Abstract
Wounds created in the oral cavity heal rapidly and leave minimal scarring. We have examined a role of a previously isolated cDNA from oral wounds encoding wound inducible transcript-3.0 (wit3.0), also known as fibroblast growth factor receptor 1 oncogene partner 2 (FGFR1OP2). FGFR1OP2/wit3.0 was highly expressed in oral wound fibroblasts without noticeable up-regulation of α-smooth muscle actin. In silico analyses, denaturing and nondenaturing gel Western blot, and immunocytology together demonstrated that FGFR1OP2/wit3.0 were able to dimerize and oligomerize through coiled-coil structures and appeared to associate with cytoskeleton networks in oral wound fibroblasts. Overexpression of FGFR1OP2/wit3.0 increased the floating collagen gel contraction of naïve oral fibroblasts to the level of oral wound fibroblasts, which was in turn attenuated by small-interfering RNA knockdown. The FGFR1OP2/wit3.0 synthesis did not affect the expression of collagen I as well as procontractile peptides such as α-smooth muscle actin, and transforming growth factor-β1 had no effect on FGFR1OP2/wit3.0 expression. Fibroblastic cells derived from embryonic stem cells carrying FGFR1OP2/wit3.0 (+/−) mutation showed significant retardation in cell migration. Thus, we postulate that FGFR1OP2/wit3.0 may regulate cell motility and stimulate wound closure. FGFR1OP2/wit3.0 was not up-regulated during skin wound healing; however, when treated with FGFR1OP2/wit3.0 -expression vector, the skin wound closure was significantly accelerated, resulting in the limited granulation tissue formation. Our data suggest that FGFR1OP2/wit3.0 may possess a therapeutic potential for wound management.
An excisional wound created in skin does not immediately close without surgical assistance and often results in scar formation.1,2 Responding to wound-induced cytokines and growth factors, a subset of skin fibroblasts undergoes transdifferentiation and acquires the expression of α-smooth muscle actin (α-SMA). These α-SMA-expressing myofibroblasts are believed to be responsible for the late stage wound contraction.3–5 On the contrary, clinical observations and animal studies consistently demonstrate that wounds in the oral cavity heal with minimal scarring. The inflammatory response in wounded oral mucosa is less intensive than in skin,6 and the expression of cytokines is also low in oral wounds during the healing period.7–11 Saliva contains a group of cytokines,12,13 which may provide a unique oral environment suitable for wound healing. However, skin grafts transposed into the oral cavity maintain the skin wound healing phenotype,14 indicating that constitutive oral cellular phenotypes, not the oral environment, may play the primary role in the oral wound healing.
It has been pointed out that there are similarities between oral and fetal skin wound healing. Uniquely, both wounds rapidly reduce the wound size through approximation of wound margins during early healing stages.11,15–18 Wounded fetal skin epithelial cells do not proliferate but exhibit the unique “purse-string” contraction, which primarily contributes to the rapid wound closure.19 Oral wound epithelium undergoes active proliferation and migration, and does not appear to initiate the “purse-string” wound closure. Thus, the rapid and spontaneous oral wound closure must be achieved by other mechanisms. A genome-wide gene expression study using cDNA microarray revealed various distinctive gene expression patterns among fetal fibroblasts as well as adult fibroblasts isolated from different sites. The supervised hierarchical clustering analyses indicated that the gene expression profile of fetal fibroblasts was closest to that of oral fibroblasts.20 Oral fibroblasts are originated from the neural crest-derived ectomesenchymal cells, and have been postulated to play a key role in the accelerated oral wound closure.
Previously, we have isolated a unique cDNA from tooth extraction wound tissue encoding wound inducible transcript-3.0 (wit3.0).21 The GenBank database search has identified that wit3.0 is fibroblast growth factor receptor-1 oncogene partner-2 (FGFR1OP2). The FGFR1OP2/wit3.0 allele is composed of seven exons and generates two isoforms of 215-amino acid (FGFR1OP2/wit3.0α) and 253-amino acid (FGFR1OP2/wit3.0β) through alternative splicing of exon 5.22 The amino-terminal sequence of FGFR1OP2/wit3.0 encoded by the first four exons has been shown to facilitate the ligand-independent dimer formation of the FGFR1 tyrosin kinase domain in the cytogenetic abnormality (12;8) (p11; p11p22) (Figure 1A), having caused acute myeloid leukemia.23
Figure 1.
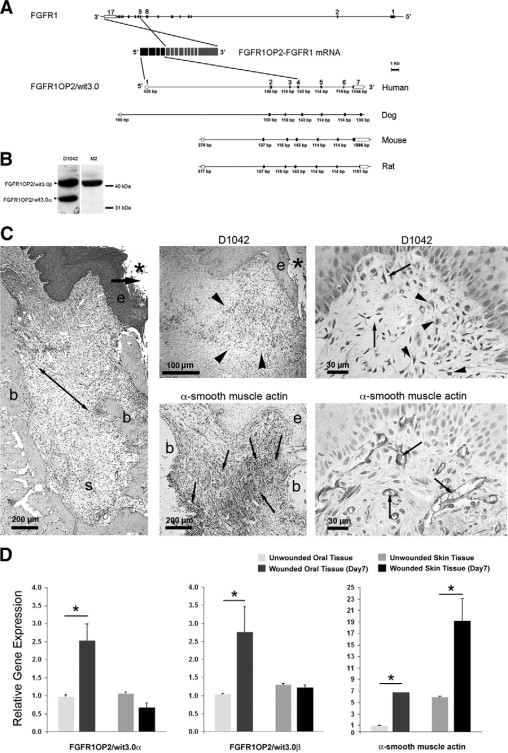
FGFR1OP2/wit3.0 and α-SMA in oral wound. A: Gene structures of FGFR1 and FGFR1OP2/wit3.0. In a case report of acute myeloid leukemia,23 the exons 1 to 4 of FGFR1OP2/wit3.0 were trans-inserted to the intron between exons 8 and 9 of FGFR1, and the resulting chimerical peptide formed a ligand-independent dimer through the FGFR1OP2/wit3.0-derived coiled-coil structure. The native FGFR1OP2/wit3.0 gene contains seven highly conserved exons. B: Western blot analysis of monospecific polyclonal antibody (D1042) recognized 41 kDa and 32 kDa bands, corresponding to FGFR1OP2/wit3.0β and FGFR1OP2/wit3.0α, respectively, in NIH fibroblasts. The transfected 3XFLAG-FGFR1OP2/wit3.0β fusion peptide was recognized by M2 monoclonal antibody against 3XFLAG epitope at 41 kDa. C: Oral open wound (asterisk) created by tooth extraction showed progressive wound contraction (arrows) by wound margin approximation (e: proliferation/migration front of oral epithelium; b: alveolar bone; s: tooth extraction socket, H&E staining). D1042 antibody stained a highly restricted zone of oral connective tissue (arrowheads) immediately adjacent to the migration/proliferation front of oral epithelium (e). α-SMA (small arrows) was identified along the ligament-like structure (double arrowhead in H&E stained section) connecting alveolar bones (b). D1042 stained the cytoplasma of fibroblasts with cuboidal shapes (arrowheads), whereas α-SMA was found in vascular smooth muscles (small arrows). D: Real-time RT-PCR revealed that FGFR1OP2/wit3.0α and FGFR1OP2/wit3.0β were up-regulated in oral wound tissues but not in skin wound tissues, whereas α-SMA was up-regulated in both oral and skin wound tissues albeit at different levels (*P < 0.05).
FGFR1OP2/wit3.0 has been identified as a small cytoplasmic peptide expressed by oral wound fibroblasts22; however, the function of native FGFR1OP2/wit3.0 has not been elucidated. We report here that FGFR1OP2/wit3.0 is a novel cytoskeleton molecule, with a regulatory role in fibroblast migration and rapid wound closure, a hallmark of oral wound healing. The transduction of FGFR1OP2/wit3.0 in mouse skin wounds appeared to induce oral wound-like healing phenotypes: rapid closure and minimal scarring. Our data suggest that FGFR1OP2/wit3.0 may play an important role in favorable oral wound healing and possess a therapeutic potential for wound management.
Materials and Methods
Generation of FGFR1OP2/wit3.0 Monospecific Antibody
The conserved sequence of human, mouse, and rat FGFR1OP2/wit3.0 [45]QYQEEIQELNEVARHRPRS [63] was selected for polyclonal antibody synthesis (Quality Controlled Biochemicals, Hopkinton, MA), resulting in the isolation of D1042. For the specificity evaluation, Western blot of cytoplasmic protein of NIH3T3 fibroblasts transfected with p3x-FLAG-human FGFR1OP2/wit3.0β expression plasmid was analyzed by D1042 antibody and M2 antibody recognizing 3x-FLAG epitope (Sigma-Aldrich, St. Louis, MO).
FGFR1OP2/wit3.0 Immunohistology in an Oral Wound
An oral wound was created by simple tooth extraction from maxilla of adult (8 to 10 weeks old) male Sprague-Dawley rats following the previously described protocol.24 Seven days after extraction of three molars, maxilla were harvested, fixed in 10% buffered formalin and decalcified (Cal-Ex, Fischer Scientific, Pittsburgh, PA). Eight-micrometer thick paraffin sections were generated through the second molar. After the epitope retrieval by using a pressure cooker and Tris Buffer (pH9.0), sections were incubated in primary antibody (D1042, 1:100 to 1:500) and then in horseradish peroxidase conjugated secondary antibody (Dako Envision, Carpinteria, CA). Sections were treated with diaminobenzamine and stained with hematoxylin. The neighboring sections were separately incubated with monoclonal antibody against α-SMA (Clone 1A4, Sigma-Aldrich). Some sections were stained with H&E for references.
