrity and loading of total RNA. Emb, pooled embryos between 0-36 hr after egg deposition; Larvae; mixed instar larvae; Pupae, mixed puape; F, adult females one to five days after emergence; M, adult males one to five days after emergence. 10 μg of total RNA were used per sample.
Elevated levels of miRNAs after blood feeding in the midgut of Ae. aegypti
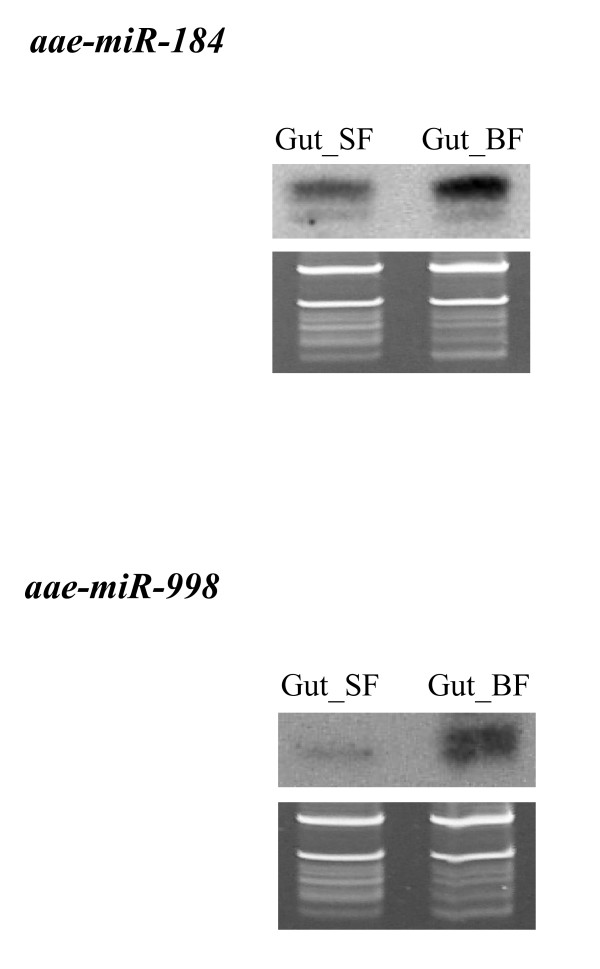
The numbers of small RNA sequences in the midgut samples from sugar-fed and blood-fed female Ae. aegypti may not be high enough for quantitative comparison. Nonetheless, we decided to compare the relative miRNA levels for miRNAs that showed more than 25 hits in at least one of the midgut samples. We used either the total number of all miRNA hits [12] or the total number of small RNA reads to normalize the data. As shown in the last two columns of Table 2, except for miR-989 and miR-281*, all miRNAs showed an increase after blood feeding. We then performed northern blots using miR-184 and miR-998 probes. It is clear that miR-998 level is higher in bloodfed samples than in sugar-fed samples. Although less obvious, miR-184 level also appears to be higher in blood-fed samples than in sugar-fed samples (Figure 4). Thus, the northern results are largely consistent with the data shown in Table 1. We have previously analyzed the level of miR-989 in the midgut before and after blood feeding [27]. The signal was too weak to confirm or rule out reduction of miR-989 after blood feeding. Some miRNAs that are expressed in the midgut samples are also found in large numbers in embryos in Ae. aegypti (Table 1).
Table 2.
Comparison of the number of miRNA sequences in sugar-fed and blood-fed midgut samples.
| miRNAs 1 | Gut_SF 6 | Gut_BF 6 | Fold Change I 7, 9 | Fold Change II 8, 9 |
|---|---|---|---|---|
| aae-miR-11 2 | 42 | 436 | 6.55 | 8.16 |
| aae-miR-1175* | 2 | 59 | 18.61 | 23.18 |
| aae-miR-13b 3 | 2 | 42 | 13.25 | 16.50 |
| aae-miR-184 | 76 | 1307 | 10.85 | 13.51 |
| aae-miR-190 | 2 | 43 | 13.56 | 16.89 |
| aae-miR-281 | 16 | 50 | 1.97 | 2.46 |
| aae-miR-281* | 5686 | 4806 | 0.53 | 0.66 |
| aae-miR-283 | 1 | 131 | 82.65 | 102.93 |
| aae-miR-2a/2b/2c 3, 4 | 80 | 249 | 1.96 | 2.45 |
| aae-miR-306 5 | 0 | 27 | NA 10 | NA 10 |
| aae-miR-317-1 | 38 | 737 | 12.24 | 15.24 |
| aae-miR-317-2 | 38 | 737 | 12.24 | 15.24 |
| aae-miR-34 | 55 | 639 | 7.33 | 9.13 |
| aae-miR-8 | 1 | 38 | 23.98 | 29.86 |
| aae-miR-989 | 33 | 3 | 0.06 | 0.07 |
| aae-miR-998 2 | 6 | 42 | 4.42 | 5.50 |
| aae-miR-9b 5 | 0 | 69 | NA 10 | NA 10 |
Notes.
