Abstract
The organotypic slice culture (Stoppini et al., 1991) has become the method of choice to answer a variety of questions in neuroscience. For many experiments however, it would be beneficial to image or manipulate a slice culture repeatedly, for example over the course of many days.
We prepared organotypic slice cultures of the auditory brainstem of P3 and P4 mice and kept them in vitro for up to 4 weeks. Single cells in the auditory brainstem were transfected with plasmids expressing fluorescent proteins by way of electroporation (Haas et al., 2001). The culture was then placed in a chamber perfused with oxygenated ACSF and the labeled cell imaged with an inverted wide-field microscope repeatedly for multiple days, recording several time-points per day, before returning the slice to the incubator.
We describe a simple method to image a slice culture preparation over to the course of multiple days and over many continuous hours, without noticeable damage to the tissue or photobleaching. Our method employs a simple, inexpensive custom-built insulator constructed around the microscope to maintain controlled temperature, and uses a perfusion chamber as used for in vitro slice recordings.
Keywords: slice culture, single-cell gene electroporation, multi-day imaging
Introduction
The investigation of brain development poses one of the great challenges of neuroscience. With the help of fluorescent markers and microscopic imaging techniques manifold insights on how the brain develops are being gained. Although in vivo models are the preferred choice, acute slice preparations and slice culture systems are often of benefit to answer specific questions that cannot be addressed using an in vivo preparation. Acute slices offer advantages for some manipulations and can allow observation for many hours (Daley and Waite, 1999). However, acute slices do not enable the scientist to modify preparations over the course of many days or weeks, nor do they allow repeated observation over the same time span. Slice culture systems do offer the possibility to manipulate organotypic neuronal connections and observe them at the same time in 3D over the course of days or weeks. Questions that require continuous imaging over long periods of time or repeated observation over the course of several days, even weeks can thus, in principle be approached by using slice culture systems.
The Stoppini organotypic slice culture method, also called membrane or filter cultures, (Stoppini et al., 1991) is used for a variety of studies (e.g. Reid et al., 2008; Mao et al., 2008; Panicker et al., 2008; Li et al., 2007) and provides a means to observe development of neuronal systems in a slice over the course of weeks. This slice culture system provides a relatively easy access to an organotypic culture system, since apart from an incubator, no special rotating devices, such as used for Gähwiler cultures (Gähwiler, 1981) are needed. It appears that there have been few efforts to use these preparations for repeated imaging over multiple days or weeks. Previous studies to observe slice cultures repeatedly over the course of several days involved special procedures, such as simulating incubator conditions on top of the microscope (Yamamoto et al., 1997).
Here we present a simple approach for long term imaging of cultured brain slices that does not require a specialized imaging environment and is easy to perform at low cost. Our method combines a standard slice culture method (Stoppini et al., 1991) and a standard in vitro perfusion chamber, identical to what is used for electrophysiological recordings in acute brain slices. Single cells in a slice cultures are transfected with plasmids leading to the expression of a fluorescent protein. We successfully image transfected cells repeatedly over the course of many days and over long continuous time spans without noticeable degeneration or photobleaching. We believe that this method can be beneficial for a variety of experiments involving slice culture systems where inter-neuronal connections can be maintained and time-lapse observation is wanted.
Materials and Methods
Animals
Male and female C57Bl/6J mice and CMV-24 mice (gift from Anthony van den Pool, van den Pol and Ghosh, 1998) were used. CMV-24 mice express the green fluorescent protein (GFP) in neurons in the auditory brainstem (provided by Anthony van den Pol, Yale University). Pups were considered 0 days (P0) on the day of birth. The University of Washington Institutional Animal Care and Use Committee approved all procedures.