Expression of FGFR1OP2/wit3.0 in the Rat Oral and Skin Wounds
Oral mucosal tissue was harvested from a tooth extraction wound (Day 7) or from unwounded alveolus of rat maxilla. The dorsal rat skin was collected from full-thickness excisional wound (Day 7) with a biopsy punch (8 mm in diameter; Skalr Tru-Punch, Sklar Instruments, West Chester, PA) or from the unwounded area. Tissue specimens were immediately frozen in liquid nitrogen and total RNA samples were prepared. The steady state mRNA levels of FGFR1OP2/wit3.0α, FGFR1OP2/wit3.0β, and α-SMA were determined by the Taqman-based quantitative real-time RT-PCR. Student's t test was performed against the unwounded tissue, and statistical significance was accepted for P < 0.05.
In Silico Search on FGFR1OP2/wit3.0 Peptide Structure
The human FGFR1OP2/wit3.0β peptide sequence was submitted to online computational software COILS: Prediction of Coiled Coil Regions in Proteins (http://www.ch.embnet.org/software/COILS_form.html, last accessed August 26, 2006) and the Human Protein Reference Database (http://www.hprd.org/summary?hprd_id=10589&isoform_id=10589_1&isoform_name=Isoform_1, last accessed August 26, 2006) for consensus functional structure searches, and the Imperial College Protein Homology/Analogy Recognition Engine (http://www.sbg.bio.ic.ac.uk/∼phyre/, last accessed August 26, 2006) for three-dimensional molecular structure prediction.
Cell Cultures of Oral Fibroblasts, Oral Wound Fibroblasts, and Skin Fibroblasts
The tissue samples of rat gingiva, tooth extraction wound, and skin were minced in PBS with 1X antibiotic-antimycotics (Invitrogen, Carlsbad, CA) and treated with Collagenase A (200 unit/ml; Sigma-Aldrich) in complete fibroblast culture medium: Dulbecco's modified Eagle's medium, antibiotic-antimycotics solution, and 10% of fetal bovine serum (FBS). After incubation for 5 days, the epithelial cells were separated by centrifugation at 100xg for 3 minutes. The pellet was resuspended with complete fibroblast medium and cultured for an additional 2 weeks. Cells from three to five passages were used in the following experiments.
Denature and Nondenature Gel Western Blot Analyses
Cytoplasmic proteins from oral fibroblasts and oral wound fibroblasts were extracted by using NE-PER Nuclear and Cytoplasmic Extraction Reagent (Pierce, Rockford, IL). For the denatured condition, 3.5 μg of protein samples were mixed with Laemmli loading buffer containing SDS and ß-mercaptoethanol and boiled at 100°C for 10 minutes and loaded on a 12% Tris-HCl Ready Gel (Bio-Rad, Hercules, CA). For the nondenatured condition, the protein sample was mixed with Native Sample Buffer (Bio-Rad) for electropheresis. The protein on the gel was transferred to polyvinylidene difluoride membrane (Bio-Rad) and incubated with D1045 antibody (1:100) for 1 hour at room temperature. After secondary antibody incubation, the membrane was visualized by using a chemiluminescence standard protocol (Pierce) and a CCD imaging system (LAS-3000, Fujifilm, Stamford, CT). In some experiments, the polyvinylidene difluoride membrane was striped and reprobed with the small ubiquitin-like modifier-1 (SUMO-1) or SUMO-2/3 antibodies (Santa Cruz Biotechnology, Santa Cruz, CA).
Immunocytology of Oral Fibroblasts
Oral fibroblasts and oral wound fibroblasts were cultured in glass chamber slides (Lab Tek, Fisher Scientific) in the complete fibroblast culture medium for 24 hours. Cells were fixed with freshly prepared 3.7% formaldehyde (methanol free) followed by the treatment with 0.2% Triton X-100 and Image-IT FX signal enhancer (Invitrogen). The slides were incubated with D1042 antibody (1:100) and then with Alexa Fluor 488-conjugated goat anti-rabbit IgG antibody (1:1000) (Invitrogen). The slides were further treated with Alexa Fluor 568-conjugated phallotoxins and 4′,6-diamidino-2-phenylindole (DAPI). A confocal laser scanning microscope (LSM 310, Carl Zeiss, Novi, MI) was used to scan 1-μm focal layers of each specimen.
Floating Collagen Gel Contraction Assay
Fibroblasts were seeded into the collagen gel and casted in 6-well plates (1.2 × 105 cells/well) following the previously established protocol.22 Once solidified at 37°C, fibroblast/gel was released from the well. The area of fibroblast/gel complex in digitized photographs was measured (Media Cybernetics, Bethesda, MD), and the ratio of collagen gel area against the culture well was calculated at each time point. Repeated measures analysis of variance was used to determine statistical significance for P < 0.05.
Small-Interfering RNA Knockdown of FGFR1OP2/wit3.0
Small-interfering RNAs (siRNAs) targeting the FGFR1OP2/ wit3.0 were designed and generated by commercially available sources (Ambion, Austin, TX; and Dharmacon, Lafayette, CO) (Table 1). Using the optimal siRNA transfection condition for primary oral wound fibroblasts determined by the glyceraldehyde-3-phosphate dehydrogenase siRNA assay (Silencer glyceraldehyde-3-phosphate dehydrogenase siRNA, Ambion), each FGFR1OP2/wit3.0 siRNA candidate was applied to oral wound fibroblasts. The efficiency and specificity of the knock-down effect was determined by Taqman-based real time RT-PCR. The siRNA (number 4 and 5)-treated oral wound fibroblasts were applied to the floating collagen gel contraction assay as described above.
Table 1.
SiRNA Target Sequences for FGFR1OP2/wit3.0
| siRNA | Sequence | Source |
|---|---|---|
| No. 1 | ||
| Sense | 5′-GCAGAGAAGGGCCAUUUAUU-3′ | Dharmacon |
| Antisense | 5′-PUUAAUGGCCCUUCUCUGCUU-3′ | |
| No. 2 | ||
| Sense | 5′-GCAGAACGAGUUGGGAAUAUU-3′ | Dharmacon |
| Antisense | 5′-PUAUUCCCAACUCGUUCUGCUU-3′ | |
| No. 3 | ||
| Sense | 5′-AUAACAAGCUCGCUGCAAAUU-3′ | Dharmacon |
| Antisense | 5′-PUUUGCAGCGAGCUUGUUAUUU-3′ | |
| No. 4 | ||
| Sense | 5′-GGACCAACAGCAUUGAAUUUU-3′ | Dharmacon |
| Antisense | 5′-PAAUUCAAUGCUGUUGGUCCUU-3′ | |
| No. 5 | ||
| Sense | 5′-GGACGACCCGGGCAUAAUA-3′ | Ambion |
| Antisense | 5′-UAUUAUGCCCGGGUCGUCC-3′ | |
| Control | ||
| Sense | 5′-GCAGCAACUGGACACGUGAU-3′ | Invitrogen |
| Antisense | 5′-UCACGUGUCCAGUUGCUGC-3′ |
Mouse Embryonic Stem Cell-Derived Fibroblastic Cells Carrying FGFR1OP2/wit3.0 (+/−) Mutation
From the International Gene Trap Consortium (www.genetrap.org, last accessed December 21, 2008), six mouse embryonic stem (ES) cell lines were identified containing a gene trap mutagenesis25 in the FGFR1OP2/wit3.0 allele (Table 2). C57Bl/6J ES cell line with IST10830D12 mutation (Texas A&M Institute for Genomic Medicine, Houston, TX) and wild-type ES cells were expanded on feeder cells following a standard protocol. The trypsinized ES cells were cultured in nonadherent plates (Corning Inc, Corning, NY) to form aggregates in the fibroblast differentiation medium consisted of knockout Dulbecco's modified Eagle's medium (Invitrogen), 20% FBS, 1 mmol/L l-glutamine, 0.1 mmol/L β-mercaptoethanol, 1% nonessential amino acids, and 1% antibiotic-antimycotics.26 The aggregates were then transferred into gelatin-coated plates and the outgrowing cells were serially passaged every 5 to 7 days.26 Most of the cells in culture appeared fibroblast-like morphology after four passages. The genotype was determined by PCR, and the mRNA levels of collagen I α 2 chain (Mm00483937_m1) and FGFR1OP2/wit3.0 (Mm00470836_m1) were determined by Taqman-based real time PCR.
Table 2.
Available Mouse ES Cell Lines Containing Disrupted FGFR1OP2/wit3.0 Allele (146526432-146547719) through the International Gene Trap Consortium
| Cell line ID | Identification status | Source | Insertion location |
|---|---|---|---|
| CA0175 | Localized + transcript | SIGTR | 146526450-146526731 |
| Q017D08 | Localized + transcript | GGTC | 146526450-146526731 |
| PST26232-NR | Localized only | ESDB | 146527049-146527145 |
| IST10830D12BBF1 | Localized only | TIGM | 146531786-146532310 |
| IST10830D12HMF1 | Localized only | TIGM | 146531799-146532314 |
| IST12138D4BBF1 | Localized only | TIGM | 146544488-146544618 |
SIGTR, Sanger Institute Gene Trap Resource (Cambridge, UK).