1. Only miRNAs that showed 25 or more sequences in one of the gut samples are shown.
2. MiR-11 and miR-998 are physically linked, less than 300 bp apart.
3. MiR-13b and miR-2a/2b/2c are in a physically linked cluster. Mir-13b is less than 200 bp apart from miR-2b.
4. It is difficult to distinguish between hits that match miR-2a, miR-2b, or miR-2c. Thus we refer to these hits miR-2a/2b/2c.
5. MiR-306 and miR-9b are physically linked, less than 500 bp apart.
6. The second and third columns are raw numbers of sequence hits in sugar-fed (Gut_SF) and blood-fed (Gut_BF) midgut samples, respectively.
7. Column four is the fold difference of Gut_BF over Gut_SF, normalized by the total miRNA hits within a sample, as suggested in reference [12]. The total miRNA hits are 6260 and 9922 in Gut_SF and Gut_BF samples, respectively. Thus, for each miRNA, the Fold Change = [(Raw Gut_BF number)/9922]/[(Raw Gut_SF number)/6260].
8. Column five is the fold difference of Gut_BF over Gut_SF, normalized by the total small RNA reads within a sample. The total small RNA reads are 33, 000 and 42, 000 in Gut_SF and Gut_BF samples, respectively. Thus, for each miRNA, the Fold Change = [(Raw Gut_BF number)/42000]/[(Raw Gut_SF number)/33000].
9. The numbers in columns 4 and 5 reflect the same trend, generally higher levels of these miRNAs in blood-fed sample than in sugar-fed sample. Although the number of small RNA sequences may not be sufficient for the analysis to be quantitative, overall trends suggest leads for further analysis.
10. NA, not applicable as the denominator is zero.
Figure 4.
Higher levels of miRNAs are observed in the female Ae. aegypti midgut 24 hrs after blood feeding (Gut_BF) compared to sugar feeding (Gut_SF). Three-day old females were either fed on blood or sugar and dissected 24 hrs later. 10 μg of total RNA were used per sample. The top panels are northern results and the bottom panels are RNA gel images for verification of small ribosomal RNA and tRNA integrity and loading of total RNA.
Blood feeding is critical for mosquito physiology and its ability to transmit disease pathogens. It is through feeding on an infected host mosquitoes acquire pathogens such as malaria parasites and dengue viruses. It is also through blood feeding by an infected mosquito these pathogens may spread to a different host. Midgut is the first barrier the pathogens have to cross before they establish infection in mosquitoes. Thus midgut is one of the most important links in the disease transmission cycle. Furthermore, blood-feeding triggers a cascade of gene regulatory events in multiple tissues including midgut through the interplay of endocrine signals and transcription factors and thus has great impact on mosquito biology [33-36]. The correlation between blood feeding and miRNA levels in Ae. aegypti midgut warrants further investigations, which may shed light on the possible roles of miRNAs in physiology related to blood feeding and perhaps in mosquito-pathogen interactions.
Multi-species survey of eight mosquito-specific miRNAs revealed conserved and lineage-specific miRNAs
Previously five miRNAs were reported to be only found in mosquitoes. These are miR-1174, miR-1175 [21], miR-1889, miR-1890, and miR-1891 [27]. As described above, we uncovered eight additional mosquito miRNAs, bringing the number of total "mosquito-specific" miRNAs to 13. We conducted a detailed multi-species expression analysis of eight of the 13 mosquito-specific miRNAs using northern blot. When appropriate, we examined the expression of these miRNAs across the life stages of four mosquito species, An. gambiae, An. stephensi, Ae. aegypti, and T. amboinensis.
