Abstract
Pigment epithelium-derived factor (PEDF) has several biological actions on tumor cells, but its effects are cell-type dependent. The aim of this study was to examine the pathophysiological role of PEDF in hepatocellular carcinoma (HCC). PEDF expression was examined in various hepatoma cell lines and human HCC tissues, and was seen in various hepatoma cell lines including HepG2 cells. In human HCC tissues, PEDF expression was higher than in adjacent non-HCC tissues. In addition, serum PEDF levels were higher in HCC patients than in non-HCC patients, and curative treatment of HCC caused significant reductions in serum PEDF levels compared with pretreatment levels. In vitro experiments, camptothecin (CPT) was used to induce apoptosis and the effect of PEDF was investigated by knockdown of the PEDF gene in CPT-treated HepG2 cells. Knockdown of the PEDF gene enhanced CPT-induced apoptosis, simultaneously down-regulating Bcl-xL expression in HepG2 cells. Expression of apoptosis-related molecules and effects of bafilomycin A1 on CPT-induced apoptosis were also examined in PEDF gene knockdown HepG2 cells. Treatment with bafilomycin A1 suppressed CPT-induced decreases in Bcl-xL expression and increases in apoptosis in PEDF gene knockdown HepG2 cells. PEDF may, therefore, exert anti-apoptotic effects through inhibition of lysosomal degradation of Bcl-xL in CPT-treated HepG2 cells.
Hepatocellular carcinoma (HCC) is a common cancer that causes nearly 1 million deaths a year worldwide.1 The incidence of HCC is predicted to continue to increase over the next 30 years.2 To develop new therapeutic strategies, it is important to elucidate molecular mechanisms underlying hepatocarcinogenesis.
Pigment epithelium-derived factor (PEDF) is a 50-kDa glycoprotein initially isolated from fetal human retinal pigment epithelial cells.3 PEDF exerts a range of biological effects depending on the type of the target cell. PEDF induces apoptosis of endothelial cells and results in inhibition of neovascularization.4 Overexpression of PEDF causes a reduction in tumor microvessel density and subsequent anti-tumor effects in pancreatic adenocarcinoma and melanoma cells.5,6 In contrast to its effects in endothelial cells, PEDF causes the opposite effect in other types of cells. PEDF protects granule cells against both natural and potassium-induced apoptosis through activation of prosurvival genes.7 In cultured retinal pericytes, PEDF inhibits oxidative stress-induced apoptosis through an increased ratio of B-cell leukemia/lymphoma 2 (Bcl-2)-associated X protein (bax) to bcl-2 mRNA levels with subsequent activation of caspase-3.8,9 PEDF also inhibits light-induced apoptotic processes in photoreceptor cells in vivo.10,11
HCC is a hypervascular solid tumor, in which neovascularization plays an important role in disease progression and prognosis. However, changes in PEDF expression in human HCC have never been investigated and therefore, it is unclear whether PEDF is a useful target for therapeutic strategies in patients with HCC. Dysregulation of apoptosis is also deeply involved in hepatocarcinogenesis. Although HCC is known to be resistant to apoptosis,12 it remains unknown whether PEDF has anti-apoptotic effects in HCC.
Thus, PEDF is a multifunctional protein with opposing activities, apoptotic and anti-apoptotic activities. These disparate effects depend on cell type. The aims of the present study were to investigate changes in PEDF expression and the role of PEDF in HCC.
Materials and Methods
Materials
All reagents were purchased from Wako Pure Chemical Industries (Osaka, Japan) unless otherwise indicated.
Cell Lines
Human hepatoma cell lines, HepG2, Hep3B, Huh-7, PLC/PRF/5, HLF and SK-Hep1, and the human hepatocyte cell line, OUMS-29,13,14 were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, penicillin (100 U/ml), and streptomycin (100 U/ml) at 37°C in a humidified atmosphere containing 5% CO2 as previously described.15 Because PEDF is present in fetal bovine serum, each cell line was incubated for 48 hours with Dulbecco's modified Eagle's medium without fetal bovine serum to examine PEDF expression in culture medium.