GGTC, German Gene Trap Consortium (Germany).
ESDB, Embryonic Stem Cell Database (University of Manitoba, Canada).
TIGM, Texas A&M Institute for Genomic Medicine (Houston, TX).
In Vitro Wound Healing Scratch Plate Assay
The ES cell-derived fibroblastic cells with wild-type and FGFR1OP2/wit3.0 (+/−) genotypes were seeded in 12-well plates and allowed to grow in the fibroblast differentiation medium to approximately 90% confluence. In vitro wounds were created by drawing lines down the well with a 20 μl plastic pipette.27 After washing, cells were maintained in the low serum medium containing 0.5% FBS. The digital images were captured after 6 and 12 hours of incubation, and the area occupied by the cells within the scratch was measured (NIH Image J version 1.42). From the Student's t test performed against the wild-type cells, statistical significance was accepted for P < 0.05.
3xFLAG-Human FGFR1OP2/wit3.0β Lentiviral Vector Carrying Wild-Type Sequence and Nonsynonymous Single Nucleotide Polymorphism (SNPs)
Two nonsynonymous SNPs (rs 1058701 and rs11613) were identified in the exon 5 from the National Center for Biotechnology Information SNP database, and were named SNP1 and SNP2, respectively. Using a site-directed mutagenesis (QuikChange Site-Directed Mutagenesis Kit, Agilent Technologies, La Jolla, CA), three additional cDNA constructs were generated each containing SNP1, SNP2, and both SNP1 and 2. The 3xFLAG-human FGFR1OP2/wit3.0β cDNAs with wild-type sequence as well as SNP1, SNP2, and SNP1/2 sequences were subcloned into the lentivirus-derived vector: pRRLsinhCMV-IG. The construct (pRRLsinCMV-hwit3.0-IG) has an internal ribosome entry site-green fluorescent protein gene downstream of 3xFLAG-human FGFR1OP2/wit3.0β fusion peptide. The construct was used to co-transfect 293T cells together with two packaging vectors, pCMVΔR8.2 and pVSVG. The lentiviral vector was concentrated to five ∼20 μg p24/ml virus stock.
The Effect of FGFR1OP2/wit3.0β Wild-Type, SNP1, SNP2, and SNP1/2 on Gel Contraction
Oral fibroblasts and skin fibroblasts (2 × 105 cells/well) were treated with 1 μg of lentivirus (1 μg p24 = 5 × 107 infectious units in 293T cells) and polybrene (8 μg/ml) for 24 hours. The lentivirus transduction efficiency to oral fibroblasts and skin fibroblasts were evaluated for the internal ribosome entry site-driven green fluorescent protein expression. The effect of FGFR1OP2/wit3.0β overexpression carrying wild-type, SNP1, SNP2, and SNP1/2 sequences on floating collagen gel contraction were determined as described above.
The Effect of Transforming Growth Factor-β1 on the Expression of FGFR1OP2/wit3.0 and α-SMA
Oral fibroblasts and skin fibroblasts were seeded on a 6-well plate (1 × 105 cells/well). After 24 hours of starvation treatment by low serum culture medium (0.5% FBS), cells were incubated in Dulbecco's modified Eagle's medium containing transforming growth factor-β1 (TGF-β1; 0, 0.1, and 0.5 ng/ml). The steady state mRNA levels of FGFR1OP2/wit3.0 and α-SMA were accessed by Taqman-based real-time RT-PCR. The experiments were triplicated. From the Student's t test performed against the untreated cells, statistical significance was accepted for P < 0.05.
Mouse Dorsal Skin Full-Thickness Excisional Wound Model
Under isofluorane inhalation anesthesia, two identical skin wounds were created on the right and left side equidistant (0.5 cm) from the dorsal mid point in male C57BL/6J mice (10 weeks old). Using an 8-mm dermal biopsy punch, full-thickness skin including panniculus carnosus layer was excised. The skin specimens containing the healing excisional wound were collected and examined for the steady state mRNA levels of FGFR1OP2/wit3.0.
Efficacy of Gene Transfer Methods to Mouse Skin Wound
The efficiency of gene transfer to mouse skin wound was compared for plasmid DNA and lentivural vector carrying firefly luciferase reporter gene, which were mixed in either type I collagen (Cell Prime, Cohesion, Palo Alto, CA) or cationized gelatin (CG) SM50 (generous gift from Dr. Tabata, Kyoto University, Japan). Cell Prime or CG mixed with β-galactosidase reporter gene plasmid DNA (1 μg/μl) served as the negative control. Four days after gene delivery, anesthesized mice were injected with 200 μl of filtered D-Luciferince firefly (15 mg/ml in PBS) and luciferase activity in skin wound area was measured by using the fluorescent single-view three-dimensional optical imaging system (Xenogen IVIS System, Caliper Life Science, Hopkinton, MA).
The Effect of FGFR1OP2/wit3.0β Treatment on Excisional Skin Wound Closure In Vivo
After the full-thickness excisional skin wound was created in C57Bl/6J mice, 80 μl of CG/3x-flag-hwit3.0β-WT lentivirus, CG/3x-flag-hwit3.0β-SNP1 lentivirus, or CG/Luciferase-GFP lentivirus was topically applied. The standardized photographs were collected 0, 4, and 7 days after wounding. The skin wound area was measured at each time point (n = 10 in each group). One cm2 of skin tissue samples framing the circular wound were collected for histological analysis (n = 6 for each time point). Six-micrometer thick paraffin sections were made through the center of the wound. The sections were stained with Goldner's trichrome staining or Sirius Red.28 Collagen fiber from the sirius red-stained sections were visualized by using confocal laser scanning microscopy. Immunohistological staining by using D1042 antibody was performed as described above.
Results
Localized Expression of FGFR1OP2/wit3.0 in the Oral Wound Margin
We generated a monospecific polyclonal antibody, D1042, against FGFR1OP2/wit3.0. Western blot analysis of NIH3T3 fibroblasts expression 3XFLAG-FGFR1OP2/wit3.0β fusion peptide indicated that both M2 antibody against 3XFLAG epitope and D1042 recognized the 41 kDa peptide. Because D1042 also recognized 32 kDa peptide, we concluded that D1042 is specific to the putative epitope of 41 kDa FGFR1OP2/wit3.0β as well as 32 kDa FGFR1OP2/wit3.0α (Figure 1B).
Tooth extraction created both oral mucosal wound and alveolar bone wound in rat maxilla. The spatial localization of FGFR1OP2/wit3.0 and α-SMA was evaluated by immunohistology. The oral fibroblasts appear to vary in the expression of FGFR1OP2/wit3.0 and α-SMA, which did not seem to overlap each other. The fibroblasts expressing FGFR1OP2/wit3.0 showed cuboidal shapes and localized within the connective tissue immediately below the epithelial proliferation and migration front (Figure 1C). On the contrary, α-SMA was strongly expressed by vascular smooth muscle cells. The α-SMA positive myofibroblasts were abundant in the bone socket created by tooth extraction and showed elongated shape along the direction of the ligament-like structure attached to the alveolar bone (Figure 1C).
The real time RT-PCR revealed that FGFR1OP2/wit3.0 was significantly up-regulated in the oral wound, whereas wounding of skin tissues did not modulate the expression of FGFR1OP2/wit3.0 (Figure 1D). In both wounded tissues, the steady state mRNA levels of α-SMA were significantly increased albeit at different levels.
FGFR1OP2/wit3.0 has the Characteristics as a Cytoskeleton Molecule
The protein structure databases, Human Protein Reference Database and COIL, predicted that FGFR1OP2/wit3.0β contains two separate coiled-coil structures flanking a center domain (Figure 2A). The Imperial College Protein Homology/Analogy Recognition Engine 3D protein structure in silico analysis29 revealed that α-spectrin/α-actinin and apolipoprotein A-I could predict the FGFR1OP2/wit3.0β structure at the estimated precision levels of 85 to 90%. Both α-spectrin/α-actinin and apolipoprotein A-I contain short antiparallel α-helices separated by reverse turns, resulting in the dimer and polymer formation through coiled-coil structures (Figure 2B).
Figure 2.