Four of the eight miRNAs (miR-M1, -1175, -1890, and -1891) are detected in all of the above four species (Figure 5). Furthermore, the expression patterns of these miRNAs are similar in the four species and expression is detected in multiple developmental stages in three of the four miRNAs. The exceptions are the relatively low embryonic expression of miR-1175 in Ae. aegypti (Figure 5B) and the low or hardly detectable embryonic expression of miR-1891 in Ae. aegypti and T. amboinensis (Figure 5D), compared to the rest of the species studied here. Overall, this is consistent with the observation that conserved miRNAs tend to be widely expressed [12,37]. On the other hand, miR-M1 is expressed only in the embryos in all four species.
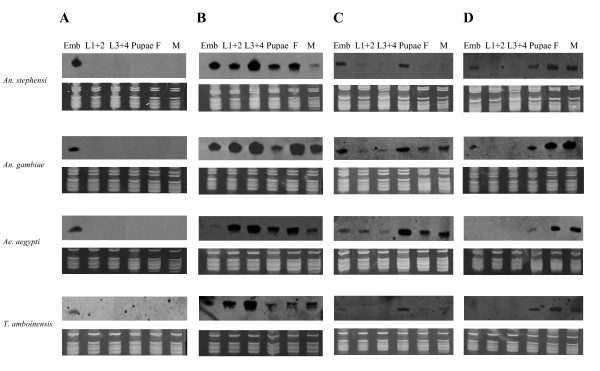
Figure 5.
Four mosquito-specific miRNAs that are expressed in all four species of three highly divergent genera. MiRNAs examined include miR-M1 (A), miR-1175 (B), miR-1890 (C), and miR-1891 (D). Expression was examined across the developmental stages of An. stephensi, An. gambiae, Ae. aegypti, and T. amboinensis. The top panels are northern results and the bottom panels are RNA gel images for verification of small ribosomal RNA and tRNA integrity and loading of total RNA. Emb, pooled embryos between 0-36 hr after egg deposition; L1+2, pooled 1st and 2nd instar larvae; L3+4, pooled 3rd and 4th instar larvae; Pupae, mixed pupae; F, adult females one to five days after emergence; M, adult males 1-5 days after emergence. 15 μg of total RNA per sample for An. stephensi, An. gambiae, and Ae. aegypti were used. 10 μg of T. amboinensis total RNA per sample were used. For T. amboinensis northerns, 3rd instar larvae were not included. For the T. amboinensis miR-M1 northern, hybridization and washes were carried out at 49°C instead of 42°C to reduce background across the membrane.
Four other miRNAs (miR-1174, miR-N1, miR-N2, and miR-N3) are only detected in a subset of the four mosquitoes. As shown in Figure 6, miR-1174 is not found in T. amboinensis but strong signals were detected in the other three species. MiR-1174 level in Ae. aegypti embryos is low or hardly detectable. It is interesting to point out that miR-1174 and miR-1175 are in the same contig separated by only ~200 bp. The expression patterns of miR-1174 and miR-1175 are similar in all three blood-feeding mosquitoes, suggesting that they may be under the same transcriptional control. MiR-1174 and miR-1175 share some sequence similarity at the 5' end. Thus it is possible that miR-1174 and miR-1175 resulted from gene duplication and miR-1174 may either have been lost in T. amboinensis or evolved beyond recognition by the miR-1174 probe. Ae. aegypti miR-1174 and An. gambiae miR-1174 differ by one nt. It is also possible that miR-1174 simply was not duplicated in T. amboinensis.
Figure 6.
MiR-1174 is expressed in An. stephensi, An. gambiae, and Ae. aegypti, but not in T. amboinensis. The top panels are northern results and the bottom panels are RNA gel images for verification of small ribosomal RNA and tRNA integrity and loading of total RNA. Emb, pooled embryos between 0-36 hr after egg deposition; L1+2, pooled 1st and 2nd instar larvae; L3+4, pooled 3rd and 4th instar larvae; Pupae, mixed puape; F, adult females one to five days after emergence; M, adult males one to five days after emergence. 15 μg of total RNA per sample for An. stephensi, An. gambiae, and Ae. aegypti were used. 10 μg of T. amboinensis total RNA per sample were used. For T. amboinensis, 3rd instar larvae were not included. "+ Cont" indicates a positive control which was An. stephensi embryos (12-24 hr).
