Abstract
An increasingly common component of studies in synthetic and systems biology is analysis of dynamics of gene expression at the single-cell level, a context that is heavily dependent on the use of time-lapse movies. Extracting quantitative data on the single-cell temporal dynamics from such movies remains a major challenge. Here, we describe novel methods for automating key steps in the analysis of single-cell, fluorescent images – segmentation and lineage reconstruction – to recognize and track individual cells over time. The automated analysis iteratively combines a set of extended morphological methods for segmentation, and uses a neighborhood-based scoring method for frame-to-frame lineage linking. Our studies with bacteria, budding yeast and human cells, demonstrate the portability and usability of these methods, whether using phase, bright field or fluorescent images. These examples also demonstrate the utility of our integrated approach in facilitating analyses of engineered and natural cellular networks in diverse settings. The automated methods are implemented in freely available, open-source software.
INTRODUCTION
Time-lapse microscopy technologies now enable detailed data generation on dynamic cellular processes at the single cell level (1, 2). Recent studies have highlighted the use and importance of this technology for investigating biological noise in the dynamics of gene regulation, competence pathways in Bacilus subtili, and aspects of cell growth and proliferation, among other areas (2,3,4,5,6,7). Mathematical and statistical models are of growing interest in describing and testing hypothesis for such systems (9,10,11), and rely on extraction of data from time-lapse microscopy imaging techniques that generate sequences of individual, pixel-based image frames.
Unlike data generated from flow cytometry, which represent snapshots of cellular states, time-lapse movies with sufficiently high temporal and spatial resolution allow time courses of gene expression dynamics to be established at the single-cell level. These time courses are critically important in studies investigating the dynamics of both natural (12,13,14,15) and synthetic (16,17,18,19) cellular networks, dynamics that are often masked at the population level. However, recognizing cells as objects in images, and tracking them from one image to the next, remain as central technical challenges (1). These tasks, segmentation and tracking, respectively, can be approached either manually or automatically. Typically, humans are good at these tasks, but automation – essential for this fast-developing technology – is difficult and poses open questions for methodology development.
Depending on the cells being segmented and tracked, various existing algorithms are available (20,21,22,23,24). A universal solution to cell segmentation and tracking, applicable across cell types, has yet to be described. Algorithms that work well on one set of images often perform poorly on another set due to differences in the features that are exploited. A major goal in algorithm design and software development is then to provide a set of tools capable of extracting cell-like objects from many different types of images. Attempts have been made in both commercial and public domain software packages to solve these issues. Commercial software, including Imaris from BitPlabe, Amira from Mercury Computer Systems, Volocity from Improvision, MetaMorph from Molecular Devices and Matlab Image Processing toolbox from MathWorks, are mainly for general purpose image analysis, not specific nor customized to microscopy images. Among public-domain tools in development, Image-J is a general purpose image analysis program, while CellProfiler (23), Cell-ID (20), CellTracker (24), and GoFigure(1) aim to address segmentation and/or tracking in more specific areas. For example, Cell-ID was developed for automatic segmentation, tracking, and quantification of fluorescence protein levels in yeast. For specific synthetic or natural biological systems, ad hoc algorithms have also been adopted for analyses of gene expression dynamics in bacteria (5), yeast (25), and mammalian cells (26).
The algorithms implemented in above-mentioned tools use a number of variations of watershed and level-set approaches for segmentation. The success of these approaches depends largely on success in defining initial cell identity and location markers. When cells are tightly clustered, as is often the case with data on E. coli and yeast, for example, it is typically difficult to correctly identify initial cell objects to avoid mis-segmentation. Also, the methods used to define cell markers vary with cell type and image type, thus reducing algorithm portability. To address these questions, we develop the concept of hybrid grey-scale/black-white images and extend existing image filters and mathematical morphological operators for grey-scale images to work with these hybrid images. This approach allows us to extract cells from an image in an iterative process that gradually converts the grey-scale image into a black-white mask thus segmenting the image into cells, without relying on initial cell markers.
Moreover, tracking algorithms used in existing tools also face portability challenges: they often accomplish the tracking task by minimizing ad hoc global energy functions that are problem-specific, and typically do not consider locally clustered cells. To overcome these complications, we incorporate neighboring cell information to compute numerical likelihood scores for cell identity between each pair of time steps t to t+1. We then apply an integer programming method to generate frame-to-frame correspondences between cells and the lineage map.
The resulting, fully automated analysis, wrapped in an open-source Matlab application, is demonstrated here on two E. coli bacteria data sets, one yeast dataset and one human cell data set. Full algorithmic, workflow and analysis details, and more extensive examples, are given in several sections of the Supplementary Materials. Additional information and examples, together with the source code, a graphical user interface, examples of alternative workflows and other supporting documents, can be found at the CellTracer website (http://www.stat.duke.edu/research/software/west/celltracer/). With this methodology and tool, we have been able to successfully analyze single-cell time-lapse movies of cellular dynamics for diverse cell types, spanning from bacteria to yeast to human cells and diverse image types, from phase to bright-field to fluorescence images. As such, we believe it will facilitate quantitative spatiotemporal analysis of synthetic and natural cellular networks.