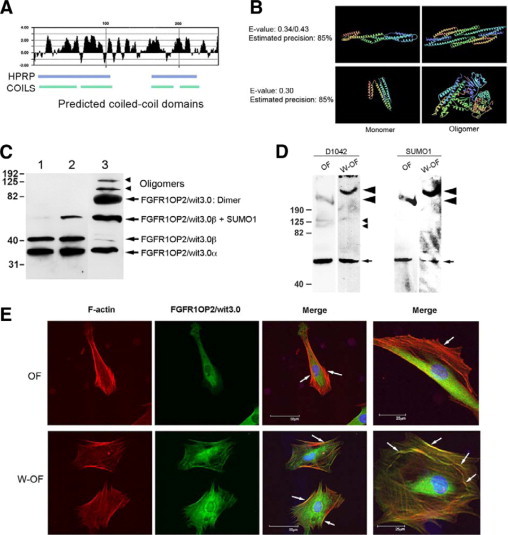
FGFR1OP2/wit3.0 coiled-coil structure and its association with cytoskeletal network in oral wound fibroblasts. A: Hydrophylicity plot of FGFR1OP2/wit3.0β and the coiled-coil domains predicted by the Human Protein Reference Database and COILS. B: The Imperial College Protein Homology/Analogy Recognition Engine predicted that three-dimensional structures of FGFR1OP2/wit3.0β would share similar structures of a-spectrin (top) and apoliprotein A-I (bottom), both molecules were known to form dimers and oligomers. C: In denatured Western blot analysis, D1042 antibody depicted FGFR1OP2/wit3.0α and FGFR1OP2/wit3.0β in NIH3T3 fibroblasts (lane 1), NIH3T3 fibroblasts overexpressing FGFR1OP2/wit3.0β (lane 2), and rat oral wound fibroblasts (lane 3). In oral wound fibroblasts, FGFR1OP2/wit3.0 molecules formed dimers and oligomers that were not affected by the reducing agent, β-melcaptoethanol. D: Nondenature gel Western blot analysis of cytoplasmic peptide of naïve oral fibroblasts (OFs) and oral wound fibroblasts (W-OFs) with D1042 indicated a single band at 50 kDa, which was found modified by SUMO-1 (small arrow). The D1042 positive FGFR1OP2/wit3.0 oligomers at 100 kDa and 160 kDa (small arrowheads) were visible in OFs but not in W-OFs. The involvement of FGFR1OP2/wit3.0 in a large molecular moiety of over 190 kDa (large arrowhead) was also noted in both OFs and W-OFs; however, the size of the large molecular moiety and D1042 signal intensity increased in W-OFs. E: Immunocytology of OFs and W-OFs with D1042 antibody (FGFR1OP2/wit3.0: green) as well as phallotoxins (F-actin: red) and DAPI (nuclei: blue). FGFR1OP2/wit3.0 appeared to aggregate in perinucleic cytoplasma. Whereas stress fibers were formed in thick bundles in OF (white arrows), cuboidal shaped W-OFs formed a thin web of cytoskeleton networks (white arrows). Scale bar = 25 μm.
The denatured gel Western blot analysis showed that NIH3T3 fibroblasts expressed FGFR1OP2/wit3.0α and FGFR1OP2/wit3.0β as single molecules; however, when 3XFLAG-FGFR1OP2/wit3.0β was overexpressed, a 50 kDa product appeared. The 50 kDa FGFR1OP2/wit3.0 positive band was about 10 kDa larger than FGFR1OP2/wit3.0β and was posttranslationally modified with SUMO-1 (data not shown). FGFR1OP2/wit3.0β in oral wound fibroblasts seemed to be extensively sumoylated. In addition, D1042 positive bands were observed at 70 to 82 kDa, 100 kDa, and 150 to 160 kDa, suggesting the formation of dimers and oligomers (Figure 2C). In the nondenature gel Western blot analysis, the D1042 antibody recognized a peptide at 50 kDa that was co-recognized with SUMO-1 antibody. There were D1042-positive 100 kDa and 160 kDa bands in oral fibroblasts but not in oral wound fibroblasts. The involvement of FGFR1OP2/wit3.0 in a large molecular moiety of over 190 kDa was also noted in both oral fibroblasts and oral wound fibroblasts; however, the size of the large molecular moiety and D1042 signal intensity increased in oral wound fibroblasts (Figure 2D).
Oral fibroblasts and oral wound fibroblasts were subjected for immunocytology. The majority of oral fibroblasts exhibited elongated cellular morphology and F-actin stress fibers that were localized in the long axis of cell periphery. FGFR1OP2/wit3.0 was primarily localized in the perinuclear cytoplasma and did not overlap with the F-actin. On the contrary, oral wound fibroblasts were more spread to form cuboidal shapes and developed a fine web of cytoskeleton fiber networks (white arrows in Figure 2E). FGFR1OP2/wit3.0 was found to localize in the wider area of cytoplasm. Both F-actin and FGFR1OP2/wit3.0 appeared to associate with the cytoskeleton fiber network; however, they do not seem to co-localize but are rather seen in a somewhat mutually exclusive pattern (Figure 2E).
siRNA FGFR1OP2/wit3.0 Knockdown Attenuated the Floating Collagen Gel Contraction
Compared with skin fibroblasts, oral fibroblasts exhibited the higher rates of gel contraction activity at 12 hours and sustained its activity until 24 hours. Oral wound fibroblasts showed the greatest gel contraction rates (repeated measure analysis of variance; P < 0.05) (Figure 3A).
Figure 3.
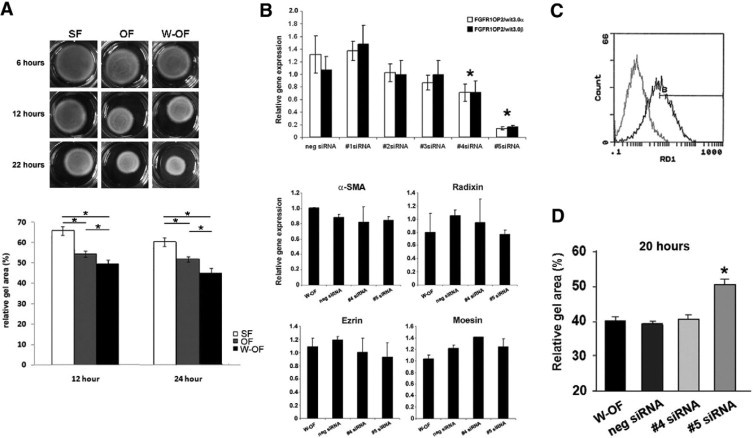
SiRNA knockdown of FGFR1OP2/wit3.0 attenuated collagen gel contraction of oral wound fibroblasts. A: Floating collagen gel contraction assay showed the increased gel contraction by oral wound fibroblasts (W-OFs) as compared with naïve oral fibroblasts (OFs) and skin fibroblasts (SFs) at 12 hours and 24 hours (*P < 0.05). B: Efficacy of five siRNA constructs was evaluated by real time RT-PCR (*P < 0.05 against negative control). The treatment by siRNAs number 4 and number 5 did not affect the expression of contractile and procontractile molecules: α-SMA, radixin, ezrin, and moesin. C: FACS analysis demonstrated the establishment of the optimal transfection protocol for primary oral wound fibroblasts achieving >80% transfection rates (black line) compared with untransfected control (gray line). D: Floating collagen gel contraction assay of oral wound fibroblasts showed that FGFR1OP2/wit3.0 siRNA number 5 significantly attenuated gel contraction (*P < 0.05).
The role of FGFR1OP2/wit3.0 in fibroblast-driven wound contraction was examined in siRNA knockdown experiments. We designed five siRNA target sequences (Table 1), out of which number 5 siRNA showed the 80 to 90% knockdown efficiency followed by number 4 siRNA, which showed 20 to 30% knockdown efficiency (Figure 3B). The treatment of number 4 and number 5 siRNAs did not affect other contractile and procontractile molecules such as α-SMA, ezrin, radixin, and moesin (Figure 3B).
Considering optimal balance factor (transfection efficiency versus cell survival) and the silencing efficiency among the three transfection reagents tested, the optimal transfection efficiency of siRNA to primary oral wound fibroblasts was established: 0.25 μl Lipofectamine 2000 with 6000 cells/well cell density, generating a transfection effect of 92% (Figure 3C).
In collagen gel contraction assay, number 5 siRNA significantly decreased gel contraction rate of oral wound fibroblasts (repeated measure analysis of variance; P < 0.05), which became equivalent to that of unwounded oral fibroblasts. The negative control siRNA and number 4 siRNA showed no effect on the floating collagen gel contraction rates (Figure 3D).
FGFR1OP2/wit3.0 (+/−) Mutation Reduced Fibroblastic Cell Migration
To elucidate the mechanism of reduced FGFR1OP2/wit3.0 expression, migration capability of fibroblasts were evaluated. We applied the available mouse ES cell line carrying the gene trap-mediated FGFR1OP2/wit3.0 knockout mutation. The mouse ES cell line IST10830D12 was generated under the International Gene Trap Consortium and a β-geo cassette was inserted in the first intron of an FGFR1OP2/wit3.0 allele (Figure 4A). Because the translational start site is located in the second exon, this gene trap mutation would create the knockout mutation. Fibroblastic cells carrying FGFR1OP2/wit3.0 (+/−) mutation (Figure 4B) did not affect the collagen I expression but reduced the FGFR1OP2/wit3.0 mRNA level to approximately 50% of the wild-type control (Figure 4C). The in vitro wound scratch plate assay demonstrated that cell migration of FGFR1OP2/wit3.0 (+/−) cells was significantly decreased compared with the wild-type control (Figure 4D).
Figure 4.