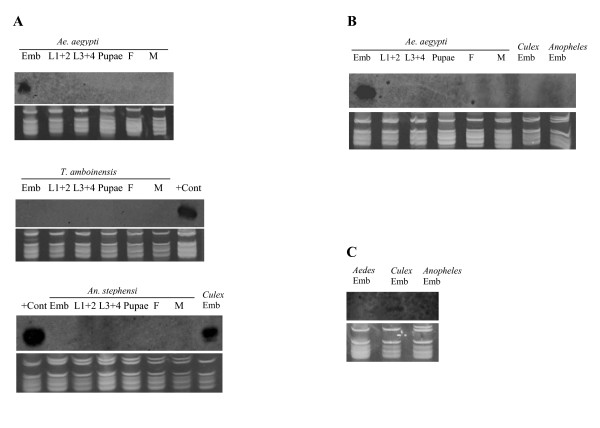
As described earlier, miR-N1, N2, and N3 are from the same intronic cluster. As shown in Figure 7, miR-N1 was abundant in both Ae. aegypti and Cx. quinquefasciatus embryos. It was undetectable in An. stephensi. MiR-N2 was abundant in Ae. aegypti embryos, but undetectable in the embryos of Cx. quinquefasciatus. MiR-N2 was also undetectable in An. stephensi. MiR-N3 was found in Cx. quinquefasciatus embryos, but not in Ae. aegypti. MiR-N3 was also undetectable in An. stephensi. The expression data are consistent with genomic sequence analysis, which is described in the previous section on the miR-N1, N2, N3 cluster.
Figure 7.
MiR-N1, miR-N2, and miR-N3 expression is restricted in particular lineages in mosquitoes. A) miR-N1 is expressed in Ae. aegypti and Cx. quinquefasciatus, but not in An. stephensi nor T. amboinensis. Emb, pooled embryos between 0-36 hr after egg deposition; L1+2, pooled 1st and 2nd instar larvae; L3+4, pooled 3rd and 4th instar larvae; Pupae, mixed pupae; F, adult females one to five days after emergence; M, adult males one to five days after emergence. Culex Emb, Cx. quinquefasciatus embryos 0-24 hrs after egg deposition. "+ Cont", positive control, Ae. aegypti embryos (12-24 hr). 15 μg of total RNA per sample for An. stephensi and Ae. aegypti were used. 10 μg of T. amboinensis total RNA per sample were used. For T. amboinensis, 3rd instar larvae were not included. B) miR-N2 is expressed in Ae. aegypti but not detected in Cx. quinquefasciatus and An. stephensi embryos. Symbols are as in A. Anopheles Emb, pooled An. stephensi embryos between 0-36 hr after egg deposition. C) miR-N3 is expressed in Cx. quinquefasciatus, but not detected in Ae. aegypti and An. stephensi embryos. Symbols are as in A and B. Aedes Emb, pooled Aedes aegypti embryos between 0-36 hr after egg deposition.
We performed northern blots using all of the above eight miRNA probes to see if any signal was detected in D. melanogaster. We used at least 5 μg of total RNA from different developmental stages or a specific stage expected for a particular miRNA. None of the eight probes produced any miRNA signal while the positive control (Ae. aegypti sample) showed intense signals (data not shown). This is consistent with these miRNAs being only found in mosquitoes.
Functions of "mosquito-specific" miRNAs
All of the eight tested mosquito-specific miRNAs showed embryonic expression in at least one mosquito species (Figures 5, 6, and 7), suggesting that these miRNAs may play important roles in mosquito embryonic development. Two of these miRNA clusters are worth noting. The first is the miR-M1-1 and miR-M1-2 cluster, which is only expressed in embryos in all four genera of mosquitoes tested. The conserved expression pattern and sequence conservation across all major branches of Culicidae suggest that miR-M1 is important during mosquito embryogenesis. Another interesting group of miRNAs are the miR-N1, -N2, and -N3 cluster. Two miR-N1 and one miR-N2 hairpins are in the first intron of a gene in Ae. aegypti encoding a putative transcription factor. Two miR-N1 hairpins and a miR-N3 hairpin are found in the orthologous gene in Cx. quinquefasciatus. None of these miRNAs are found in An. gambiae. In addition, miR-N2 is only found in Ae. aegypti while miR-N3 exists only in Cx. quinquefasciatus. These miRNAs share the same 7-8 bp 5' sequences in the seed regions important for target recognition. MiR-N1, N2, and N3 are all expressed in the embryos. Thus it is possible that these miRNAs derive from duplication events and the duplicated miRNAs may evolve into new sequences that acquire new functions. We postulate that, given their abundance and their lineage specificity, the N1/N2/N3 cluster may play a role in determining important lineage specific traits in mosquitoes. It will be important to determine the targets of these miRNAs to truly understand their function. Currently, the annotation of the 3'-UTRs of Ae. aegypti genes is limited. As these annotations improve, miRNA target prediction will likely be fruitful.