RESULTS
Image preprocessing and hybrid filters on hybrid grey-scale/black-white image
Preprocessing begins with a three-step partition of an initial grey-scale image into three distinctive regions: background, border, and undecided. The first step identifies the background using a modified nonlinear range filter followed by a standard morphological dilation. This recognizes regions that do not vary much and yet are large enough to be part of a cell. The second step identifies border regions that are not background or part of a cell, using an extended high-pass filter that operates on the masked image from the first step. Undecided regions are regions yet to be classified. It is also sometimes useful to re-label undecided pixels with extreme values as border regions by global thresholding.
Recognizing that bacterial, yeast and human cells typically have smooth borders, we used a nonlinear range filter with a disk-shaped neighborhood, or a structure element. The disk size has to be larger than the maximum width of cells from all images. Once these range values are calculated, a threshold is chosen such that a pixel with range value less than the threshold will be labeled as 0, an indicator for background. The resulting images are grey-scale with zero values indicating background. If we also assign special meaning to the largest values in the grey-scale image (255 for 8-bit images), as we do (below) for cell borders, we create a hybrid grey-scale/black-white image. In contrast, most other filters will end up with either black-white or grey-scale images only. Consequently, we also have to modify the usual high-pass filter not to use the pixels that take zero values when identifying the border regions in the initial partition. This can be done with a masked neighborhood approach. That is, we exclude all pixels that are masked to be the black-white during filtering, and use only pixels in a neighborhood that are part of the grey-scale image. This mask-based strategy, including the modified range-filter in the previous step, can be applied to other filters (Supplementary Note 2).
In general, we can extend the above ideas to design a class of mask-based filters on hybrid images, as follows. Let H = (I,M) be a hybrid image comprised of a grey-scale image I and a black-white mask M. Let H’= F(H) be a function where the output H’= (I’,M’) is also a hybrid image, created via
| (1) |
Then F is called a hybrid filter if
| (2) |
for all I and M, where.* represents the element-wise product. In other words, H’ only depends on pixel intensity values in I that have corresponding non-zero values in M. With this notation, given any hybrid image H = (I,M), we formally define a specific class of hybrid filters, called hybrid range filters (or HRFs), as follows. Each HRF is defined by a specified range parameter r > 0. For each image pixel (i,j), denote by Ei,j the structure element centered at the pixel, and define Ui,j = I(Ei,j ∩ M) where ∩ is the set intersection operator. Then the HRF map (I’,M’) = F(I,M) is given by
| (3) |
Hence, to calculate the output gray-scale intensity value Ii,j’, the HRF first extracts all the pixels from its neighborhood as defined by the structure element, and then these pixels are further selected according to the corresponding binary mask. The output intensity is then kept the same as input only when the intensity values from the above selected neighborhood fall into the range defined by r. The output binary mask Mi,j’ is simply set to 1 when the corresponding grey-scale intensity is non-zero.
In addition to range filters, we can design hybrid median, mean, or rank filters, in an obvious extension of the above definition of range filters. There are several advantages to these hybrid filters. First, both black-white and grey-scale images can be treated as special cases of the hybrid images by using proper masks, thus avoiding the need to distinguish the types of input images. For a pure grey-scale image, the mask M is a matrix of ones and for a pure black-white image M is a zero matrix of the same size as I. Second, as the input and the output images are always of the same type, we can combine various types of filters sequentially to address different filtering needs. Third, instead of being forced to threshold the grey-scale image into black-white at some point of the image processing all at once, we can gradually threshold the image in an iterative and selective way and slowly turn an input grey-scale image to an output black-white image. This advantage will be demonstrated in the segmentation step below.
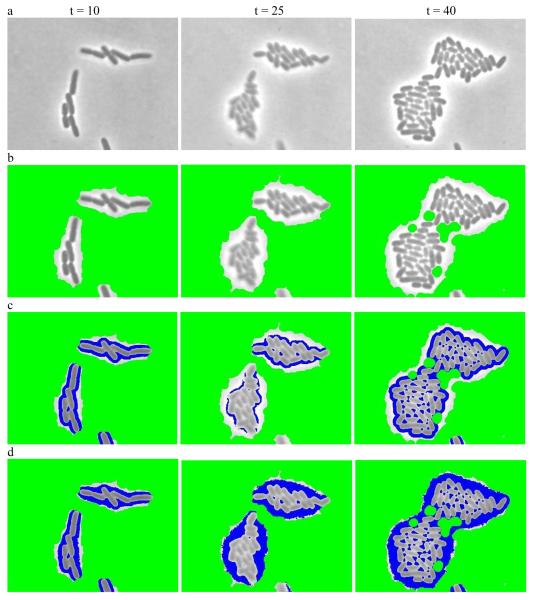
Figure 1 gives an example of applying hybrid filters to preprocess a raw, grey-scale image. This process generates a hybrid image by classifying some pixels as background and others as cell borders while keeping remaining regions for further segmentation. Figure 1a shows three images of engineered E. coli bacteria programmed via a synthetic gene circuit (Supplementary Note 1). The cells are generally represented by darker pixels and surrounded by bright borders while the grey regions are background. Figure 1b is the result of applying the hybrid range filter to identify background as colored in green. Figure 1c is the result of applying the hybrid high-pass range filter to Figure 1b to identify border regions, colored in blue. Figure 1d is the result of further applying a global threshold filter to mark more pixels as borders. Note that this preprocessing just gives an initial partition for the input images, and may or may not be needed for every data set analysis.