Slice cultures
We prepare organotypic slice cultures of the auditory brainstem of P3 or P4 mice following procedures established by Stoppini (Stoppini et al., 2001). Mice are decapitated and the brain removed from the skull. 300 μm thick brain slices containing the auditory brainstem were prepared with a vibratome (Vibratome 1000 Classic, Vibratome, St. Louis, MO) in ice-cold ACSF. The ACSF contains (in mM) 130 NaCl, 3 KCl, 1.2 KH2PO4, 20 NaHCO3, 2.4 CaCl2, 1.3 MgSO4, 3 HEPES, 10 dextrose and was constantly gassed with 95% O2–5% CO2 (modified after Tzounopoulos et al., 2004). After slicing, the sections in which auditory brainstem nuclei can be identified, typically 2 to 3 per brain, are immediately transferred under a sterile tissue culture hood with the help of a glass pipette (custom made from Pasteur pipettes) on Millicell-CM cell culture inserts (PICMORG50, Millipore, Bedford, MA) in a six-well plate (6 Well Cell Culture Cluster, Corning Incorporated, Lowell, MA) containing 1 ml of a standard culture medium with increased potassium concentration. The medium consists of 48% Advanced minimum essential medium (with NEAA, with sodium pyruvate at 110 mg/l, without L-glutamate, Gibco Invitrogen, Carlsbad, CA), 24% Earl's balanced salt solution, 1% L-glutamine (200 mM), 24% horse serum (heat inactivated, sterile filtered), 1% penicillin-streptomycin optional (all Sigma, St. Louis, MO), 1.375% glucose solution (200g/l) and 25 mM K-gluconate, (Lohmann et al., 1998). Cultures are incubated at 36.5°C and 5% CO2 in an incubator (Forma Scientific CO2 Water Jacketed Incubator, Marietta, OH). We use a maximum of three membranes per six-well plate and change culture medium three times per week under a hood. To this end, the membrane inserts are taken out with a forceps and the old medium is sucked out with a sterile 1 ml pipette tip attached to a vacuum. Then 1 ml of fresh medium is then transferred into the empty well and the membrane containing the slice cultures is put back in the well. We do not preheat or pre-incubate the culture medium we are about to use for changing the medium in the six well plates.
Electroporation
After 2 to 4 days in vitro (DIV), we electroporate single cells in the superior olivary complex with plasmids leading to an expression of fluorescent proteins (procedure modified from Haas et al., 2001). We use glass electrodes (Glass, Capillary, Filament, 1.2 mm × .68 mm, 4", A-M Systems, Inc., Carlsborg, WA) with a tip diameter of 0.5–2 μm, filled with 1 μl of a 0.9 % NaCl solution containing plasmid at a concentration of 1–2 μg/μl. The glass electrode is filled with the plasmid by a custom made syringe pulled over the flame of a Bunsen burner. We use a plasmid leading to the expression of a red fluorescent protein throughout the cell (td-Tomato) and plasmids that lead to an expression of GFP- or CFP-tagged synaptophysin, a presynaptic vesicle protein (Richard Tsien, UCSD, created the original td-Tomato plasmid, which was supplied by Rachel O. L. Wong, University of Washington). Jane Sullivan (University of Washington) supplied the Synaptophysin GFP plasmid. The modified CFP-version of the same plasmid was a gift from Hollis Cline (Cold Spring Harbor Laboratory).
Immediately before electroporation, the six-well plates containing the slice cultures are removed from the incubator. The culture to be transfected is exposed simply by removing the lid of the six-well plate. Then single cells are targeted with the electrode tip guided visually with the help of a stereo microscope allowing epifluorescent illumination (Leica MZ 16 FA, Leica Microsystems GmbH, Wetzlar, Germany) and the combination of two micromanipulators, one manual for coarse electrode adjustment and one hydraulic for the fine x-,y- and z-axis positioning (M3301 Manual Micromanipulator, World Precision Instruments, Inc., Sarasota, FL and MX630R, Siskiyou, Inc., Grants Pass, OR). The plasmids are electroporated with a current of 3–4 μA with 1 ms square pulses at 200 Hz for several seconds. The current pulse is delivered by a square pulse stimulator (SD9 Square Pulse Stimulator, Grass Technologies, West Warwick, RI) and monitored with a voltmeter connected in series (24-Range Digital Multimeter, RadioShack Corporation, Fort Worth, TX). After cells in each slice culture are electroporated, the six-well plate is put back in the incubator.
After 24 h, slice cultures are inspected under fluorescent illumination to identify transfected cells. If transfection was successful, fluorescence could be detected as early as 24 h after transfection, the earliest time point checked.