Conclusion
We report the first systematic analysis of miRNAs in Ae. aegypti during which 98 pre-miRNAs were uncovered. Thus we substantially expanded the list of miRNAs found in mosquitoes. We also provided experimental evidence for 89 of the miRNA and miRNA* sequences. We also uncovered highly abundant and conserved miRNA* sequences, which is consistent with the suggestion that some miRNA* are functional. There are 14 miRNA clusters and 17 cases where more than one pre-miRNA hairpin produces the same or highly similar mature miRNAs in Ae. aegypti . These miRNAs are a rich source for future comparative analysis to uncover the evolutionary patterns of miRNA duplication and the process of creating new miRNAs in mosquitoes. Perhaps most importantly, we identified eight novel miRNAs that are potentially specific to mosquitoes. This discovery expanded the list of mosquito-specific miRNAs from five [21,27] to 13. Four of the 13 miRNAs are specific to certain lineages within mosquitoes. The expression profiles of a few miRNAs suggest stage-specific functions and functions related to embryonic development or blood feeding. A better understanding of the functions of these miRNAs will offer novel insights in mosquito biology and may lead to novel approaches to combat mosquito-borne infectious diseases.
Methods
Insects
Ae. aegypti (Liverpool strain), An. gambiae (G3 strain), An. stephensi (Indian wild type strain), and T. amboinensis (CDC strain originally from San Juan, PR)mosquitoes were reared in a humidified insectary at 27°C on a 12 hour light:dark cycle. Cx. quinquefasciatus embryos at 0-24 hrs post-egg-deposition were kindly provided by Drs Aaron Brault and David Clark at the University of California, Davis. D. melanogaster wild type(Catalina strain, stock number 14021 -0231.47) samples were provided by the Tucson Drosophila Stock Center (Tucson, AZ). D. melanogaster W1118 eggs at 0-24 hrs post-egg-deposition were provided by Duke University Model Systems (Duke University, Durham, NC).
Ae. aegypti sample preparation for 454 pyrosequencing
Three samples were prepared for 454 sequencing. Female Ae. aegypti were either fed on mice or kept on sugar water, three days post-emergence. Midguts were dissected at 24 hours post blood meal and midguts from sugar-fed females were also dissected at the same time interval. To collect the third sample , which was mixed-age embryos, all eggs were laid on filter papers during one-hour intervals. The filter papers were then kept in an incubator (27°C, ~70% Relative Humidity) before collection at appropriate hours and stored in RNAlater (Ambion, Austin, Texas). The same volumes of eggs from different time periods were then mixed. This collection design ensured that embryos between 0-48 hrs post-egg-deposition were collected in similar quantities. All three samples were sent to vertis Biotechnologie AG (Freising-Weihenstephan, Germany) for small RNA cloning. All samples were ground in liquid nitrogen and RNA smaller than 200 bp were enriched with the mirVana miRNA isolation kit (Ambion). The population of miRNAs with a length of 15-30 bp was passively eluted from polyacrylamide gels. The RNA was then precipitated with ethanol and dissolved in water. The small RNAs collected had a poly(A)-tail added to their 3'-OH by poly-(A) polymerase. The 5'-phosphate of the small RNAs were ligated to an RNA adapter. First-strand cDNA synthesis was then performed using an oligo(dT)-linker primer and MMLV-RNase H reverse transcriptase. The resulting cDNAs were PCR-amplified to about 20 μg/μl. Primers used for PCR amplification were designed for amplicon sequencing according to the instructions of 454 Life Sciences (Branford, CT). The PCR-amplified cDNAs were size-selected using electroelution to obtain products of 119-134 bp. These cDNAs were then sequenced by 454 Life Sciences. The total small RNA reads are 55, 000, 33, 000 and 42, 000 in embryos, sugar-fed midguts (Gut_SF), and blood-fed midguts (Gut_BF), respectively.