Human Samples
We obtained six pairs of HCCs and adjacent non-HCC liver tissues at the time of surgical resection for HCC. All tissues were stored at −80°C until used. Serum samples were obtained from cirrhotic patients with non-HCC (n = 25) or HCC (n = 110). Paired serum samples were also obtained from patients with HCC before and after complete treatment with surgical resection or percutaneous radiofrequency ablation (n = 15). All serum samples were stored at −80°C until used. Informed consent in writing was obtained from each patient and the study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki as reflected in a prior approval by the institutional review committee.
RT-PCR
From each cell line or each tissue, total RNA was isolated with TRIzol reagent (Invitrogen, Carlsbad, CA). Two hundred fifty nanograms of RNA was used as a template for RT-PCR as previously described.16 Expression of mRNA was evaluated by using a pair of unique primers for human PEDF (sense 5′-CCGGGCTCTCTACTATGACTTGAT-3′ and antisense 5′-ACGGTCCTCTCTTGATCCAAGTAG-3′), β-actin primer pair (Promega, Madison, WI), or cyclophilin (sense 5′-CCCACCGTGTTCTTCGAC-3′ and antisense 5′-ATCTTCTGCTGGTCTTGCC-3′). The cycle numbers (24 cycles for PEDF; 20 cycles for β-actin; 22 cycles for cyclophilin) for amplification were chosen in the linear range. The above conditions were determined by plotting signal intensities as functions of the template amounts and cycle numbers, and reactions proceeded linearly.
Immunohistochemistry
Cell lines were fixed with 100% acetone at −20°C for 20 minutes. Nonfixed human tissues were sectioned at a thickness of 6 μm and fixed with 100% acetone at −20°C for 20 minutes. Cells or sections were washed three times for 5 minutes each in PBS (pH 7.4, 130 mmol/L NaCl, 2 mmol/L NaH2PO4, and 7 mmol/L Na2HPO4) and then blocked with 10% skim milk in PBS for 30 minutes. Cells or sections were incubated overnight at 4°C with the monoclonal antibody to human PEDF (Chemicon International, Temecula, CA or TransGenic, Kumamoto, Japan) diluted 1:100 in PBS. After several washes with PBS, cells or sections were incubated for 1 hour with the secondary antibody, horseradish peroxidase-labeled anti-mouse IgG (Amersham Biosciences, Piscataway, NJ), or a fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgM (Cappel, Aurora, OH) diluted 1:100 in PBS at room temperature. Subsequently, cells or sections were washed with PBS. Immunostaining for PEDF was developed with 3,3′-diaminobenzidine or a confocal laser scanning microscope (FluoView FV 300; Olympus, Tokyo, Japan). In addition, propidium iodide was used concomitantly for nuclear staining. The stored images were overlaid to create a single integrated image referred to as a “volume projection” by using the manufacturer's proprietary software (Olympus) as previously described.17
Immunoblotting and Quantitation
Conditioned media were quantitated by using DC protein assay kit (Bio-Rad Laboratories, Hercules, CA) according to the manufacturer's instruction. Proteins were resolved by using 10% SDS-polyacrylamide gel electrophoresis gel. Following electrophoresis, proteins were transblotted onto polyvinylidene difluoride membranes (PolyScreen, PerkinElmer Life Sciences, Waltham, MA). The membranes were blocked with 5% fat-free skim milk powder in Tris-buffered saline containing 0.02% (v/v) Tween 20. The primary antibodies were monoclonal antibodies to human PEDF (Chemicon or TransGenic), p53 (Cell Signaling Technology, Danvers, MA), phospho-p53 (Ser46) (Cell Signaling Technology), Akt (Cell Signaling Technology), phospho-Akt (Cell Signaling Technology), cyclin D1 (Cell Signaling Technology), Bax (Cell Signaling Technology), survivin (Cell Signaling Technology), or Bcl-xL (Cell Signaling Technology). The bound antibodies were detected with horseradish peroxidase-labeled anti-mouse or anti-rat IgG (Amersham Biosciences) by using an enhanced chemiluminescence detection system (ECL advanced kit, Amersham Biosciences) as previously described.18 A positive signal from the target proteins was visualized by using an image analyzer LAS-1000 plus (Fujifilm, Tokyo, Japan). Band intensities were determined by using the Scion Image (Scion Corporation, Frederick, MD). Values were based on four different experiments.