Imaging
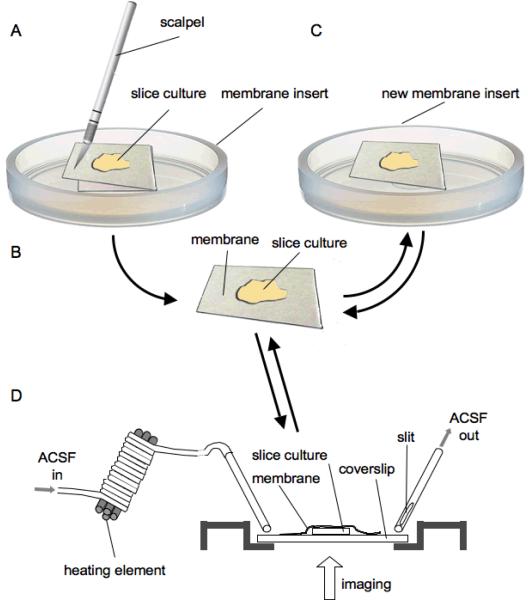
Once transfected cells are identified, a patch of the Millicell membrane containing successfully transfected slices is cut out with a scalpel (Fig. 1A). To this end we remove the complete membrane insert from the six-well plate and put it onto a sterile Petri dish. The membrane is held in place with a Dumont forceps while the patch of membrane with the culture is cut out. The resulting rectangular piece of membrane with the transfected slice (Fig. 1B) is then put with the old membrane on the lower side onto a new, second membrane insert (i.e. the slice is sitting on a double layer of insert membrane, Fig. 1C) and moved back into the six-well plate. From this point on, the slice culture is mobile and can be used for repeated high-resolution imaging.
Figure 1.
Slice culture explant and imaging technique
A The culture membrane insert was put on a sterile surface, e.g. Petri dish, and the part of the membrane containing the slice culture was cut out with a scalpel. B, C This explant was then transferred to an intact, fresh membrane insert with a Dumont forceps. The slice culture is now mobile and can be used in an imaging chamber. D The schematic shows a standard perfusion chamber, as used for in-vitro electrophysiology. The chamber is fitted with a coverslip at the bottom, through which the specimen can be imaged with an inverted microscope. The membrane with the attached slice culture is put onto the coverslip with the slice facing down. The membrane and slice are held in place with a platinum harp (not shown). Oxygenated ACSF is flowing into the chamber through a metal tube and sucked out by another metal tube attached to a vacuum. Notice the slit in the OUT-tube to prevent fluctuations in the ACSF level in the chamber (see methods).
For imaging, the membrane with the cultured slice is removed from the incubator and transferred to the imaging microscope in fresh, sterile medium. The membrane with the slice is then placed upside down in an open perfusion chamber (brass, custom made, gift from Tobias Bonhoeffer. Fig. 1D) on a coverslip, which is attached to the perfusion chamber with dental wax (Modern Materials, Wax Square Ropes, Heraeus Kulzer, Dormagen, Germany). The slice with the membrane, with the membrane now on the upper side (Fig. 1D), is held in place by a titanium harp with three single nylon fibers glued on (not shown). The harp provided enough pressure to hold the slice in place and the membrane on the slice's back ensured that there would be no tissue damage caused by the nylon fibers. The perfusion chamber is perfused with oxygenated ACSF through a gravity-pump and vacuum suction system for about 10 min prior to introducing the slice culture. The ACSF level in the perfusion chamber is determined by the tube connected to the vacuum suction system and is set so that the slice culture is fully immersed in ACSF. The perfusate is running at all times when the slice is placed in the chamber. The perfusion rate is around 2.5 ml/min, which is similar to what has been described by others (Dailey and Smith, 1996). The outgoing ACSF-tube has a slit in its side through which the ACSF is sucked out of the perfusion chamber (Fig. 1D). The slit is only half covered by ACSF. This ensures constant suction and avoids fluctuation of the ACSF-level, as the opening in the suction tube is never fully blocked ACSF. Additionally, the incoming ACSF is preheated to 35°C with the incoming ACSF-tube being wrapped around a heating element and temperature-controlled with a sensor directly attached to the perfusion chamber (Fig. 1D). For additional reading about live cell imaging see Daley et al. (2005).
The photomicrographs shown were imaged on a Marianas imaging system from Intelligent Imaging Innovations, Inc. (Denver, CO) incorporating a Zeiss Axiovert 200M microscope with an X,Y motorized stage (ASI, Eugene, OR), shuttered 175 W xenon lamp coupled with a liquid light guide (Sutter, Novato, CA) and a CoolSnap HQ digital camera (Princeton Instruments, Trenton, NJ) (Fig. 2). The microscope is housed in a rigid foam insulation-enclosure (Foamular® 250, Owens Corning, Toledo, OH) heated by a recirculating warm air heating system controlled by a proportional voltage thermostat (Omega Electronics, Knightdale, NC) utilizing a thermistor mounted on the microscope stage (custom made by Glen MacDonald and CHHD Instrument Development Laboratory, University of Washington, Fig. 2). The temperature of the entire chamber containing the microscope is controlled and maintained at 35°C to promote cell viability and optical stability. Heating the entire setup makes sure the microscope has a uniform temperature, avoiding any temperature gradient that might cause focal drift due to small movements in the system caused by heat expansion or retraction. It is beneficial to configure the enclosure as close as possible to the microscope in order to avoid creating unnecessary air space that has to be heated (Fig. 2). The rigid foam insulation-enclosure is easy to build and it should now take more than half a day to assemble such a heating-enclosure. To ensure thermal stability, the enclosure is heated to 35°C for 4 h before the start of an imaging session.