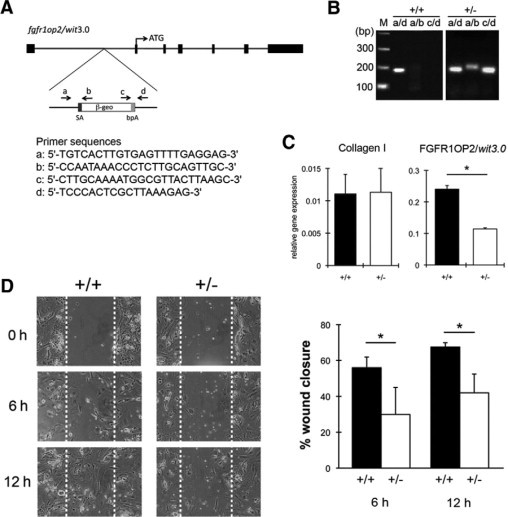
FGFR1OP2/wit3.0 (+/−) mutation reduced the migration ability of mouse ES cell-derived fibroblastic cells. A: Gene Trap mutation was accomplished by inserting a fusion construct of β-galactosidase and neomysin resistant genes (β-geo), which contains splice acceptance sequence (SA) and bovine fibroblast growth factor poly A signal (bpA) into the first intron of mouse fgfrop2/wit3.0 allele. B: Genotype of wild-type (+/+) and FGFROP2/wit3.0 heterogyzous null mutant (+/−) mouse ES cells was determined by PCR. Primers a and d flanked the β-geo construct (Figure 4A) giving rise to the wild-type PCR product (a/d), whereas the combinations of primers a and b (a/b) and c and d (c/d) recognized the β-geo insert. C: Fibroblastic cells derived from wild-type (+/+) and FGFR1OP2/wit3.0 (+/−) mutant ES cells expressed the comparative mRNA level of collagen 1 α 1 chain, whereas the FGFR1OP2/wit3.0 mRNA level was significantly decreased in the mutant cells (*P < 0.5). D:In vitro scratch test revealed that FGFR1OP2/wit3.0 (+/−) mutation significantly decreased fibroblastic cell migration.
The Human FGFR1OP2/wit3.0β Carrying Wild-Type Sequence as well as SNP1, SNP2, and SNP1/2 Sequences Increased Floating Collagen Gel Contraction
We generated three additional FGFR1OP2/wit3.0β cDNAs representing the nonsynonymous SNP sequences in exon 5 (Figure 5A) and subcloned to a third generation lentivirus vector containing internal ribosome entry site-green fluorescent protein (Figure 5B). The transduction efficiencies of the lentivirus to primary oral fibroblasts and primary skin fibroblasts were confirmed over 80% by Fluorescence Activated Cell Sorting (FACS) (Figure 5C). The transduction of the wild-type FGFR1OP2/wit3.0β to both oral and skin fibroblasts significantly increased the floating collagen gel contraction when compared with their untransduced control fibroblasts (repeated measure analysis of variance; P < 0.05) (Figure 5D). In addition, the SNP1 human FGFR1OP2/wit3.0β carrying 154E->D peptide further increased floating collagen gel contraction rates (repeated measure analysis of variance; P < 0.01), whereas SNP2 and SNP1/2 variants showed a significant, but a less drastic, increase (repeated measure analysis of variance; P < 0.05) (Figure 5E).
Figure 5.
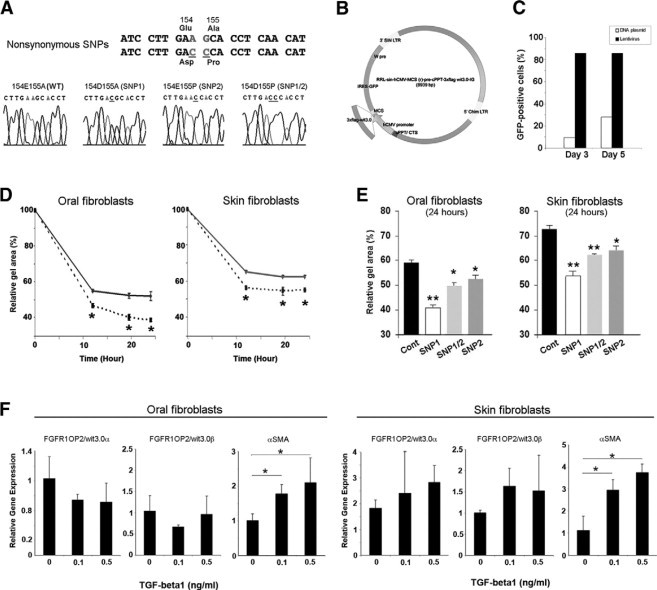
FGFR1OP2/wit3.0β overexpression increased collagen gel contraction by both oral and skin fibroblasts. A: Nonsynonymous SNPs in human FGFR1OP2/wit3.0β sequence. B: A schematic structure of lentiviral vector carrying humanized FGFR1OP2/wit3.0β (wild-type, SNP1, SNP2, or SNP1/2) and internal ribosome entry site-green fluorescent protein. C: FACS analysis of the transduction rate of the lentiviral vector for primary oral fibroblasts assessed by green fluorescent protein. D: Floating collagen gel contraction assay revealed that both oral and skin fibroblasts transduced by FGFR1OP2/wit3.0β lentiviral vectors (dotted lines) increased gel contraction rates. E: SNP1, SNP2, and SNP1/2 of FGFR1OP2/wit3.0β increased collagen gel contraction rates. In both oral and skin fibroblasts, SNP1 FGFR1OP2/wit3.0β showed the largest gel contraction rate. F: The treatment of TGF-β1 did not modulate the expression of FGFR1OP2/wit3.0 in oral and skin fibroblasts, whereas it increased the α-SMA expression. *P < 0.05, **P < 0.01.
TGF-β1 Did Not Affect the Expression of FGFR1OP2/wit3.0
Both in oral fibroblasts and skin fibroblasts, when treated with TGF-β1, the α-SMA expression was dose-dependently increased. However, TGF-β1 treatment did not affect the FGFR1OP2/wit3.0 expression (Figure 5F).
FGFR1OP2/wit3.0β Treatment to Mouse Excisional Wound Accelerated Early Wound Closure In Vivo
A full-thickness excisional wound was created through the panniculus carnosus layer in the dorsal skin of C57Bl/6J mice (Figure 6A). Unlike oral wound, the expression level of FGFR1OP2/wit3.0 was unchanged during early healing stages of the skin (Figure 6B). To address the effect of FGFR1OP2/wit3.0β overexpression in this model, we first tested the in vivo gene delivery systems suitable for mouse skin wound. Plasmid DNA with collagen carriers and CG carrier did not generate sufficient reporter gene delivery to the skin wound; however, the delivery of lentivirus vector combined with the CG SM50 carrier showed the promising result (one-way analysis of variance; P < 0.05) (Figure 6C). This method was applied to deliver the FGFR1OP2/wit3.0 expression vector to mouse full-thickness excisional wound.
Figure 6.
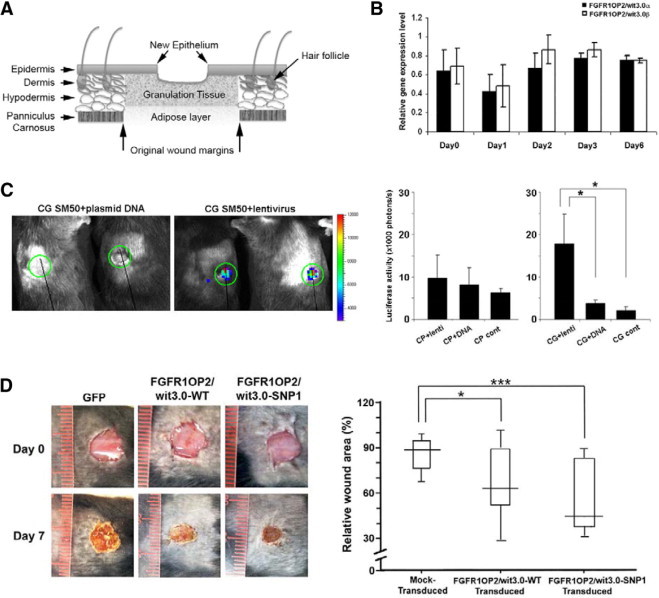
FGFR1OP2/wit3.0β induced skin wound closures in vivo. A: Schematic diagram of full-thickness wound created in mouse dorsal skin. B: Unlike in oral wounds, the mRNA levels of FGFR1OP2/wit3.0α and FGFR1OP2/wit3.0β remained unchanged in mouse skin excisional wounds during the healing period of day 0 to day 6 (P > 0.05). C: The single-view three-dimensional optical imaging depicting the luciferase reporter gene derived biofluorescence at the mouse dorsal skin excisional wound with plasmid DNA or lentiviral vectors mixed with CG SM50. The luciferase assay measuring the transfection/transduction efficiency of plasmid DNA or lentiviral vectors mixed with collagen gel carrier (Cell Prime, CP) or CG. Lentiviral vectors mixed with CG showed the highest transfection/transduction efficiency. D: Mouse dorsal skin excisional wound treated with wild-type or SNP1 FGFR1OP2/wit3.0β lentiviral vectors mixed with CG carrier. The wound areas contraction measured at day 7 indicated the significant closure of wounds treated with FGFR1OP2/wit3.0β or SNP1 FGFR1OP2/wit3.0β. *P < 0.05, ***P < 0.001.
Compared with control wounds received green fluorescent protein-expression vector, FGFR1OP2/wit3.0β-treated wounds exhibited accelerated closure (Mann-Whitney test; P < 0.05) (Figure 6D). It was further observed that the SNP1 FGFR1OP2/wit3.0β-treated wound showed greater wound closure (Mann-Whitney test; P < 0.001) (Figure 6D).
The histological observation revealed that FGFR1OP2/wit3.0β was mostly expressed by fibroblasts in granulation tissue juxtaposing dermis wound margins (Figure 7A). In the mock-treated group, while epithelialized, the dermis and panniculus carnosus wound edges remained patent. The wound area was primarily filled with highly cellular granulation tissue (Figure 7B). In the FGFR1OP2/wit3.0β treated wounds, dermis wound margins appeared to be approximated toward the center of the wound, resembling the oral wound healing. Consequently, the size of granulation tissue was found smaller in the FGFR1OP2/wit3.0β treated skin wound than the untreated wound (Figure 7C).