Assay for Serum or Medium PEDF
Serum or medium PEDF measurements were performed with a modified competitive enzyme-linked immunosorbent assay (ELISA) as previously described.19 To dissociate PEDF from binding-proteins, 50 μl of serum or medium was pretreated with 200 μl of 8 M urea for 1 hour. Then, 100-μl aliquots of standard recombinant human PEDF proteins (Chemicon International) or 50-fold diluted serum were added to wells that had been precoated with an anti-PEDF monoclonal antibody (TransGenic). Then, 50 μl of biotinylated anti-human PEDF polyclonal antibody (R and D Systems, Minneapolis, MN) was added to each well, and the plate was read at 450 nm by using a microplate reader.
Small Interfering RNA-Mediated Gene Knockdown and Evaluation of Apoptosis
PEDF gene was knocked down by using small interfering RNA (siRNA) in HepG2 cells. HepG2 cells were plated together with siRNA (individual siRNA serpinF1 or negative control number 2; Ambion, Austin, TX)-siPORT NeoFX complexes. Twenty-four hours later, siRNA-siPORT NeoFX complexes were removed by replacing them with basal medium containing 1% serum and an apoptosis inducer, camptothecin (CPT; 2 μmol/L) and then cells were incubated for 24 hours. In some experiments, 10 μmol/L of carbobenzoxy-L-leucyl-L-leucyl-L-leucinal (MG132; Peptide Institute, Osaka, Japan), a proteasomal proteolysis inhibitor or 100 nmol/L of bafilomycin A1, a lysosomal proteolysis inhibitor was mixed with CPT. Apoptosis was evaluated by visualization of caspase activity by using a FITC-labeled carbobenzoxy-valyl-alanyl-aspartyl-fluoromethylketone (CaspACE FITC-VAD-fmk in situ marker; Promega) according to the manufacturer's instructions and quantified by terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling assay by using the Cell Death Detection ELISA kit (Roche Applied Science, Mannheim, Germany) according to the manufacturer's instructions. Knockdown of the PEDF gene was verified by semiquantitative RT-PCR and by measuring medium PEDF levels.
Statistical Analysis
All data are expressed as mean ± SD. Comparisons between any two groups were performed by using the Mann-Whitney U test. Comparisons among multiple groups were analyzed by using the Kruskal-Wallis analysis of variance. A P value <0.05 was considered statistically significant.
Results
PEDF mRNA and PEDF Expression in Human Hepatoma Cell Lines
No expression and weak expression of PEDF mRNA were seen in HLF and SK-Hep1 cells, respectively. However, expression of PEDF mRNA was seen in OUMS-29, HepG2, Hep3B, Huh-7, and PLC/PRF/5 cells (Figure 1A). Intracellular expression of PEDF was seen in OUMS-29, HepG2, Hep3B, Huh-7, and PLC/PRF/5 cells, but not in HLF and SK-Hep1 cells (Figure 1B). Secreted PEDF in the culture medium was evaluated by using two different primary antibodies. Expression of PEDF was seen in the culture medium of HepG2, Hep3B, Huh-7, and PLC/PRF/5 cells, but not in that of HLF, OUMS-29, and SK-Hep1 cells (Figure 1C, upper column). Similar results were obtained when a different primary antibody was used (Figure 1C, lower column).
Figure 1.