Figure 2.
Marianas imaging system with heating enclosure
The photograph shows the Marianas imaging system microscope in the heating enclosure. The microscope is housed in a rigid foam insulation-enclosure heated by a recirculating warm air heating system controlled by a proportional voltage thermostat utilizing a thermistor mounted on the microscope stage. The temperature of the entire chamber containing the microscope is controlled and maintained at 35°C to promote physiological conditions and optical stability. The front part of the enclosure is taken off for clarity. With the front removed, the slice culture can be inserted into the perfusion chamber and fluorescent cells focused. The front cover is brought back into position for imaging to ensure temperature stability and exclusion of ambient light.
Image acquisition is controlled and images are saved by Slidebook software (Intelligent Imaging Innovations, Inc., Santa Monica, CA). The Slidebook software package allows multichannel fluorescence and RGB DIC image capture, time-lapse, 3D and 4D-imaging.
After a successful imaging session the piece of membrane with the attached slice culture is transferred back onto the second, intact membrane insert and put back to its original six-well plate with culture medium Fig. 1C). The same slice can be used for repeated imaging sessions.
Image processing
Some of the image stacks were deconvolved using Huygens Essential (Scientific Volume Imaging, Hilversum, The Netherlands) for better visibility of the synaptophysin puncta and the fine structures of cells. 3D-image stacks were converted to maximum intensity projection images and contrast enhanced using ImageJ (National Institutes of Health, Bethesda, MD).
Results
We imaged a total of 52 cells from 44 animals. 28 cells were used in single session time-lapse imaging and 24 cells were used in multi-day imaging. Cells that were imaged on multiple days were also imaged in a time-lapse manner. We were able to repeatedly image slice cultures in a standard perfusion chamber perfused with oxygenated ACSF. Individual cells were imaged until they were no longer identifiable.
Time-lapse experiments
The cells in this group were imaged between 9 and 258 min (mean 47 min) with 63% of the cells imaged for more than 30 min. We imaged the cells for 2 to 18 time-points (TP, mean 6 TP). The slice cultures we used had not previously been submerged in ACSF, yet they survived well at 35°C without noticeable decrease in viability, i.e. bloating of cells or cessation of synaptophysin movement.
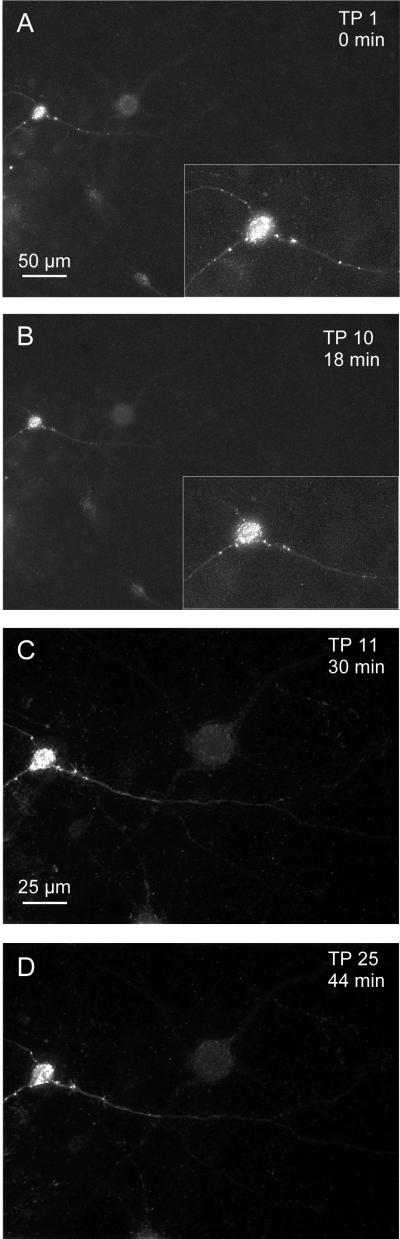
Figure 3 shows a cell transfected with a plasmid expressing CFP-tagged synaptophysin that was 3D-imaged 25 times in 44 min. The intervals between the acquired z-stacks of images were 120 s for the first 10 TPs and 60 s for the next 15 TPs. The first 10 TPs were imaged with a 20x lens and the next 15 TPs with a 40x lens. Shown are TP 1 (0 min, Fig. 3A), 10 (18 min, Fig. 3B), (magnification/lens change) 11 (30 min, Fig. 3C) and 25 (44 min, Fig. 3D). At the beginning of the imaging session, the cell body and several dendrites can be identified (Fig. 3A). Images in figure 3 show deconvolved versions of maximum intensity projections of the acquired 3D-images. Additionally, in Figures 3A and B the inserts show a close-up of the cell body. Bright synaptophysin puncta lighten the cell body and a dendrite with puncta can be identified. Synaptophysin puncta can be seen at different locations along the dendrite at the four time-points. No change in tissue quality or photobleaching could be detected. Two time-lapse movies (TPs 1–10 and TPs 11–25) are available as supplemental data (Fig. 3s).