Figure 1.
Preprocessing to convert the input grey-scale phase image into a hybrid image. (a) Sample grey-scale input images from E. coli experiment at time point t = 10,t = 25 and t = 40 respectively. (b) Resulting hybrid images after applying the modified range filter to identify initial backgrounds (colored in green). (c) Resulting hybrid images after applying the high-pass filter to (b) to further identif border regions (colored in blue). (d) the resulting hybrid images after applying a threshold filter to (c) to mark more pixels as borders.
Segmentation with iterative/selective masked erosion and dilatation
Segmentation identifies groups of pixels as cell objects. A common challenge with any algorithm is over- or under- segmentation (27,28,29). We approach this issue using an iterative algorithm that couples our hybrid filters with hybrid smoothing and dilation. We extend the concept of hybrid filtering to image smoothing and erosion/dilation (Supplementary Note 2). The hybrid image from the preprocessing step is input to analysis that applies a hybrid filter, or a combination of filters, to erode the undecided regions, followed by hybrid dilation and smoothing to restore most of the pixel values changed by the filter. The overall result is a subtle change in the mask associated with the hybrid image. We iteratively repeat this process, possibly with slightly more aggressive parameter settings, to gradually change the associated mask until a stopping rule is met (Supplementary Note 4). This final mask associated with the hybrid image is the resulting segmented image.
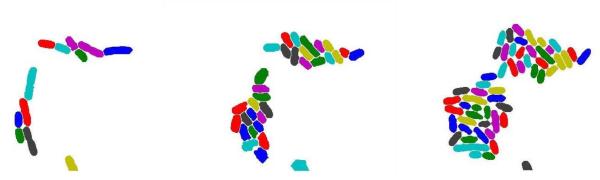
We enhance the above strategy by adding a cell object modeling and selection step at the end of each iteration. Recognizing that cells have common morphological features (shape, size, smoothness) we define cell or object shape models. Disconnected segmented regions – referred to as blobs – in the binary mask are labeled, evaluated, scored and then classified into two groups based on the object shape model. Here we apply this method to set of data generated with engineered E. coli carrying a population control circuit (31) (Supplementary Note 1). This data set is challenging in that many bacteria are elongated due to the circuit function and that there is great diversity in the sizes of closely clustered individual bacteria. With E. coli data, the cell object model assumes a straight or curved cigar shape and a minimum pixel volume; in the example of Figure 1, all blobs are classified as either cells or undecided. Undecided blobs are subject to further segmentation and cells are considered done with no further segmentation performed. This approach proves to be very useful especially when the cells exhibit diversity in scale, e.g., cells differing in diameter, in which case cells at different scales will be picked up at different iterations. More details appear in the Methods section and Supplementary Note 4.
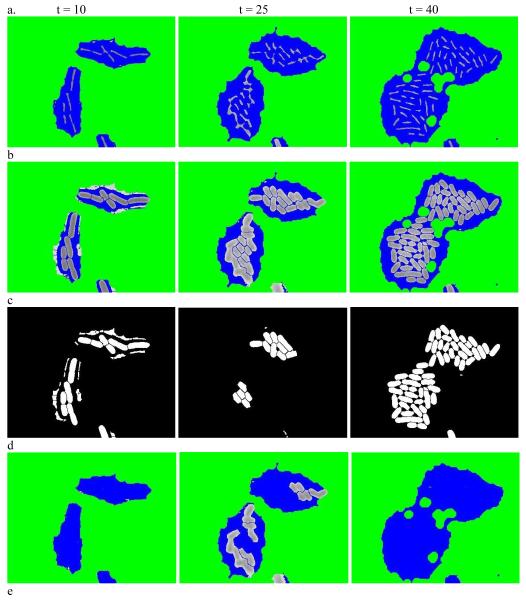
We illustrate this approach in Figure 2 for the dataset used in the preprocessing step above. Figure 2a shows the result of using a hybrid rank filter with disk-shape structure element to the hybrid images in Figure 1d. Figure 2b shows the hybrid image after further smoothing, thickening, and re-smoothing to Figure 2a. Many larger blobs in Figure 1d are broken down into smaller blobs. By thresholding, all blobs in Figure 2b are classified into two subsets as shown in Figure 2c and Figure 2d. Figure 2c shows the binary masks of the blobs with lower scores that will not be further segmented and Figure 2d shows the blobs with higher scores and will be subject to further segmentation. Figure 2e is the final segmentation (cell coloring is arbitrary and only for visuals).