Figure 7.
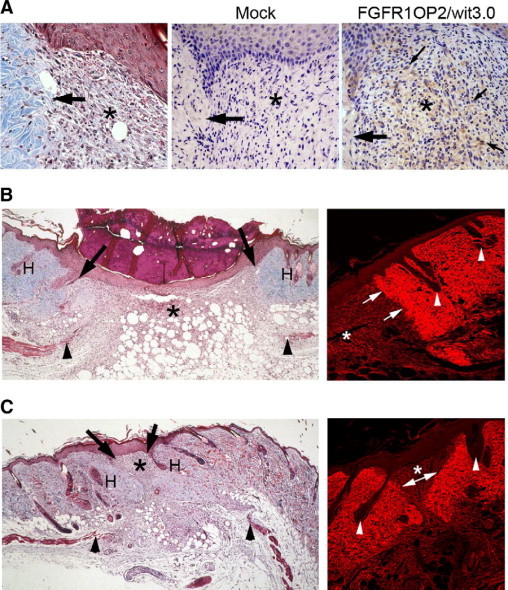
Decreased granulation tissue and scarring after FGFR1OP2/wit3.0β treatment. A: Histological specimen of mouse dorsal skin showed a clear demarcation of excisional wounding (arrow) flanking the highly cellular granulation tissue (asterisk, left; 7 days after wounding Goldner's trichrome staining). Immunostaining with D1042 was negative in the Mock-transducation group using green fluorescent protein lentiviral vector. FGFR1OP2/wit3.0β lentiviral vector treatment resulted in the strong D1042 immunostaining in fibroblasts of the granulation tissue juxtaposing the excisinal wound site (arrow). B: Mock-treated mouse skin excisional wound healing (day 7). While the blunted panniculus carnosus layer (arrowheads) remained patent, the granulation tissues asterisk were formed between the dermis wound edges (arrows) and covered by new epithelial keratinocytes. The expenditures of skin such as hair follicles (H) were not observed in the granulation tissue (left; Goldner's trichrome staining). Confocal laser scanning micrograph of skin wound stained by Sirius Red showed that collagen fibers were strongly stained in the dermis containing hair follicles (arrowheads). The dermis wound edge was clearly visible (arrows) after 7 days of wounding. The granulation tissue asterisk contained less organized collagen fibers (right). C: Histological specimen of mouse dorsal skin excisional wound treated with FGFR1OP2/wit3.0β. The blunted panniculus carnosus layer indicates the original wound edge (arrowheads). The dermis was significantly approximated toward the wound center (arrows) leaving a small area of granulation tissue asterisk covered by the keratinocyte layer. The hair follicles (H) were present in the wound area following the dermis closure (left; Goldner's trichrome staining). Confocal laser scanning micrograph of the Sirius Red stained mouse skin wound treated with FGFR1OP2/wit3.0β. The dermis wound edges (arrows) were approximated toward the wound center, surrounding a small granulation tissue (asterisk, right).
Discussion
FGFR1OP2/wit3.0 was initially identified as a differentially expressed cDNA in the rat oral wound.21 The present immunohistological study revealed that FGFR1OP2/wit3.0 was selectively expressed by a cluster of fibroblasts in the oral wound located immediately below the epithelial proliferation and migration front of wound margins that were actively approximating over the open wound (Figure 1C). Therefore, we have postulated that FGFR1OP2/wit3.0 may play a role in oral wound closure.
Oral fibroblasts isolated from gingiva or palatal mucosa have been shown strong contraction characteristics.6,9,30–34 The present study confirmed this phenotype and further revealed that oral wound fibroblasts isolated from the tooth extraction wound exhibited significantly higher collagen gel contraction ability than naïve oral fibroblasts (Figure 2A). FGFR1OP2/wit3.0 overexpression in naïve oral fibroblasts increased the collagen gel contraction to the level of that of oral wound fibroblasts (Figure 5D), which was, in turn, attenuated by the siRNA knockdown (Figure 3D). These data suggest that FGFR1OP2/wit3.0 may contribute to the fibroblast-embedded gel contraction. Wound closure is a complex physiological phenomenon, in which the mechanistic regulation may be accounted for by (1) the mediation of proinflammatory cytokines and growth factors; (2) cell migration and cell traction force; and (3) collagen and extracellular matrix (ECM) reorganization. The present study further examined the mechanistic model of oral wound closure by using FGFR1OP2/wit3.0 as a clue.
Wound contraction has been shown to be stimulated by wound-associated growth factors such as TGF-β35 and platelet-derived growth factor.36 TGF-β-stimulated collagen gel contraction appears to be primarily mediated by transdifferentation of myofibroblasts and up-regulation of α-SMA,3,37,38 whereas platelet-derived growth factor-stimulated contraction may be contributed by cytoskeleton rearrangement.39,40 These studies were conducted by using skin fibroblasts. We demonstrated that oral fibroblasts similarly responded to TGF-β1 and increased the expression of α-SMA (Figure 5F). However, TGF-β1 treatment did not modulate the steady state mRNA levels of FGFR1OP2/wit3.0 in both skin and oral fibroblasts, suggesting that FGFR1OP2/wit3.0-stimulated collagen gel contraction may not be explained by the mediation of TGF-β1. In the tooth-extraction-induced oral wound, myofibroblasts expressing α-SMA were found in the ligament-like structure connecting alveolar bone of the extraction socket (Figure 1C). After tooth extraction, periodontal ligament often remains in the socket, and oral myofibroblasts may be derived from periodontal ligament fibroblasts.41 FGFR1OP2/wit3.0-expressing oral wound fibroblasts did not appear to express α-SMA, and thus may not be considered as “myofibroblasts.” The origin of FGFR1OP2/wit3.0-expressing oral wound fibroblasts has not been determined in the scope of this study.
TGF-β1 not only up-regulates the α-SMA expression but also significantly increases collagen synthesis causing fibrotic disorders.42 Inhibition of matrix metalloproteinases by ilomastat has been shown to significantly decrease collagen production, resulting in the reduction in fibroblast-embedded collagen gel contraction.43 The reorganizing ECM has been shown to affect the wound contraction.38 To further elucidate the functional role of FGFR1OP2/wit3.0, we have developed fibroblastic cells that were derived from mouse ES cells carrying FGFR1OP2/wit3.0 (+/−) mutation. ES cell-derived fibroblastic cells with wild-type and FGFR1OP2/wit3.0 (+/−) mutation expressed collagen I at the equivalent level (Figure 4C). Our data, although limited, did not immediately support the involvement of ECM reorganization in FGFR1OP2/wit3.0-stimulated collagen gel contraction.
In the present study, fibroblastic cells derived from ES cells were further tested for the scratch plate assay, in which FGFR1OP2/wit3.0 (+/−) mutation exhibited the unexpectedly robust impairment in cell migration (Figure 4D). Fibroblast migration is an essential process during wound healing, and may, in part, result in the transmission of intracellular forces to the ECM.44,45 FGFR1OP2/wit3.0 appears to be involved in the mechanism of cell migration, which may influence the collagen gel contraction and wound closure.
The up-regulation of α-SMA dose-dependently correlated with the magnitude of cell traction force46; however, the α-SMA knockout mutation resulted in the increased cell motility.47 Myosin IIB null mutation did not affect the generation of traction force but caused the defect in directional movement resulting in the impairment of cell migration.48 It has been well established that cytoskeleton molecules such as α-SMA and myosin primarily regulate cell migration and traction force generation.48,49 FGFR1OP2/wit3.0 isoforms are small peptides possessing N-terminal and C-terminal coiled-coil domains (Figure 3A), which may facilitate the self-dimerization and self-oligomerization through anti-parallel coiled-coil domains similar to α-actinin,50 α-spectrin,51 and apolipoprotein A-I52 (Figure 2B). The dimer formation of FGFR1OP2-FGFR1 fusion peptide in the patient with the cytogenetic abnormality (12;8)(p11;p11p22)23,53 further support that FGFR1OP2/wit3.0 peptide can be dimerized and oligomerized through the coiled-coil domains. The nondenature gel Western blot suggested the incorporation of FGFR1OP2/wit3.0 in larger molecular moieties in oral wound fibroblasts (Figure 2D). Confocal laser scanning microscopy revealed the association of FGFR1OP2/wit3.0 with thin web of cytoskeleton networks of oral wound fibroblasts. F-actin was also found in the cytoskeleton network; however, F-actin and FGFR1OP2/wit3.0 appeared to be localized in a mutually exclusive fashion (Figure 2E). Taken together, FGFR1OP2/wit3.0 possesses characteristic features as cytoskeleton molecule, which may be suitable to participate in the regulation of cell migration and potentially wound closure.