Figure 3.
Time-lapse imaging
Time-lapse images of a neuron transfected with synaptophysin-CFP. A neuron in a cultured slice was transfected with a plasmid causing the expression of CFP-tagged synaptophysin. The cell was imaged at 25 TPs over 44 min. A Maximum intensity projections of the 3D-images at the beginning of the imaging session. Synaptophysin puncta outline the cell body and dendrite. The insert shows the cell body in detail. B, C, D TPs 10, 11 and 25. Not the lens change after TP 10. Puncta can be localized at different positions at different time-points. No blebbing in the cell body is detectable. (See also time-lapse movie of the 25 TPs in supplemental data, Fig. 3s).
Multi-day imaging
We imaged 24 cells from 23 animals repeatedly over the course of multiple days. 14 cells were imaged on 2 days, with the imaging sessions being at least 48 h apart. We imaged one cell where the 2 imaging session were only 7 h apart. 4 cells were imaged for three days, another 4 for 4 days and one cell was imaged on 5 days. For all cells, the first day of imaging was between 8 and 19 DIV and the last imaging day was between 11 and 20 DIV. On individual days the cells were often time-lapse imaged. On the first imaging day cells were imaged over a period from 10 to 208 min, with a mean of 50 min.
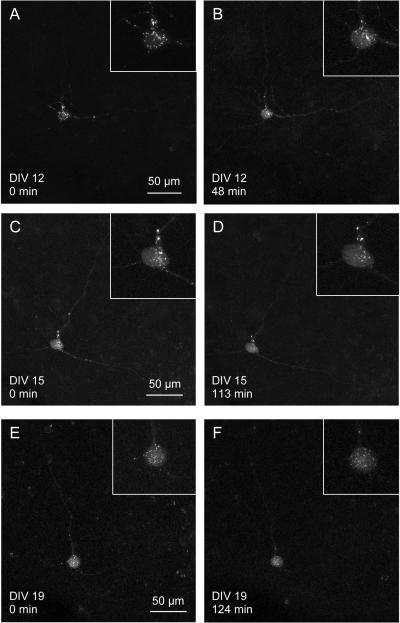
Figure 4 shows a cell that was imaged repeatedly on three different days. The cell was transfected with a plasmid labeling a presynaptic protein, synaptophysin, with CFP. The cell was transfected by electroporation on DIV 8 and first imaged in 3D on DIV 12 (Fig. 4A, B). The next two imaging days were DIV 15 (Fig. 4 C, D) and DIV 19 (Fig. 4 E, F). The same cell could be identified on the three imaging days and CFP-tagged synaptophysin can be seen being transported along the projections of the cell (see supplemental data for movies of each of the imaging days). On DIV 12, the cell was imaged for 30 TP (48 min), on DIV 15 it was imaged for 20 TP (113 min) and for 25 TP (124 min) on DIV 19. Figure 4 shows deconvolved versions of the maximum intensity projections of the first and the last 3D-image taken on the respective day. The inserts in those figures show a close-up of the cell body with its synaptophysin puncta. The inserts in figure 4 show no blebbing of the cell body, often a sign of an unhealthy cell. Time-lapse movies of the three imaging days are available as supplemental data (Fig. 4s).
Figure 4.