Figure 2.
Iterative segmentation. (a) Result of using a hybrid quantile filter on images in Figure 1d. (b) Hybrid images after further smoothing, thickening and re-smoothing to (a). (c) The binary masks of the blobs with lower scores in (b). (d) The blobs with higher scores in (b). (e) The final segmentation result with (arbitrary) cell coloring for visual clarity.
The same approach for yeast and human cells uses a slightly different cell object shape model. The main difference is that we assume cells have convex or nearly convex shapes, instead of the “bendy” cigar shape. Detailed description of this model can be found in Supplementary Note 6. Though designed for yeast and human cells, this model works equally well on bacterial examples, illustrating the robustness of the method.
Once the iterations terminate, cells are labeled by recognizing disconnected blobs in the binary mask image. Basic quantities such as shape, size, boundary, and centroid can then be measured using techniques such as described in (20).
Cell lineage reconstruction through neighborhood-based scoring
Segmentation generates a sequence of images with objects segmented in each. To track cells over time and to reconstruct lineage trees, we use a two-step algorithm to first construct score matrices for all cells in each pair of consecutive images and then apply an optimization algorithm to obtain the frame-to-frame correspondence matrices. Suppose that image t has m cells and image t+1 has n cells. A forward score matrix (m by n) and backward score matrix (n by m) describe the likelihood of a cell in image t corresponding to a cell in image t+1 and that of a cell in image t+1 corresponding to a cell in image t, respectively. These matrices are further combined and transformed into a 0-1 correspondence matrix with 1 indicating a correspondence between a cell in image t and a cell in image t+1. Due to cell growth, death, or division, a cell in image t can correspond to 0, 1 or 2 cells in image t+1.
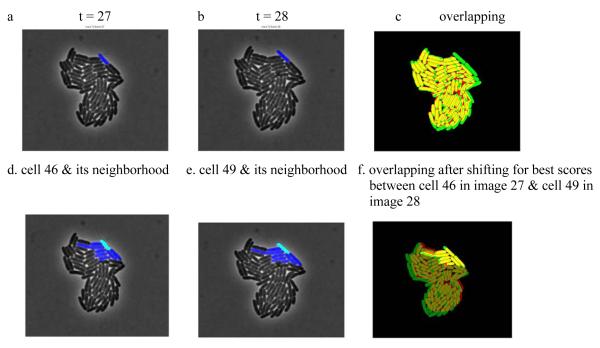
To construct forward and backward scores for each pair of cells, we calculate geometric overlapping scores for the pair. We then locate all neighbors of each cell in each image and compute a pair of neighborhood scores over the neighborhood of each cell using the overlapping scores. By repeatedly shifting one image slightly, we recalculate overlapping and neighborhood scores for cell pairs and record the final scores as the largest combined neighborhood score over all shifts within a predefined distance. We make these scores more robust by using a threshold when calculating the neighborhood scores so that small overlapping scores are not used to calculate final scores. Note that we do not specify a cell object model for the tracking algorithm. Instead, only the area representing a cell is used in the algorithm. By design, this neighborhood-based approach introduces robustness to image shifting during image capturing or cell movement. Figure 3 provides an example for calculating the score for a pair of E. coli cells, using data of Rosenfeld et al. (4). Figures 3a and 3b show two images at consecutive times with the pair of cells labeled in blue. Figure 3c is the result if we overlay the segmented images. This will not yield an acceptable score as two highlighted cells do not overlap at all. Instead, to construct a pair of scores for the cell at these two times, the neighborhoods for these cells are first located as shown in Figure 3d and 3e respectively. Then by shifting image 3b around, the best scores are obtained at the overlapping position as shown in Figure 3f. The bright yellow indicates the cells used to construct the scores.
Figure 3.
Neighborhood based cell tracking. (a,b) Two images at consecutive times with the same cell labeled in blue.
(c) Result of overlaying the segmented images from a and (b). (d,e) Neighborhood of the cell identified in (a) and (b) respectively, labeled in blue and cyan. (f) The overlapping position where the best scores are obtained for the labeled cells in and (b).
To transform the forward and backward score matrices into a single correspondence matrix, we start by noting that a cell in image t can disappear (die), grow, or divide into two cells in image t+1. So a cell in image t can only correspond to 0, 1, or 2 cells in image t+1. We also assume that a cell in image t+1 can correspond to at most 0 or 1 cells in image t, which restates the fact that a cell is an orphan or a descendent of a cell in image t. We then combine the two score matrices using dot product and calculate the row and column maxima in the combined score matrix; our algorithm then scans the combined score matrix to locate values that are both row and column maxima and assigns matches for corresponding cell pairs. During this process, if more than two cells in image t+1 can be assigned to one cell in image t the backward score matrix is referenced so that the two cells with best scores are used to assign the matches. We repeat this process while excluding the cells in image t+1 that are already matched and enforcing the cell growth event restraints until no more correspondences can be found. Once the correspondence matrices for all consecutive images are generated, we reconstruct the cell lineage trees by linking the sequence of correspondence matrices. Individual tracks can be further obtained by back-tracking the cells identified in the last frame to the cells in the first frame from the lineage trees.