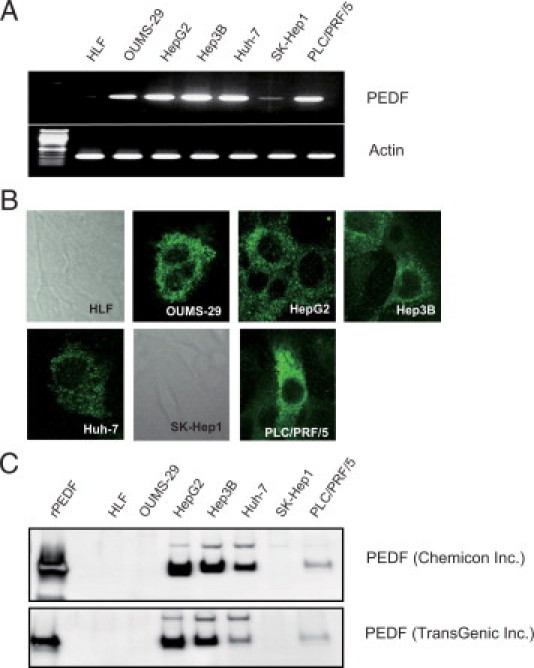
Expression of PEDF mRNA and actin mRNA as a control in various human hepatoma cell lines (A), intracellular expression of PEDF (B), and secreted PEDF into the culture medium (C) were evaluated by RT-PCR, immunohistochemistry, and immunoblotting, respectively. In HLF and SK-Hep1 cells, PEDF expression was negative and cell morphology was demonstrated by phase contrast (B).
PEDF mRNA and PEDF Expression in Human Liver Tissues and Serum Samples
Expression of PEDF mRNA was higher in HCC tissue than in non-HCC tissue in all cases (Figure 2A). Similar to expression of PEDF mRNA, expression of PEDF was higher in HCC tissue than in non-HCC tissue in all cases (Figure 2B). When PEDF was visualized by 3,3′-diaminobenzidine, expression of PEDF was seen in nuclei of hepatocytes and mainly in the cytoplasm of HCC where no lipid accumulation was seen by Sudan IV staining (Figure 2C). Similar results were obtained by confocal laser scanning microscopy. In hepatocytes, PEDF (green color) was colocalized with propidium iodide (red color), showing a yellow color in the overlay image (Figure 2D, upper column). However, in HCC, PEDF (green color) was dissociated with propidium iodide (red color; Figure 2D, lower column). Serum PEDF levels were significantly higher in patients with HCC than in patients with non-HCC (Figure 2E). After complete treatment of HCC such as surgical resection or percutaneous radiofrequency ablation, serum PEDF levels were significantly lower than before treatment (Figure 2F).
Figure 2.
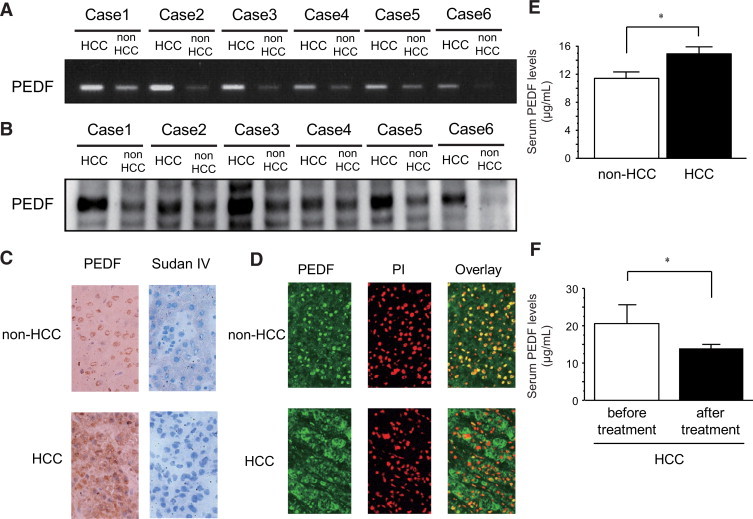
In human HCC tissues and adjacent non-HCC liver tissues, expression of PEDF mRNA (n = 6) (A) and PEDF (n = 6) (B) were evaluated by RT-PCR and immunoblotting, respectively. The distribution of PEDF was examined by immunostaining. Immunostaining for PEDF was developed with 3,3′-diaminobenzidine (C) and lipid accumulation was evaluated by Sudan IV staining (C). The distribution of PEDF was examined by a confocal laser scanning microscopy. Green color indicates expression of PEDF and red color indicates propidium iodide, a marker for the nucleus. Yellow color indicates co-localization of PEDF and propidium iodide (D). Serum PEDF levels were measured in non-HCC patients (n = 25) or HCC patients (n = 110) with a competitive ELISA (E). Changes in serum PEDF levels after HCC treatment were examined in HCC patients treated with surgical resection or percutaneous radiofrequency ablation (n = 15) (F). *P < 0.01.