Identification of pre-miRNA sequences in Ae. aegypti
Sequences that match known mosquito and D. melanogaster pre-miRNAs (miRBase v.12.0, September 2008) were used to identify miRNAs in Ae. aegypti. Potential homologues were identified in the genome assemblies of Ae. aegypti, An. gambiae and Cx. quinquefasciatus. The homologous sequences plus 200 bp flanking sequences were retrieved from three mosquito genomes and aligned using Clustalx [38] with a gap open penalty of five and a gap extension penalty of 0.05. The alignments were manually inspected and pre-miRNAs were identified and confirmed by RNAfold http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi using default settings. Eighty-nine pre-miRNAs were identified in Ae. aegypti including pre-miRNAs for five previously reported miRNAs that have only been found in mosquitoes so far. Expanding this homologous search approach by using all miRNA and miRNA* sequences in miRBase (September 2008 version) as queries did not yield additional pre-miRNAs in mosquitoes.
To identify novel miRNAs (or pre-miRNAs), we searched the Ae. aegypti 454 small RNA sequence libraries for sequences that matched both Ae. aegypti and Cx. quinquefasciatus genome assemblies by BLAST (Ref [39]; e-value cut-off is 0.01). We focused on the comparison between Ae. aegypti and Cx. quinquefasciatus because these two species are more closely related to each other than to An. gambiae. The basic approach is to use sequences in the Ae. aegypti small RNA libraries that showed conservation to Cx. quinquefasciatus non-coding sequences as leads to find novel miRNAs. Briefly, we first filtered the 454 small RNA sequence libraries by removing sequences that match previously characterized miRNAs, Ae. aegypti transposable elements (TEfam: http://tefam.biochem.vt.edu), and other known mosquito non-coding RNAs. We also masked all cDNA transcripts from the Cx. quinquefasciatus genome assembly using RepeatMasker http://www.repeatmasker.org/ on a Linux server. We then identified sequences in the masked Cx. quinquefasciatus genome that matched the filtered Ae. aegypti small RNA libraries using BLASTN (e-value cut-off is 0.01). We then retrieved these matched Cx. quinquefasciatus sequences with their flanking genomic sequence as well as the homologous sequences in the Ae. aegypti assembly. The homologous sequences from the two species were aligned and folded as described in the above paragraph. Nine novel pre-miRNAs that could produce 7 distinct miRNAs were identified in Ae. aegypti during this study (Table 1). Efforts to identify novel miRNAs simply by whole-genome comparison between Ae. aegypti and Cx. quinquefasciatus did not yield additional miRNAs, nor did efforts based on comparison of 454 small RNA sequences to Ae. aegypti genome alone.
To determine whether the novel miRNAs discovered in this study are indeed novel, we used BLAST [39] under low stringent conditions as well as oligomap [31], a program designed for comparisons of short sequences such as miRNAs, allowing gaps and mismatches. BLAST searches were done using low stringent parameters (word size at seven, e-value cut-off at 10) against miRBase and non-redundant GenBank sequences. Oligomap [31] was performed against all miRBase sequences using default parameters.
454 sequence count
To determine the number of miRNA and miRNA* hits per sample, 98 Ae. aegypti pre-miRNAs identified above were used as query for BLAST analysis. We require 100% match in at least 18 bp for a sequence to be counted. This approach does not distinguish between paralogous pre-miRNAs that produce the same miRNAs, nor does it distinguish between miRNAs that share at least 18 bp perfect match.
Sample collection for northern blots
For Ae. aegypti midgut samples, the sample collection was done the same way as described for preparing samples for 454 sequencing. Sample collections from different developmental stages of Ae. aegypti, An. stephensi, and An. gambiae are briefly described below. Embryo collections were made at 0-12, 12-24, and 24-36 hours after placing a damp collection cup within a cage. To generate points after 12 hours, egg containers were set aside and allowed to incubate at 27°C in a damp collection cup. 0-36 hour samples represent equal mixed pools of 0-12, 12-24 and 24-36 hour samples. Larval samples were collected at each instar, and pooled to generate early (I and II instars) and late (III and IV instars) larvae as listed in each figure. Pupal samples were collected from a pool of varied ages. Adults one to five days following eclosion were collected. In some cases, we did not separate early and late larval samples and used one mixed larval sample instead. These variations are specified in the figure legends of the northern blots. For T. amboinensis, embryos were collected at 0-24 hours post-oviposition. Samples were also collected for 1st and 2nd instar larvae and pooled to generate an early larvae sample, and a separate collection of 4th instar larvae was collected for late larvae. Pupae were collected from a pool of varied ages, and male and female adults were collected at two to five days post emergence.