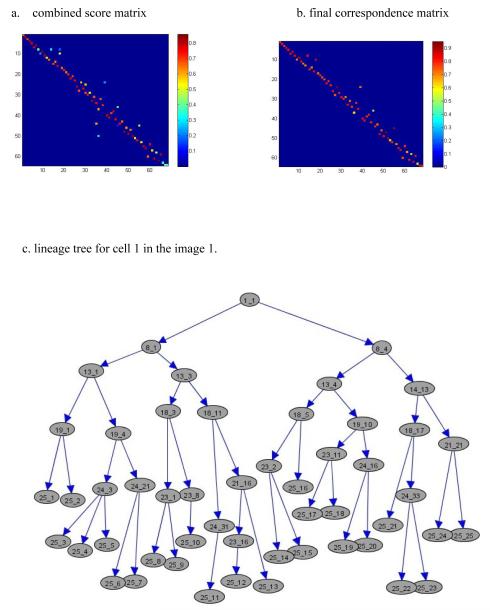
Figure 4a shows the combined score matrix for Figure 3, displayed as a heat map. Figure 4b shows the resulting correspondence matrix heat map after applying our optimization algorithm. This can be converted into a 0-1 correspondence matrix by identifying those scores above a specified threshold. From the image, we notice from the columns that each cell corresponds to only one cell from the previous image; similarly, from the rows we see that each cell corresponds to one or two cells (cell splitting) in the following image. Figure 4c shows a complete lineage tree for cell 1 in images 1 through 25 (displayed using GraphExplore: www.stat.duke.edu/research/software/west/graphexplore.html). The numbers in each node represent the image index and the cell index, respectively.
Figure 4.
Cell tracking and lineage tree reconstruction. (a) The combined score matrix displayed as a heat map, with warm color indicating good matching. (b) Heat map for the final correspondence matrix. (c) Complete lineage tree for cell number 1 tracked in images 1-25.
Comparison with existing algorithms
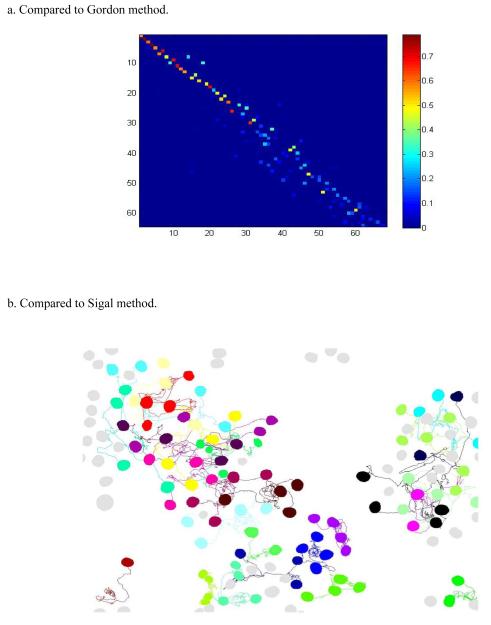
Sigal et al. (26), Gordon et al. (20), and Charvin et al. (30) demonstrated segmentation/tracking results on human and yeast cells using the Cell-ID, CellProfiler and other tools. While complete yeast segmentation/tracking results are not available from (20), Figure 5a provides the correspondence heat map analogous to Figure 4a using the methods in (20). Compared to Figure 4a, it shows more small but non-zero values (in light blue) and fewer large values (in bright red). It is evident that our neighborhood score based tracking method provides a much sharper score matrix with substantially improved cell correspondences as indicated by red/orange pixels. The Sigal et al. study provided human cell imaging data in terms of supplementary movies as well as summaries of cell tracking results. This human data example provides a nice context for a side-by-side comparison with our method. Figure 5b demonstrates all 97 tracks reconstructed using our approach as compared with only 57 reported in (26). Further investigation based on our full tracking result suggests that there are at most 99 actual tracks from the last frame back to the first frame in the movie. i.e., our algorithm reconstructs almost 98% of actual tracks in the movie. Importantly, our analysis was a direct application of the automated method applied to bacterial data with no additional parameter tuning. More details on analysis of this data set, as well as additional examples using budding yeast data, can be found in Supplementary Note 6.
Figure 5.
Comparison with other leading algorithms. (a) Heat map of score matrix analogous to Figure 4(a) using the method of (20). (b) Tracking result for human cells compared to that of (26).
DISCUSSION
Our results demonstrate the utility and portability of our cell segmentation and tracking analysis for single-cell images of bacterial, yeast and human cells. The method applies similarly to other cell types using cell object shape models. Whatever the cell type, the automated, iterative algorithm for segmentation provides a novel and accurate way to extract cell information from time-course images. The concept of hybrid images and hybrid image based filters can be readily ported to existing tools that are designed purely for grey scale or black-white images. The neighborhood-based score function provides an efficient way to track cells through time. By using very common cell features and avoiding modeling cell growth transformations, our approach is robust and has good portability for deployment in other studies. By thresholding and using only higher scores when calculating correspondence, we can easily make the tracking algorithm iterative to deal more complicated cases and design algorithms that allowing skipping a frame for cell correspondence.