Effects of PEDF Gene Knockdown on CPT-Induced Apoptosis in HepG2 Cells
Knockdown of the PEDF gene was verified by semiquantitative RT-PCR (Figure 3A) and PEDF levels in the culture medium (Figure 3B).
Figure 3.
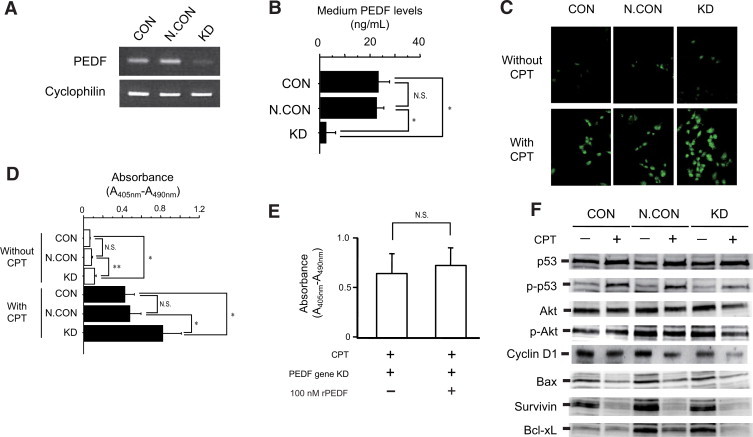
In PEDF gene knockdown HepG2 cells, expression of PEDF mRNA (A) and medium PEDF levels (B) were evaluated by semiquantitative RT-PCR and a competitive ELISA (n = 4), respectively. Effects of PEDF gene knockdown on CPT-induced apoptosis was examined in HepG2 cells. Apoptosis was visualized by FITC-conjugated VAD-FMK, which binds with activated caspase and green color indicated apoptotic cells (C). Apoptosis was quantified by using Cell Death Detection ELISA kits (Roche Applied Science) and increases in absorbance (A405 nm-A490 nm) indicate increased number of apoptotic cells (n = 4, D). Effects of rPEDF on CPT-induced apoptosis was examined in PEDF gene knockdown HepG2 cells. Apoptosis was quantified by using Cell Death Detection ELISA kits (Roche Applied Science) (n = 4, E). Effects of PEDF gene knockdown on cell cycle-related molecules (p53, Akt, and cyclin D1) and apoptosis-related molecules (Bax, survivin, and Bcl-xL) were evaluated by immunoblotting in HepG2 cells (F). *P < 0.01, **P < 0.05.
Apoptosis was visualized by FITC-conjugated VAD-FMK, which binds with activated caspase. Laser scanning microscopic images showed a slight increase of apoptosis in PEDF gene knockdown HepG2 cells compared with CON and N.CON (Figure 3C, upper column). By treatment with CPT, an apoptotic inducer, marked increase of apoptosis was seen in PEDF gene knockdown HepG2 cells compared with CON and N.CON (Figure 3C, lower column).
In addition, apoptosis was quantified by terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling-based ELISA. PEDF gene knockdown increased apoptotic cell numbers in HepG2 cells compared with CON and N.CON (Figure 3D, white bars). By treatment with CPT, a significant increase in apoptotic cell number was seen in knockdown compared with CON and N.CON (Figure 3D, black bars). Similar findings were found in Hep3B and Huh7 cells (Supplemental Figure 1, see http://ajp.amjpathol.org).