The relationship between cell motility, migration, and ECM remodeling relevant to wound healing should occur in the three-dimensional ECM environment.54 Grinnell55 has proposed that the strained collagen gel may provide a more suitable experimental model to investigate the role of fibroblasts in wound healing. In the present study, we further evaluated the role of FGFR1OP2/wit3.0 in the wound healing in vivo by using the mouse dorsal skin wound model. The full-thickness excisional wound created in dorsal skin of C57Bl/6J mice underwent slow onset of wound closure during the first week followed by the progressive wound contraction during the second week. The topical application of FGFR1OP2/wit3.0β-expression vector significantly accelerated the wound closure as compared with controls (Figure 6D). The FGFR1OP2/wit3.0β-treated wound exhibited the unique approximation of dermis wound margins toward the center of excisional wound, resulting in the limited area of granulation tissue (Figure 7C). Because the blunted panniculus carnosus remained patent, we postulate that the unique approximation of dermis wound margins was likely to be contributed by FGFR1OP2/wit3.0-induced fibroblast activities within the granulation tissue and/or dermis wound margins.
The primary goals of skin wound management are to achieve rapid wound closure and to minimize the scar formation.2,56 This study demonstrated that a previously unknown molecule, FGFR1OP2/wit3.0, possessed the characteristics consistent with a cytoskeleton molecule and played a role in the wound closure. Our data further suggest that the single molecule treatment of FGFR1OP2/wit3.0 seems to be sufficient for inducing the accelerated skin wound closure, similar to the oral wound healing phenotype. As such FGFR1OP2/wit3.0 therapy may offer a novel approach for wound management, in which the critical initial wound closure may be better achieved.
Acknowledgements
We thank Dr. Matt Schibler, University of California, Los Angeles, Brain Research Institute for confocal laser scanning microscopy, Mr. Edward Wu, University of California, Los Angeles, College of Letters and Sciences, for his capable assistance, and Dr. Rong Qian, University of California ES Cell Core Facility for technical assistance and suggestions for the mouse ES Cell Study.
Footnotes
Supported in part by University of California, Los Angeles, Academic Senate Faculty Research Grants and Sumitomo Chemical Corporation. Conducted in part in a facility constructed with support from Research Facilities Improvement Program grant C06 RR014529 from the National Center for Research Resources, NIH.
References
- 1.Martin P. Wound healing: aiming for perfect skin regeneration. Science. 1997;276:75–81. doi: 10.1126/science.276.5309.75. [DOI] [PubMed] [Google Scholar]
- 2.Singer AJ, Clark RAF. Cutaneous wound healing. New Eng J Med. 1999;341:738–746. doi: 10.1056/NEJM199909023411006. [DOI] [PubMed] [Google Scholar]
- 3.Desmouliere A, Chaponnier C, Gabbiani G. Tissue repair, contraction, and the myofibroblast. Wound Repair Regen. 2005;13:7–12. doi: 10.1111/j.1067-1927.2005.130102.x. [DOI] [PubMed] [Google Scholar]
- 4.Darenfed H, Mandato CA. Wound-induced contractile ring: a model for cytokinesis. Biochem Cell Biol. 2005;83:711–720. doi: 10.1139/o05-164. [DOI] [PubMed] [Google Scholar]
- 5.Hinz B. Formation and function of the myofibroblast during tissue repair. J Invest Dermatol. 2007;127:526–537. doi: 10.1038/sj.jid.5700613. [DOI] [PubMed] [Google Scholar]
- 6.Szpaderska AM, Zuckerman JD, DiPietro LA. Differential injury responses in oral mucosal and cutaneous wounds. J Dent Res. 2003;82:621–626. doi: 10.1177/154405910308200810. [DOI] [PubMed] [Google Scholar]
- 7.Irwin CR, Myrillas T, Smyth M, Doogan J, Rice C, Schor SL. Regulation of fibroblast-induced collagen gel contraction by interleukin-1beta. J Oral Pathol Med. 1998;27:255–259. doi: 10.1111/j.1600-0714.1998.tb01952.x. [DOI] [PubMed] [Google Scholar]
- 8.Chaussain Miller C, Septier D, Bonnefoix M, Lecolle S, Lebreton-Decoster C, Coulomb B, Pellat B, Godeau G. Human dermal and gingival fibroblasts in a three-dimensional culture: a comparative study on matrix remodeling. Clin Oral Investig. 2002;6:39–50. doi: 10.1007/s00784-001-0143-2. [DOI] [PubMed] [Google Scholar]
- 9.Stephens P, Davies KJ, al-Khateeb T, Shepherd JP, Thomas DW. A comparison of the ability of intra-oral and extra-oral fibroblasts to stimulate extracellular matrix reorganization in a model of wound contraction. J Dent Res. 1996;75:1358–1364. doi: 10.1177/00220345960750060601. [DOI] [PubMed] [Google Scholar]
- 10.Lorimier S, Hornebeck W, Godeau G, Pellat B, Gillery P, Maquart FX, Laurent-Maquin D. Morphometric studies of collagen and fibrin lattices contracted by human gingival fibroblasts; comparison with dermal fibroblasts. J Dent Res. 1998;77:1717–1729. doi: 10.1177/00220345980770090801. [DOI] [PubMed] [Google Scholar]
- 11.Stephens P, Davies KJ, Occleston N, Pleass RD, Kon C, Daniels J, Khaw PT, Thomas DW. Skin and oral fibroblasts exhibit phenotypic differences in extracellular matrix reorganization and matrix metalloproteinase activity. Br J Dermatol. 2001;144:229–237. doi: 10.1046/j.1365-2133.2001.04006.x. [DOI] [PubMed] [Google Scholar]
- 12.Yang J, Tyler LW, Donoff RB, Song B, Torio AJ, Gallagher GT, Tsuji T, Elovic A, McBride J, Yung CM, Galli SJ, Weller PF, Wong DT. Salivary EGF regulates eosinophil-derived TGF-alpha expression in hamster oral wounds. Am J Physiol. 1996;270:G191–G202. doi: 10.1152/ajpgi.1996.270.1.G191. [DOI] [PubMed] [Google Scholar]
- 13.Ohshima M, Sato M, Ishikawa M, Maeno M, Otsuka K. Physiologic levels of epidermal growth factor in saliva stimulate cell migration of an oral epithelial cell line. HO-1-N-1. Eur J Oral Sci. 2002;110:130–136. doi: 10.1034/j.1600-0722.2002.11179.x. [DOI] [PubMed] [Google Scholar]
- 14.Bussi M, Valente G, Curato MP, Carlevato MT, Cortesina G. Is transposed skin transformed in major head and neck mucosal reconstruction? Acta Otolaryngol. 1995;115:348–351. doi: 10.3109/00016489509139327. [DOI] [PubMed] [Google Scholar]
- 15.McCluskey J, Martin P. Analysis of the tissue movements of embryonic wound healing: DiI studies in the limb bud stage mouse embryo. Dev Biol. 1995;170:102–114. doi: 10.1006/dbio.1995.1199. [DOI] [PubMed] [Google Scholar]
- 16.Martin P, Lewis J. Actin cables and epidermal movement in embryonic wound healing. Nature. 1992;360:179–183. doi: 10.1038/360179a0. [DOI] [PubMed] [Google Scholar]
- 17.Sciubba JJ, Waterhouse JP, Meyer J. A fine structural comparison of the healing of incisional wounds of mucosa and skin. J Oral Pathol. 1978;7:214–227. doi: 10.1111/j.1600-0714.1978.tb01596.x. [DOI] [PubMed] [Google Scholar]
- 18.Walsh LJ, L'Estrange PR, Seymour GJ. High magnification in situ viewing of wound healing in oral mucosa. Aust Dent J. 1996;41:75–79. doi: 10.1111/j.1834-7819.1996.tb05916.x. [DOI] [PubMed] [Google Scholar]
- 19.Brock J, Midwinter K, Lewis J, Martin P. Healing of incisional wounds in the embryonic chick wing bud: characterization of the actin purse-string and demonstration of a requirement for Rho activation. J Cell Biol. 1996;135:1097–1107. doi: 10.1083/jcb.135.4.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang HY, Chi JT, Dudoit S, Bondre C, van de Rijn M, Botstein D, Brown PO. Diversity, topographic differentiation, and positional memory in human fibroblasts. Proc Natl Acad Sci USA. 2002;99:12877–12882. doi: 10.1073/pnas.162488599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sukotjo C, Abanmy AA, Ogawa T, Nishimura I. Molecular cloning of wound inducible transcript (wit 3.0) differentially expressed in edentulous oral mucosa undergoing tooth extraction wound-healing. J Dent Res. 2002;81:229–235. doi: 10.1177/154405910208100402. [DOI] [PubMed] [Google Scholar]
- 22.Sukotjo C, Lin A, Song K, Ogawa T, Wu B, Nishimura I. Oral fibroblast expression of wound-inducible transcript 3.0 (wit3.0) accelerates the collagen gel contraction in vitro. J Biol Chem. 2003;278:51527–51534. doi: 10.1074/jbc.M309616200. [DOI] [PubMed] [Google Scholar]
- 23.Grand EK, Grand FH, Chase AJ, Ross FM, Corcoran MM, Oscier DG, Cross NC. Identification of a novel gene. FGFR1OP2, fused to FGFR1 in 8p11 myeloproliferative syndrome. Genes Chromosomes Cancer. 2004;40:78–83. doi: 10.1002/gcc.20023. [DOI] [PubMed] [Google Scholar]