Common methods such as background filtering, contrast enhancement, iterative morphological operations are of proven utility in image segmentation. However, effectively combining such methods, as several prior studies have done, is challenging as these operators each apply to the entire image under investigation without discrimination and thus often create undesired side effects. Our hybrid image-based operators allow us to take advantage of these useful methods but in such a way as to minimize these undesired side effects.
Depending on the types of images to be analyzed, the image preprocessing step can be vital to the success of segmentation. In the contexts here, image preprocessing is tuned to work specifically with cells that have smooth shapes and that are generally represented by pixels that are darker or brighter than the surrounding environment, which for example, might not be the case for DIC images. However, the general strategy can be ported to deal with other cell types based on alternative assumptions about distinctive border and inner regions.
Our iterative approach for cell segmentation gives another example of deploying different operators defined on hybrid images to selectively target the unclassified regions of images while leaving well classified regions untouched. This approach is especially useful when the cells in the images have different scales or distinctive subclass features. Compared to commonly used global and adaptive threshold methods, iterative morphological erosion/dilation, or watershed based methods for segmentation, our analysis across a range of examples is uniformly effective in terms of overcoming the common pitfalls of over- or under- segmentation.
A core issue of cell tracking is to determine the correspondences between cells in two consecutive images. Some assumptions have to be made about the image acquisition rate and also about the cell growth rate to have a well defined problem. Low image sampling rate, high cell growth rate, or both will make tracking nearly impossible without strong assumptions. Even for well-behaved data, however, finding correspondences between cells across images is an NP-complete problem. This problem is further complicated by poor segmentation and various transformations and spatial mappings due to cell growth such as change of shapes, cell division and also imperfect image acquisition techniques. Some of the best methods currently available use joint estimation of correspondence and spatial mappings via optimization of energy functions (32). The success of such approaches is heavily dependent on the design of energy function and the choice of optimization technique as well as the choice of mapping functions and cell features. Other approaches use more formal statistical models to estimate the likelihoods of cell death and division between frames and consolidate these likelihood values into correspondence using integer programming optimizations (33).
Our approach has built on the best characteristics of these general strategies. We first derive a score that uses only one robust feature extracted from cells together with the neighborhood information. Then our greedy search algorithm consolidates the scores into cell correspondences. To construct the score, we intentionally avoid using the popular cell feature candidates such as volume, orientation, centroid, and major and minor axis. Instead, we used geometric overlapping as a basis for our score. However, some obstacles have to be overcome to use this feature successfully. First, the standard overlapping measure is symmetric for two involved images and does not deal with cell division events correctly. We address this issue by using forward and backward overlapping measures, which turns out to be critical for successful tracking of budding yeast cells. Second, systematic shifts of image or local movements of cells can cause standard overlapping measure to be unstable. We address this issue by shifting one image around and finding the best local neighborhood match before deciding the actual overlapping measure. The resulting overlapping measure is robust to these obstacles and can be easily applied to other cell images. As a result, there is very little parameter tuning for three different types of cells we analyzed here. To further test our tracking algorithm, we have rerun analyses by skipping every other frame in the image sequences and have found that there is little change in tracking results for E. coli and human cells. In the case of budding yeast cells, however, we did observe more tracking mistakes at times when daughter cells were just separated from mother cells.
METHODS
Initial partition to convert grey-scale image into hybrid image
The input grey-scale phase images are first preprocessed to generate hybrid images for further segmentation. To identify the initial background, we customized the range filter from the Matlab image processing toolbox to use a disk shaped structure element in observing the fact that all the cells under consideration have rather smooth shapes. We then scan all image frames to generate an initial estimate of the maximum half width, r, of all cells and choose the radius of the disk structure element to be between r and 2r. After applying the range filter, a fixed threshold is chosen so that in any image, a pixel with range value less than the threshold will be marked as 0 for background. While there might be optimal values for such parameters to get best initial background partition, we find this usually gave satisfying initial results after a few preprocessing attempts. The resulting hybrid images are then fed into a modified high-pass range filter to identify the cell border regions. The high-pass range filter is modified in two ways: we use the disk shaped structure element as we did for background, and we exclude all background pixels when recording the minimum value within the neighborhood of each pixel. We choose a value for this threshold so that only the very obvious border pixels are marked as 255 for border in our 8-bit input images.