Effects of Recombinant PEDF on CPT-Induced Apoptosis in PEDF Gene Knockdown HepG2 Cells
After addition of recombinant PEDF (rPEDF) (100 nmol/L) into the culture medium, no significant changes were seen in CPT-induced apoptotic cell numbers in PEDF gene knockdown HepG2 cells (Figure 3E).
Effects of PEDF Gene Knockdown on Expression of Cell Cycle- and Apoptosis-Related Molecules in CPT-Treated HepG2 Cells
Although there was no significant decrease of p53 expression on CPT-treated knockdown cells compared with that in CPT-treated CON and CPT-treated N.CON cells, significant decrease was seen in phospho-p53, Akt, phospho-Akt, and cyclin D1 in CPT-treated knockdown cells compared with those in CPT-treated CON and CPT-treated N.CON cells (Figure 3F and Table 1). Treatment with CPT decreased expression of Bax and survivin in all groups (Figure 3F and Table 1). However, expression of Bcl-xL was markedly decreased in CPT-treated knockdown cells compared with that in CPT-treated CON and CPT-treated N.CON cells (Figure 3F). Decreased expression of Bcl-xL was also found in Hep3B and Huh7 cells (Supplemental Figure 2, see http://ajp.amjpathol.org).
Table 1.
Densitometric Analysis of Expression of Cell Cycle- and Apoptosis-Related Molecules in CPT-Treated HepG2 Cells
| Molecule | CON (n = 4) | N.CON (n = 4) | kd (n = 4) | |||
|---|---|---|---|---|---|---|
| CPT | − | + | − | + | − | + |
| P53 | 223 ± 24 | 235 ± 19 | 221 ± 21 | 232 ± 20 | 229 ± 21 | 238 ± 21 |
| p-p53 | 136 ± 15 | 193 ± 22 | 106 ± 17 | 195 ± 18 | 101 ± 15 | 124 ± 26*† |
| Akt | 173 ± 16 | 169 ± 19 | 186 ± 16 | 157 ± 17 | 182 ± 18 | 120 ± 25*† |
| p-Akt | 194 ± 18 | 197 ± 21 | 242 ± 23 | 236 ± 22 | 244 ± 21 | 160 ± 28*† |
| cyclin D | 215 ± 29 | 194 ± 31 | 209 ± 41 | 164 ± 43 | 193 ± 37 | 118 ± 39*† |
| Bax | 132 ± 15 | 75 ± 11 | 156 ± 18 | 85 ± 11 | 149 ± 12 | 81 ± 10 |
| Survivin | 194 ± 18 | 56 ± 7 | 226 ± 21 | 61 ± 10 | 210 ± 18 | 54 ± 9 |
| Bcl-xL | 111 ± 13 | 130 ± 11 | 215 ± 19 | 159 ± 15 | 205 ± 21 | 87 ± 9‡§ |
Data presented as mean ± SD.
P < 0.05 compared with CON with CPT.
P < 0.05 compared with N.CON with CPT.
P < 0.01 compared with CON with CPT.
P < 0.01 compared with N.CON with CPT.
Effects of PEDF Gene Knockdown on Bcl-xL mRNA in HepG2 Cells
Expression of Bcl-xL mRNA was higher in CPT-treated knockdown cells compared with that in CPT-treated CON and CPT-treated N.CON cells at 6 hours and 12 hours after PEDF gene knockdown (Figure 4A).
Figure 4.