Northern blot
All samples were either directly processed for RNA isolation or flash frozen on liquid nitrogen immediately following collection, then stored at -80°C. Total RNA isolation was carried out using a mirVana miRNA isolation kit (Ambion, Austin, TX). The amount of total RNA used for each sample is specified in the relevant figure legend. Northern blots were carried out based upon [27]. Briefly, total RNA were loaded onto 15% denaturing polyacrylamide gels, and run beside 19 and 23 nucleotide long ssDNA markers. The RNA gels were transferred to BrightStar-Plus nylon membranes (Ambion), crosslinked using a UV crosslinker, and prehybridized, then hybridized overnight in the ULTRAhyb-Oligo Hybridization Buffer (Ambion) with the appropriate DIG-labeled probe at 42°C. Wash conditions were the same as described in [27]. Antisense 5' digoxigenin-labeled miRCURY LNA probes were purchased from Exiqon (Vedbaek, Denmark). Probe sequences were reverse-complementary to sequences shown in Table 1. The probe for miR-1174 is derived from aga-miR-1174 (miRBase) and it had one base difference to aae-miR-1174.
Authors' contributions
SLi prepared samples for 454 sequencing, conducted the first northern blot using DIG-labeled probes, performed bioinformatics analysis, provided relevant tables, and wrote a draft of part of the methods section. EAM performed northern analysis on all mosquito-specific miRNAs, worked on the initial bioinformatics analysis on the miR-N1, N2, and N3 cluster, provided relevant figures, assisted in sample dissections, wrote a draft of parts of this manuscript and assisted in revisions. SLiang performed northern analysis on all conserved miRNAs, performed comparisons of miRNA expression in midgut samples, and provided relevant figures. ZT designed and oversaw the project, performed bioinformatics analysis, and wrote most of the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Supplementary table S1. Supplementary table S1 contains information provided in Table 1 as well as the entire hairpin sequence of each pre-miRNA.
Contributor Information
Song Li, Email: soli@vt.edu.
Edward A Mead, Email: emead@vt.edu.
Shaohui Liang, Email: wzliangshaohui@yahoo.com.cn.
Zhijian Tu, Email: jaketu@vt.edu.
Acknowledgements
We thank Randy Saunders for mosquito-rearing and for help with sample preparations. We thank the Culex genome consortium for making the genome assembly available before publication. This work is supported by NIH grant AI070854, FNIH grant GC7 #316, and the Virginia Experimental Station.
References
- Bushati N. MicroRNA Functions. Ann Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. NuclAcids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Shen Y, Wu Q, Kumar S, He B, Shi S, Carthew RW, Wang SM, Wu CI. The birth and death of microRNA genes in Drosophila. Nat Genet. 2008;40:351–355. doi: 10.1038/ng.73. [DOI] [PubMed] [Google Scholar]
- Zhang R, Wang YQ, Su B. Molecular evolution of a primate-specific microRNA family. Mol Biol Evol. 2008;25:1493–1502. doi: 10.1093/molbev/msn094. [DOI] [PubMed] [Google Scholar]
- Miska EA. How microRNAs control cell division, differentiation and death. Curr Opin Genet Dev. 2005;15:563–568. doi: 10.1016/j.gde.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Kim YK, Kim VN. Processing of intronic microRNAs. EMBO J. 2007;26:775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Okamura K, Tyler DM, Phillips MD, Chung WJ, Lai EC. The evolution and functional diversification of animal microRNA genes. Cell Res. 2008;18:985–996. doi: 10.1038/cr.2008.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Saito K, Ishizuka A, Siomi H, Siomi MC. Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol. 2005;3:e235. doi: 10.1371/journal.pbio.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler DM, Okamura K, Chung WJ, Hagen JW, Berezikov E, Hannon GJ, Lai EC. Functionally distinct regulatory RNAs generated by bidirectional transcription and processing of microRNA loci. Genes Dev. 2008;22:26–36. doi: 10.1101/gad.1615208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Belasco JG. Let me count the ways: mechanisms of gene regulation by miRNAs and siRNAs. Mol Cell. 2008;29:1–7. doi: 10.1016/j.molcel.2007.12.010. [DOI] [PubMed] [Google Scholar]