Multi-day imaging
A neuron transfected with a synaptophysin-CFP plasmid was imaged on 3 different days. The cell was transfected by electroporation on DIV 8. A, B The cell is shown at the beginning (TP 1) and at the end (TP 30) of the first imaging session on DIV 12. The two 3D-images are taken 48 min apart. Inserts show the cell body containing synaptophysin puncta. C, D, E, F The next two imaging days were DIV 15 (D, E) and DIV 19 (G, H). The cell could be identified by its three dendrites. The cell appears healthy, as the synaptophysin puncta are moving about in the cell body and along the dendrites (see movie in supplemental data, Fig. 4s). The inserts show the cell bodies at the respective time points, which show no sign of blebbing.
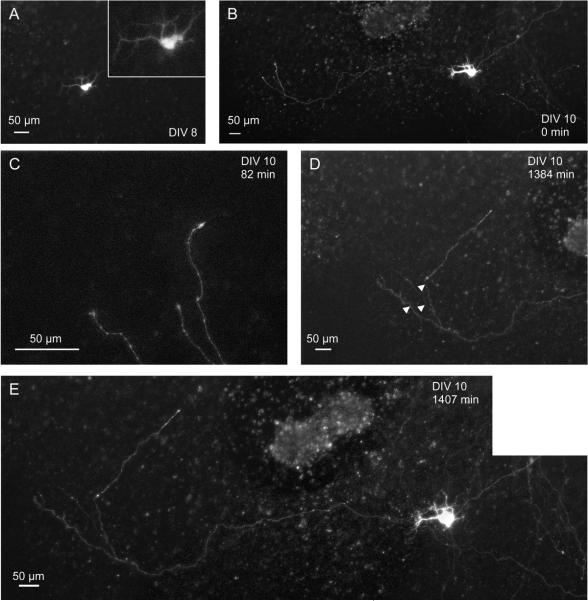
Another example is shown in Figure 5. A slice culture was electroporated with a GFP-plasmid after 4 DIV. After 8 DIV, 4 d later, a cell is clearly labeled (maximum intensity projection of 3D-image, Fig. 5A). Just 2 days later, after 10 DIV, the cell is still present and an axon extending to the left of the transfected cell can be seen (Fig 5B). At the same day (DIV 10), the axon was observed to be growing and its extending tips were imaged 86 times for almost 22 h (Fig. 5C, D). The intervals between acquisitions of the 3D-images ranged from 10 to 30 min (5 min for one interval). Images of the growing axon were taken with a 10x, a 20x and a 40x dry lens. The images were recorded in blocks of 30 and 60 min and in one case in a block of 600 min. During those blocks the lens was not readjusted and the tissue remained in focus. Figure 5C shows the tip of the axon at the beginning of the imaging session (TP indicated above). Three processes can be identified. 22 h and 51 min later, the axon has grown significantly (Fig. 5D, the white arrowheads indicate the extent of the axon in Fig. 5C). One branch of the axon grew over 200 μm during the imaging period. Figure 5E shows the cell and the axon at the end of the imaging session. Over multiple days and almost 23 hours of continuous imaging, cell viability was not impaired and no photobleaching was detectable. A movie of 78 TPs covering 1302 min is available as supplemental data (Fig. 5s).
Figure 5.
Multi-day and time-lapse imaging combined
A neuron was transfected with a GFP-plasmid after 4 DIV. A At 8 DIV the neuron was identified in the slice culture. B Two days later, at 10 DIV, the neuron could be identified again and an growing axon lateral to the cell body was detected (composite image). C, D The tip of the axon was imaged over almost 22 h, while the axon was continuously growing (see movie in supplemental data). C shows the axon at the first of 78 TPs and D shows the last TP. The two 3D-images are taken 1302 min apart. The arrowheads in D indicate the position of the axon tips in C. E Composite image of the cell at the end of the imaging session. The cell did no show any apparent change when compared to B.
Discussion
We present a simple method for multi-day imaging of cultured brainstem slices. To our knowledge no such method has been reported previously. We used slice cultures containing cells expressing a fluorescent protein and time-lapse imaged the cells at each imaging day over long time periods and over the course of several days. Phototoxicity seemed to be reduced when only certain parts of a labeled cell (e.g. growing axon) were imaged and not the whole neuron including cell body.
A total of 52 cells from 44 animals were imaged for this report, either in a time-lapse fashion or on multiple days. We found the fluorescent proteins, once a cell was transfected, to be remarkably stable and slice culture viability was quite satisfactory.