Iterative cell segmentation
Iterative cell segmentation in general involves the following steps: (1) Label disconnected blobs in the associated binary masks from hybrid images. (2) Filter each labeled blob using hybrid filters to obtain a refined estimate, which usually will break down some undesired blobs into smaller blobs. (3) Score the refined blobs based on the cell shape model assumptions. Some example cell shape models can be found in Supplementary Notes 3 and 6. (4) Threshold all blobs into two subsets using a user chosen score threshold. (5) Keep the subset of blobs with lower scores unchanged and repeat (1) through (4) until no more blobs can be broken down and the resulting binary mask is taken as the segmentation result. For our example datasets, step (2) is further detailed as follows: (2a) For each blob in the subset with higher scores, a modified rank filter with disk-shape structure element is applied to the corresponding grey-scale blob to identify the pixels with higher intensity values and to erode them from the corresponding binary mask; (2b) The eroded mask blob is then further smoothed by applying a distance transform to the inverted mask,which caculates the distance between each pixel that is set to zero and the nearest nonzero pixel in the mask, and then thresholding it; (2c) The smoothed mask is dilated through a morphological thickening with respect to the original binary mask in (2a). As a result of (2a) through (2c), many blobs with higher scores will break down into smaller blobs and yield further segmented hybrid images. We have also developed alternative procedures in the software for step 2 that user might find more effective for specific dataset or cell types.
Construction of forward and backward score matrices
We use the strategy described earlier to construct the forward and backward score matrices. Our realization of this strategy uses the following implementations. At time t, we define the forward geometric overlapping score for cell i in image t and cell j in image t+1 as the ratio of number of pixels in cell i divided by the total number of pixels in both cell i and cell j. Similarly, we define the backward geometric overlapping score for cell j in image t+1 and cell i in image t as the ratio of number of pixels in cell j divided by the total number of pixels in both cell i and cell j. We regard a cell i is in the neighborhood of a cell j if they overlap at least once if we shift cell i within given pixels p, where p controls the maximum neighborhood size. The threshold we chose to decide if a geometric overlapping score will be counted as valid is 0.2. This threshold can be set to a higher value if cells do not move or change shape a lot. The combined neighborhood score is the square of the sum of the forward and backward scores.
Correspondence matrix
We transform the score matrices into correspondence matrix using the greedy approximation algorithm implemented in CellTracer, with implementation details provided in Supplementary Note 5.
Alternative implementations
We have presented one particular implementation of our overall strategy and resulting algorithm, in a form that works well on all three types of testing data sets – bacterial, yeast and mammalian cells – and based on phase, bright field or fluorescent images. It is however understood that improved segmentation results, or more efficient algorithms, may be achieved by tailoring the workflow to more specific features of a data set. Our emphasis is on robustness and portability, but we have detailed the opportunities to further customize the approach, if desired, in supporting material and describe some such customization in explicit detail in examples at the software website.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to the editor and anonymous referees for constructive comments; to M. Elowitz and N. Rosenfeld for discussion and provision of a data set (Figure 3, Supp. Note 2); to Philippe Marguet for a data set (Supp. Note 4); and to Eric Siggia for a yeast data set (Supp Note 6). Research was partially supported by the National Science Foundation (DMS-0342172, BES- 0625213) and National Institutes of Health (NCI U54-CA-112952, P50 GM081883-01). Any opinions, findings and conclusions or recommendations expressed in this work are those of the authors and do not necessarily reflect the views of the NSF or NIH.
REFERENCES
- 1.Megason SG, Fraser SE. Imaging in Systems Biology. Cell. 2007;130:784–795. doi: 10.1016/j.cell.2007.08.031. [DOI] [PubMed] [Google Scholar]
- 2.Longo D, Hasty J. Dynamics of single-cell gene expression. Molecular Systems Biology. 2006;2:64. doi: 10.1038/msb4100110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression on a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 4.Raser JM, O’Shea EK. Control of stochasticity in eukaryotic gene expression. Science. 2004;304:1811–1814. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenfeld N, Young JW, Alon U, Swain PS, Elowitz MB. Gene regulation at the single-cell level. Science. 2005;307:1962–1965. doi: 10.1126/science.1106914. [DOI] [PubMed] [Google Scholar]
- 6.Levine M, Davidson EH. Gene regulatory networks for development. Proc. Natl. Acad. Sci. USA. 2005;91:4936–4942. doi: 10.1073/pnas.0408031102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Süel GM, Gracia-Ojalvo J, Liberman LM, Elowitz MB. An excitable gene regulatory circuit induces transient cellular differentiation. Nature. 2006;440:545–550. doi: 10.1038/nature04588. [DOI] [PubMed] [Google Scholar]