- Caldas C, Brenton JD. Sizing up miRNAs as cancer genes. Nat Med. 2005;11:712–714. doi: 10.1038/nm0705-712. [DOI] [PubMed] [Google Scholar]
- Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, Saib A, Voinnet O. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308:557–560. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- Sullivan CS, Gründhoff AT, Tevethia S, Pipas JM, Ganem D. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature. 2005;435:682–686. doi: 10.1038/nature03576. [DOI] [PubMed] [Google Scholar]
- Winter F, Edaye S, Huttenhofer A, Brunel C. Anopheles gambiae miRNAs as actors of defence reaction against Plasmodium invasion. Nucl Acids Res. 2007;35:6953–6962. doi: 10.1093/nar/gkm686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carissimi C, Fulci V, Macino G. MicroRNAs: novel regulators of immunity. Autoimmun Rev. 2009;8:520–524. doi: 10.1016/j.autrev.2009.01.008. [DOI] [PubMed] [Google Scholar]
- Lai EC, Tomancak P, Williams RW Rubin GM. Computational identification of Drosophila microRNA genes. Genome Biol. 2003;4:R42. doi: 10.1186/gb-2003-4-7-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhang J, Li F, Gu J, He T, Zhang X, Li Y. MicroRNA identification based on sequence and structure alignment. Bioinformatics. 2005;21:3610–3614. doi: 10.1093/bioinformatics/bti562. [DOI] [PubMed] [Google Scholar]
- Chatterjee R, Chaudhuri K. An approach for the identification of microRNA with an application to Anopheles gambiae. Acta Biochim Pol. 2006;53:303–309. [PubMed] [Google Scholar]
- Xu Y, Zhou X, Zhang W. MicroRNA prediction with a novel ranking algorithm based on random walks. Bioinformatics. 2008;24:i50–58. doi: 10.1093/bioinformatics/btn175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead EA, Tu Z. Cloning, Characterization, and Expression of microRNAs from the Asian Malaria Mosquito, Anopheles stephensi. BMC Genomics. 2008;9:244. doi: 10.1186/1471-2164-9-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell CL, Black WC 4th, Hess AM, Foy BD. Comparative genomics of small RNA regulatory pathway components in vector mosquitoes. BMC Genomics. 2008;9:425. doi: 10.1186/1471-2164-9-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucl Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berninger P, Gaidatzis D, van Nimwegen E, Zavolan M. Computational analysis of small RNA cloning data. Methods. 2007;44:13–21. doi: 10.1016/j.ymeth.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Sempere LF, Sokol NS, Dubrovsky EB, Berger EM, Ambros V. Temporal regulation of microRNA expression in Drosophila melanogaster mediated by hormonal signals and broad-Complex gene activity. Dev Biol. 2003;259:9–18. doi: 10.1016/S0012-1606(03)00208-2. [DOI] [PubMed] [Google Scholar]
- Raikhel AS, Kokoza VA, Zhu J, Martin D, Wang SF, Li C, Sun G, Ahmed A, Dittmer N, Attardo G. Molecular biology of mosquito vitellogenesis: from basic studies to genetic engineering of antipathogen immunity. Insect Biochem Mol Biol. 2002;32:1275–1286. doi: 10.1016/S0965-1748(02)00090-5. [DOI] [PubMed] [Google Scholar]
- Hagedorn HH. In: The Biology of Disease Vectors. Marquardt WC, editor. Vol. 2. Burlington: Elsevier Academic Press; 2005. Mosquito Endocrinology. [Google Scholar]
- Zhu J, Chen L, Raikhel AS. Distinct roles of Broad isoforms in regulation of the 20-hydroxyecdysone effector gene, Vitellogenin, in the mosquito Aedes aegypti. Mol Cell Endocrinol. 2007;267:97–105. doi: 10.1016/j.mce.2007.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieglaff DH, Dunn WA, Xie XS, Megy K, Marinotti O, James AA. Comparative genomics allows the discovery of cis-regulatory elements in mosquitoes. Proc Natl Acad Sci. 2009;106:3053–3058. doi: 10.1073/pnas.0813264106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Kheradpour P, Parts L, Brennecke J, Hodges E, Hannon GJ, Kellis M. Systematic discovery and characterization of fly microRNAs using 12 Drosophila genomes. Genome Res. 2007;17:1865–1879. doi: 10.1101/gr.6593807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary table S1. Supplementary table S1 contains information provided in Table 1 as well as the entire hairpin sequence of each pre-miRNA.