For an efficient gene-electroporation the exact placement of the electroporation electrode close to the neuron seems to be crucial for transfection (see also Rathenberg et al., 2003). We tried a variety of different settings for plasmid concentration, electrode tip size and stimulus parameters for electroporation, but found no reliable differences in transfection efficiency. We found efficiency to be low (<5 % of electroporation sites had transfected cells). Since the electroporation success rate was very low, we generally aimed to electroporate at several spatially different sites in a slice culture to increase the probability of a cell being transfected with a plasmid. Others have reported low success rates with single-cell gene transfer by electroporation as well (Haas et al., 2001; Rathenberg et al., 2003), especially for transfection in slice cultures (Kurt Haas, personal communication). One group has achieved high transfection efficacy by guiding the position of the electroporation electrode with a multiphoton microscope (Rathenberg et al., 2003).
For this study we used a standard perfusion chamber, similar to what is used for in-vitro electrophysiology. We used also standard oxygenated ACSF, which was preheated to physiological conditions. We had twofold control of the temperature, since the microscope was housed in a heated enclosure. Having the whole system at the same temperature avoided thermal drift. Once held down by the harp, our samples stayed optically stable for many hours (Fig. 5C, D).
Of special interest is the fact that we were able to repeatedly switch our slice cultures from Stoppini-type incubator conditions (Stoppini et al., 1991) to being submerged in oxygenated ACSF. We did not find this to impair the viability of our slice cultures, as plasmid expression remained strong, and in many case we could observe either synaptophysin being transported along dendrites or axons growing (Figs. 3–5). We did not employ any antibiotic supplement to avoid the infection or contamination problems once the cultures have been taken out of the incubator. We assume that after a few days in culture, our brainstem slice cultures have become robust enough to survive the change between incubator conditions and oxygenated ACSF.
Others have imaged organotypic cultures over several days as well (e.g., Uesaka et al., 2005; Yasumatsu et al., 2008). However their methods sometimes involve an elaborate setup to mimic incubator-like conditions at the microscope (e.g., Yamamoto et al., 1997) or do not observe the cultures for as long time-spans as we did (Yasumatsu et al., 2008). A recent publication presents a detailed protocol for long term imaging of hippocampus slice cultures (Gogolla et al., 2006). However, this protocol focuses on a singular observation at each imaging session, rather than time-lapse imaging at each imaging day as we describe it in this report.
The method presented here does not employ any special equipment other than the microscope housing, but rather used a conventional in-vitro setup, available in many laboratories. We were able to successfully image labeled cells in slice cultures for up to 20 DIV. We believe that this simple method to image a slice culture repeatedly over the course of multiple days up to weeks will be useful to many laboratories.
Supplementary Material
Acknowledgements
We would like to thank Eckhard Friauf (TU Kaiserslautern) for suggesting using K-gluconate. Florence Reyes and Kevin Witham supported the artwork, Glen MacDonald designed and built the heating enclosure for the microscope and helped with imaging. We also thank Rachel O. L. Wong, Kathryn M. Tabor, Glen MacDonald and Jason T. Sanchez, and also the anonymous reviewers for critical comments on the manuscript. Support was provided by NIDCD grants DC03829 and DC04661. Armin H. Seidl was supported by the Leopoldina Fellowship Program of the Deutsche Akademie der Naturforscher Leopoldina (German Academy of Natural Scientists Leopoldina, grant number BMBF-LPD 9901/8-109) while he carried out the experiments.