- 8.Colman-Lerner A, et al. Regulated cell-to-cell variation in a cell-fate decision system. Nature. 2005;437:699–706. doi: 10.1038/nature03998. [DOI] [PubMed] [Google Scholar]
- 9.Rosenfeld N, Perkins TJ, Alon U, Elowitz MB, Swain PS. A fluctuation method to quanlity In Vovo Fluorescence Data. Biophysical Journal. 2006;91:759–766. doi: 10.1529/biophysj.105.073098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kask P, Palo K, Ullmann D, Gall K. Fluorescence-intensity distribution analysis and ites application in biomolecular detection technology. Porc. Natl. Acad. Sci. USA. 1999;96:13756–13761. doi: 10.1073/pnas.96.24.13756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Golding I, Paulsson J, Zawilski SM, Cox EC. Real-time kinetics of gene activity in individual bacteria. Cell. 2005;123:1025–1036. doi: 10.1016/j.cell.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 12.Mihalcescu I, Hsing W, Leibler S. Resilient circadian oscillator revealed in individual cyanobacteria. Nature. 2004;430:81–85. doi: 10.1038/nature02533. [DOI] [PubMed] [Google Scholar]
- 13.Chabot JR, Pedraza JM, Luitel P, van Oudenaarden A. Stochastic gene expression out-of-steady-state in the cyanobacterial circadian clock. Nature. 2007;450:1249–1252. doi: 10.1038/nature06395. [DOI] [PubMed] [Google Scholar]
- 14.Süel GM, Kulkarni RP, Dworkin J, Garcia-Ojalvo J, Elowitz MB. Tunability and Noise Dependence in Differentiation Dynamics. Science. 2007;23:1716–1719. doi: 10.1126/science.1137455. [DOI] [PubMed] [Google Scholar]
- 15.Weinberger LS, Dar RD, Simpson ML. Transient-mediated fate determination in a transcriptional circutof HIV. Nature Genetics. 2008;40:466–470. doi: 10.1038/ng.116. [DOI] [PubMed] [Google Scholar]
- 16.Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 17.Kaufmann BB, Yang Q, Mettetal JT, van Oudenaardem A. Heritable stochastic switching revealed by single-cell genealogy. PloS Biol. 2007;5(9) doi: 10.1371/journal.pbio.0050239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guido NJ, et al. A bottom-up approach to gene regulation. Nature. 2006;439:856–860. doi: 10.1038/nature04473. [DOI] [PubMed] [Google Scholar]
- 19.Austin DW, et al. Gene network shaping of inherent noise spectra. Nature. 2006;439:608–611. doi: 10.1038/nature04194. [DOI] [PubMed] [Google Scholar]
- 20.Gordon, et al. Single-cell quantification of molecules and rates using open-source microscope-based cytometry. Nat. Methods. 2007;4:175–181. doi: 10.1038/nmeth1008. [DOI] [PubMed] [Google Scholar]
- 21.Bao Z, et al. Automated cell lineage tracking in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2006;103:2707–2712. doi: 10.1073/pnas.0511111103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gor V, Elowitz MB, Bacarian T, Mjolsness E. Tracking cell signals in fluorescent Images. Proc. IEEE Computer society conference on Computer Vision and pattern recognition. 2005 [Google Scholar]
- 23.Carpenter AE, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wessels D, Kuhl S, Soll DR. Applications of 2D and 3D DIAS to motion analysis of live cells in transmission and confocal microscopy imaging. Methods Mol. Biol. 2006;346:261–279. doi: 10.1385/1-59745-144-4:261. [DOI] [PubMed] [Google Scholar]
- 25.DiTalia S, Skotheim JM, Bean JM, Siggia ED, Cross FR. The effects of molecular noise and size control on variability in budding yeast cell cycle. Nature. 2007;488:947–951. doi: 10.1038/nature06072. [DOI] [PubMed] [Google Scholar]
- 26.Sigal A, et al. Dynamic proteomics in individual human cells uncovers widespread cell-type dependence of nuclear proteins. Nat. Methods. 2006;3:525–531. doi: 10.1038/nmeth892. [DOI] [PubMed] [Google Scholar]
- 27.Chen X, Zhou X, Wong STC. Automated segmentation, classification, and tracking of cancer cell numlei in time-lapse microscopy. IEEE Trans. Biomed. Eng. 2006;53:762–766. doi: 10.1109/TBME.2006.870201. [DOI] [PubMed] [Google Scholar]
- 28.Soille P, Vincent L. Watersheds in digital spaces: an efficient algorithm based on immersion simulations; IEEE Trans. Patt. Anal. Mach. Intell; 1991. pp. 583–598. [Google Scholar]
- 29.Dzyubachyk O, Niessen W, Meijering E. Advanced Level-Set Based Multi-Cell Segmentation and Tracking in Time-Lapse Fluorescence Microscopy Images; IEEE International Symposium on Biomedical Imaging --ISBI 2008; 2008. pp. 185–188. [Google Scholar]
- 30.Charvin G, Cross FR, Siggia ED. A microfluidic device for temporally controlled gene expression and long-term fluorescent inaging in unperturbed dividing yeast cells. PloS One. 2008 doi: 10.1371/journal.pone.0001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.You L, Cox RS, III, Weiss R, Arnold FH. Programmed population control by cell-cell communication and regulated killings. Nature. 2004;428:868–871. doi: 10.1038/nature02491. [DOI] [PubMed] [Google Scholar]
- 32.Al-Kofahi O, et al. Automated cell lineage construction: a rapid method to analyze clonal development established with murine neural progenitor cells. Cell Cycle. 2006;5(3):327–335. doi: 10.4161/cc.5.3.2426. [DOI] [PubMed] [Google Scholar]
- 33.Shen H, et al. Automated tracking of gene expression in individual cells and cell compartments. Journal of The Royal Society Interface. 2006;3:787–794. doi: 10.1098/rsif.2006.0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.