- 24.Nishimura I, Damiani PJ, Atwood DA. Resorption of residual ridges (RRR) in rats. J Dent Res. 1987;66:1753–1757. doi: 10.1177/00220345870660121101. [DOI] [PubMed] [Google Scholar]
- 25.Friedrich G, Soriano P. Insertional mutagenesis by retroviruses and promoter traps in embryonic stem cells. Methods Enzymol. 1993;225:681–701. doi: 10.1016/0076-6879(93)25044-3. [DOI] [PubMed] [Google Scholar]
- 26.Xu C, Jiang J, Sottile V, McWhir J, Lebkowski J, Carpenter MK. Immortalized fibroblast-like cells derived from human embryonic stem cells support undifferentiated cell growth. Stem Cells. 2004;22:972–980. doi: 10.1634/stemcells.22-6-972. [DOI] [PubMed] [Google Scholar]
- 27.Nanney LB, Woodrell CD, Greives MR, Cardwell NL, Pollins AC, Bancroft TA, Chesser A, Michalak M, Rahman M, Siebert JW, Gold LI. Calreticulin enhances porcine wound repair by diverse biological effects. Am J Pathol. 2008;173:610–630. doi: 10.2353/ajpath.2008.071027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eilber KS, Sukotjo C, Raz S, Nishimura I. Alteration of collagen three-dimensional architecture in noncompliant human urinary bladder. Adv Exp Med Biol. 2003;539:791–801. doi: 10.1007/978-1-4419-8889-8_49. [DOI] [PubMed] [Google Scholar]
- 29.Moreland JL, Gramada A, Buzko OV, Zhang Q, Bourne PE. The Molecular Biology Toolkit (MBT): a modular platform for developing molecular visualization applications. BMC Bioinformatics. 2005;6:21. doi: 10.1186/1471-2105-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Irwin CR, Picardo M, Ellis I, Sloan P, Grey A, McGurk M, Schor SL. Inter- and intra-site heterogeneity in the expression of fetal-like phenotypic characteristics by gingival fibroblasts: potential significance for wound healing. J Cell Sci. 1994;107(Pt 5):1333–1346. doi: 10.1242/jcs.107.5.1333. [DOI] [PubMed] [Google Scholar]
- 31.Shah M, Foreman DM, Ferguson MW. Neutralising antibody to TGF-beta 1,2 reduces cutaneous scarring in adult rodents. J Cell Sci. 1994;107(Pt 5):1137–1157. doi: 10.1242/jcs.107.5.1137. [DOI] [PubMed] [Google Scholar]
- 32.Shah M, Foreman DM, Ferguson MW. Neutralisation of TGF-beta 1 and TGF-beta 2 or exogenous addition of TGF-beta 3 to cutaneous rat wounds reduces scarring. J Cell Sci. 1995;108(Pt 3):985–1002. doi: 10.1242/jcs.108.3.985. [DOI] [PubMed] [Google Scholar]
- 33.Lee HG, Eun HC. Differences between fibroblasts cultured from oral mucosa and normal skin: implication to wound healing. J Dermatol Sci. 1999;21:176–182. doi: 10.1016/s0923-1811(99)00037-7. [DOI] [PubMed] [Google Scholar]
- 34.Shannon DB, McKeown ST, Lundy FT, Irwin CR. Phenotypic differences between oral and skin fibroblasts in wound contraction and growth factor expression. Wound Repair Regen. 2006;14:172–178. doi: 10.1111/j.1743-6109.2006.00107.x. [DOI] [PubMed] [Google Scholar]
- 35.Montesano R, Orci L. Transforming growth factor beta stimulates collagen-matrix contraction by fibroblasts: implications for wound healing. Proc Natl Acad Sci USA. 1988;85:4894–4897. doi: 10.1073/pnas.85.13.4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clark RA, Folkvord JM, Hart CE, Murray MJ, McPherson JM. Platelet isoforms of platelet-derived growth factor stimulate fibroblasts to contract collagen matrices. J Clin Invest. 1989;84:1036–1040. doi: 10.1172/JCI114227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amadeu TP, Coulomb B, Desmouliere A, Costa AM. Cutaneous wound healing: myofibroblastic differentiation and in vitro models. Int J Low Extrem Wounds. 2003;2:60–68. doi: 10.1177/1534734603256155. [DOI] [PubMed] [Google Scholar]
- 38.Carlson MA, Longaker MT. The fibroblast-populated collagen matrix as a model of wound healing: a review of the evidence. Wound Repair Regen. 2004;12:134–147. doi: 10.1111/j.1067-1927.2004.012208.x. [DOI] [PubMed] [Google Scholar]
- 39.Grinnell F, Ho CH, Tamariz E, Lee DJ, Skuta G. Dendritic fibroblasts in three-dimensional collagen matrices. Mol Biol Cell. 2003;14:384–395. doi: 10.1091/mbc.E02-08-0493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rhee S, Grinnell F. P21-activated kinase 1: convergence point in PDGF- and LPA-stimulated collagen matrix contraction by human fibroblasts. J Cell Biol. 2006;172:423–432. doi: 10.1083/jcb.200505175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bildt MM, Bloemen M, Kuijpers-Jagtman AM, Von den Hoff JW. Matrix metalloproteinase inhibitors reduce collagen gel contraction and alpha-smooth muscle actin expression by periodontal ligament cells. J Periodontal Res. 2009;44:266–274. doi: 10.1111/j.1600-0765.2008.01127.x. [DOI] [PubMed] [Google Scholar]
- 42.Leask A, Abraham DJ. TGF-beta signaling and the fibrotic response. FASEB J. 2004;18:816–827. doi: 10.1096/fj.03-1273rev. [DOI] [PubMed] [Google Scholar]
- 43.Daniels JT, Cambrey AD, Occleston NL, Garrett Q, Tarnuzzer RW, Schultz GS, Khaw PT. Matrix metalloproteinase inhibition modulates fibroblast-mediated matrix contraction and collagen production in vitro. Invest Ophthalmol Vis Sci. 2003;44:1104–1110. doi: 10.1167/iovs.02-0412. [DOI] [PubMed] [Google Scholar]
- 44.Shreiber DI, Barocas VH, Tranquillo RT. Temporal variations in cell migration and traction during fibroblast-mediated gel compaction. Biophys J. 2003;84:4102–4114. doi: 10.1016/S0006-3495(03)75135-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Munevar S, Wang Y, Dembo M. Traction force microscopy of migrating normal and H-ras transformed 3T3 fibroblasts. Biophys J. 2001;80:1744–1757. doi: 10.1016/s0006-3495(01)76145-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen J, Li H, SundarRaj N, Wang JH. Alpha-smooth muscle actin expression enhances cell traction force. Cell Motil Cytoskeleton. 2007;64:248–257. doi: 10.1002/cm.20178. [DOI] [PubMed] [Google Scholar]
- 47.Takeji M, Moriyama T, Oseto S, Kawada N, Hori M, Imai E, Miwa T. Smooth muscle alpha-actin deficiency in myofibroblasts leads to enhanced renal tissue fibrosis. J Biol Chem. 2006;281:40193–40200. doi: 10.1074/jbc.M602182200. [DOI] [PubMed] [Google Scholar]
- 48.Lo CM, Buxton DB, Chua GC, Dembo M, Adelstein RS, Wang YL. Nonmuscle myosin IIb is involved in the guidance of fibroblast migration. Mol Biol Cell. 2004;15:982–989. doi: 10.1091/mbc.E03-06-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Even-Ram S, Doyle AD, Conti MA, Matsumoto K, Adelstein RS, Yamada KM. Myosin IIA regulates cell motility and actomyosin-microtubule crosstalk. Nat Cell Biol. 2007;9:299–309. doi: 10.1038/ncb1540. [DOI] [PubMed] [Google Scholar]
- 50.Djinovic-Carugo K, Young P, Gautel M, Saraste M. Structure of the alpha-actinin rod: molecular basis for cross-linking of actin filaments. Cell. 1999;98:537–546. doi: 10.1016/s0092-8674(00)81981-9. [DOI] [PubMed] [Google Scholar]
- 51.Kusunoki H, Minasov G, Macdonald RI, Mondragon A. Independent movement, dimerization and stability of tandem repeats of chicken brain alpha-spectrin. J Mol Biol. 2004;344:495–511. doi: 10.1016/j.jmb.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 52.Ajees AA, Anantharamaiah GM, Mishra VK, Hussain MM, Murthy HM. Crystal structure of human apolipoprotein A-I: insights into its protective effect against cardiovascular diseases. Proc Natl Acad Sci USA. 2006;103:2126–2131. doi: 10.1073/pnas.0506877103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53.Gu TL, Goss VL, Reeves C, Popova L, Nardone J, Macneill J, Walters DK, Wang Y, Rush J, Comb MJ, Druker BJ, Polakiewicz RD. Phosphotyrosine profiling identifies the KG-1 cell line as a model for the study of FGFR1 fusions in acute myeloid leukemia. Blood. 2006;108:4202–4204. doi: 10.1182/blood-2006-06-026666. [DOI] [PubMed] [Google Scholar]
- 54.Tamariz E, Grinnell F. Modulation of fibroblast morphology and adhesion during collagen matrix remodeling. Mol Biol Cell. 2002;13:3915–3929. doi: 10.1091/mbc.E02-05-0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grinnell F. Fibroblast biology in three-dimensional collagen matrices. Trends Cell Biol. 2003;13:264–269. doi: 10.1016/s0962-8924(03)00057-6. [DOI] [PubMed] [Google Scholar]
- 56.Clark RA, Ghosh K, Tonnesen MG. Tissue engineering for cutaneous wounds. J Invest Dermatol. 2007;127:1018–1029. doi: 10.1038/sj.jid.5700715. [DOI] [PubMed] [Google Scholar]