References
- Dailey M, Smith S. The dynamics of dendritic structure in developing hippocampal slices. J Neurosci. 1996;16(9):2983–94. doi: 10.1523/JNEUROSCI.16-09-02983.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dailey M, Waite M. Confocal imaging of microglial cell dynamics in hippocampal slice cultures. Methods. 1999;18(2):222–30. doi: 10.1006/meth.1999.0775. 177. [DOI] [PubMed] [Google Scholar]
- Dailey ME, Marrs GS, Kurpius D. Maintaining Live Cells and Tissue Slices in the Imaging Setup. In: Yuste R, Konnerth A, editors. Imaging in Neuroscience and Development. Lab Manual edition. Cold Spring Harbor Laboratory Press; 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehay C, Kennedy H. Cell-cycle control and cortical development. Nat Rev Neurosci. 2007;8(6):438–50. doi: 10.1038/nrn2097. [DOI] [PubMed] [Google Scholar]
- Gogolla N, Galimberti I, DePaola V, Caroni P. Long-term live imaging of neuronal circuits in organotypic hippocampal slice cultures. Nat Protoc. 2006;1(3):1223–6. doi: 10.1038/nprot.2006.169. [DOI] [PubMed] [Google Scholar]
- Gähwiler B. Organotypic monolayer cultures of nervous tissue. J Neurosci Methods. 1981;4(4):329–42. doi: 10.1016/0165-0270(81)90003-0. [DOI] [PubMed] [Google Scholar]
- Haas K, Sin WC, Javaherian A, Li Z, Cline HT. Single-cell electroporation for gene transfer in vivo. Neuron. 2001;29(3):583–91. doi: 10.1016/s0896-6273(01)00235-5. [DOI] [PubMed] [Google Scholar]
- Hafidi A, Sanes D, Hillman D. Regeneration of the auditory midbrain intercommissural projection in organotypic culture. J Neurosci. 1995;15(2):1298–307. doi: 10.1523/JNEUROSCI.15-02-01298.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Woo R, Mei L, Malinow R. The neuregulin-1 receptor erbB4 controls glutamatergic synapse maturation and plasticity. Neuron. 2007;54(4):583–97. doi: 10.1016/j.neuron.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann C, Bonhoeffer T. A role for local calcium signaling in rapid synaptic partner selection by dendritic filopodia. Neuron. 2008;59(2):253–60. doi: 10.1016/j.neuron.2008.05.025. [DOI] [PubMed] [Google Scholar]
- Lohmann C, Ehrlich I, Friauf E. Axon regeneration in organotypic slice cultures from the mammalian auditory system is topographic and functional. J Neurobiol. 1999;41(4):596–611. doi: 10.1002/(sici)1097-4695(199912)41:4<596::aid-neu14>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- Lohmann C, Ilic V, Friauf E. Development of a topographically organized auditory network in slice culture is calcium dependent. J Neurobiol. 1998;34(2):97–112. doi: 10.1002/(sici)1097-4695(19980205)34:2<97::aid-neu1>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Mao T, O'Connor D, Scheuss V, Nakai J, Svoboda K. Characterization and subcellular targeting of GCaMP-type genetically-encoded calcium indicators. PLoS ONE. 2008;3(3):e1796. doi: 10.1371/journal.pone.0001796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panicker S, Brown K, Nicoll R. Synaptic AMPA receptor subunit trafficking is independent of the C terminus in the GluR2-lacking mouse. Proc Natl Acad Sci U S A. 2008;105(3):1032–7. doi: 10.1073/pnas.0711313105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathenberg J, Nevian T, Witzemann V. High-efficiency transfection of individual neurons using modified electrophysiology techniques. J Neurosci Methods. 2003;126(1):91–8. doi: 10.1016/s0165-0270(03)00069-4. [DOI] [PubMed] [Google Scholar]
- Reid C, Adams B, Myers D, O'Brien T, Williams D. Sub region-specific modulation of synchronous neuronal burst firing after a kainic acid insult in organotypic hippocampal cultures. BMC Neurosci. 2008;9:59. doi: 10.1186/1471-2202-9-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoppini L, Buchs P, Muller D. A simple method for organotypic cultures of nervous tissue. J Neurosci Methods. 1991;37(2):173–82. doi: 10.1016/0165-0270(91)90128-m. [DOI] [PubMed] [Google Scholar]
- Tzounopoulos T, Kim Y, Oertel D, Trussell L. Cell-specific, spike timing-dependent plasticities in the dorsal cochlear nucleus. Nat Neurosci. 2004;7(7):719–25. doi: 10.1038/nn1272. [DOI] [PubMed] [Google Scholar]
- Uesaka N, Hirai S, Maruyama T, Ruthazer E, Yamamoto N. Activity dependence of cortical axon branch formation: a morphological and electrophysiological study using organotypic slice cultures. J Neurosci. 2005;25(1):1–9. doi: 10.1523/JNEUROSCI.3855-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Pol A, Ghosh P. Selective neuronal expression of green fluorescent protein with cytomegalovirus promoter reveals entire neuronal arbor in transgenic mice. J Neurosci. 1998;18(24):10640–51. doi: 10.1523/JNEUROSCI.18-24-10640.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto N, Higashi S, Toyama K. Stop and branch behaviors of geniculocortical axons: a time-lapse study in organotypic cocultures. J Neurosci. 1997;17(10):3653–63. doi: 10.1523/JNEUROSCI.17-10-03653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasumatsu N, Matsuzaki M, Miyazaki T, Noguchi J, Kasai H. Principles of long-term dynamics of dendritic spines. J Neurosci. 2008;28(50):13592–608. doi: 10.1523/JNEUROSCI.0603-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.