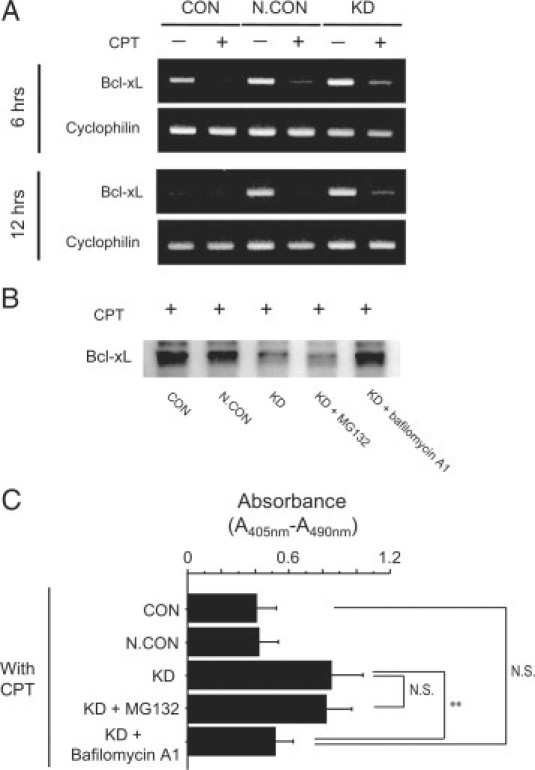
Effects of PEDF gene knockdown on Bcl-xL mRNA and cyclophilin mRNA as a control were evaluated by semiquantitative RT-PCR 6 and 12 hours after PEDF gene knockdown in CPT-treated HepG2 cells (A). In CPT-treated PEDF gene knockdown HepG2 cells, effects of 10 μmol/L of MG132 or 100 nmol/L of bafilomycin A1 on expression of Bcl-xL (B) and on CPT-induced apoptosis were evaluated by immunoblotting and Cell Death Detection ELISA kits (Roche Applied Science), respectively. Increases in absorbance (A405 nm-A490 nm) indicate increases in number of apoptotic cells (n = 5, C). **P < 0.05.
Effects of MG132 or Bafilomycin A1 on Decreases in Expression of Bcl-xL and Increases in Apoptosis in CPT-Treated PEDF Gene Knockdown HepG2 Cells
After treatment with 10 μmol/L of MG132, a proteasomal proteolysis inhibitor, no marked change in expression of Bcl-xL was seen in CPT-treated knockdown cells (Figure 4B). Treatment with 100 nmol/L of bafilomycin A1, a lysosomal proteolysis inhibitor, inhibited decreases in expression of Bcl-xL in CPT-treated knockdown cells (Figure 4B). Treatment with bafilomycin A1, but not MG132, also inhibited an increase of apoptosis in CPT-treated PEDF gene knockdown HepG2 cells (Figure 4C).
Discussion
In this study, we demonstrated that PEDF was overexpressed not only in human hepatoma cell lines, but also in human HCC tissues. Knockdown of the PEDF gene enhanced CPT-induced apoptosis with down-regulation of Bcl-xL expression. Treatment with bafilomycin A1, a lysosomal proteolysis inhibitor, inhibited the CPT-induced decreases in expression of Bcl-xL and increases in apoptotic cell number in PEDF gene knockdown HepG2 cells. Thus, PEDF may exert anti-apoptotic effects by inhibiting lysosomal-mediated degradation of Bcl-xL in HepG2 cells.
Decreased expression of PEDF was reported in various cancers such as prostate,20 pancreas,21 breast,22 and ovarian cancers.23 PEDF has anti-angiogenic24 and tumorcidal effects through induction of both apoptosis and differentiation.25 In fact, treatment with exogenous PEDF has been shown to inhibit neovascularization and proliferation of malignant melanoma in vivo.5 Generally, substitution of PEDF is considered as a novel therapeutic strategy against several cancers. However, there is some controversy about the pathophysiological role of PEDF in cancers because PEDF levels have been increased in some types of cancers.26,27 Indeed, in this study, we found that PEDF was overexpressed in most of the human hepatoma cell lines and HCC tissues. Although PEDF mRNA was not seen only in HLF cell line, HLF cell line resembles fibroblast in morphology and does not produce alpha-fetoprotein.28 Thus, HLF cell line shows different characteristics from other hepatoma cell lines and this may be a reason why PEDF mRNA was not seen only in HLF cell line.
More than 99% of PEDF cDNA from HepG2 cells was identical to human PEDF cDNA (data not shown) and therefore, PEDF in HepG2 cells seems to have biological activity. Although treatment with exogenous PEDF is reported as a potential therapeutic strategy for HCC,29 endogenous expression of PEDF in human HCC has never been investigated and the validity of treatment strategies targeting PEDF has not been confirmed. Expression of PEDF was increased in all six HCC tissues that we examined, although all six HCCs were typical hypervascular tumors. PEDF is known to localize regions of steatosis.30,31 However, no lipid accumulation was seen in the cytoplasm of HCC and therefore, high expression of PEDF seems to be a character of HCC. Moreover, serum PEDF levels were significantly higher in HCC patients than in non-HCC patients or in HCC patients who had complete treatment. Recently, we, along with others, have found that adipocytes secrete PEDF and serum PEDF level is associated with metabolic syndrome.19,32–34 However, in this study, enrolled patients were not obese and there was no significant difference in body mass index between HCC and non-HCC patients (22.5 ± 2.1 vs. 22.3 ± 1.9; not significant). Taken together, PEDF expression levels were increased in HCC as a countersystem against neovascularization. Further, our present results suggest that elevation of PEDF may have other biological effects on HCC, ie, promotion of HCC growth and expansion by suppressing apoptosis.
Because PEDF was expressed in human hepatoma cell lines, knockdown of the PEDF gene seemed to be a better strategy for examining the role of PEDF in HCC than overexpression of PEDF or treatment with recombinant PEDF. Knockdown of the PEDF gene itself induced apoptosis and the apoptotic effect was enhanced by the co-treatment with CPT, an inducer of apoptosis, in various hepatoma cell lines. These findings indicate that PEDF has a protective role against CPT-induced apoptosis in HepG2 cells. Cao et al35 reported that PEDF protects cultured retinal neurons against hydrogen peroxide-induced apoptosis. In good accordance with our results, a similar protective role of PEDF against apoptosis has been found in cerebellar granule cells,7,36–39 retinal pericytes,8,9 photoreceptor cells,10,11 and retinal ganglion cells.40 Addition of rPEDF into the culture medium did not protect CPT-induced apoptosis in PEDF gene knockdown HepG2 cells. Although PEDF is a secretory protein,41,42 these data indicate that PEDF exerted its anti-apoptotic activity through intracellular interaction between PEDF and anti-apoptotic molecules, and not through autocrine or paracrine pathways in HepG2 cells. Precise mechanisms for PEDF-induced protection against apoptosis are unknown. However, several pathways have been suggested. PEDF is known to activate cAMP-responsive element binding protein, nuclear factor κB, extracellular signal-regulated kinases 1/2, and glutathione peroxidase, resulting in protection against apoptosis.8,38–40 In this study, we found that cell cylcle-related molecules such as phospho-p53, Akt, phospho-Akt, and cyclin D1 were down-regulated by knockdown of the PEDF gene in CPT-treated HepG2 cells. These changes were in good accordance with previous reports43–47 and suggest that cell cycle arrest may enhance CPT-induced apoptosis. In addition to these pathways, we first found that knockdown of the PEDF gene down-regulated Bcl-xL, an anti-apoptotic molecule, in CPT-treated HepG2 cells. Thus, PEDF may exert anti-apoptotic effects in HCC through up-regulation of Bcl-xL.
Then, we investigated mechanisms for down-regulation of Bcl-xL by PEDF gene knockdown. Expression of Bcl-xL mRNA was decreased in CPT-untreated CON cells at 12 hours after PEDF gene knockdown compared with that at 6 hours. Although the reason for down-regulation of Bcl-xL mRNA expression at 12 hours is unclear, the following is a possible reason: HepG2 cells were plated together with siRNA for PEDF. Twenty-four hours later, siRNA for PEDF were removed by replacing them with basal medium containing 1% fetal bovine serum. Fetal bovine serum contains various apoptosis-inducing cytokines including TNF-α. Those apoptosis-inducing cytokines might temporary up-regulate Bcl-xL mRNA expression at 6 hours after PEDF gene knockdown and apoptotic stimuli might not last until 12 hours.
Expression of Bcl-xL mRNA was higher in CPT-treated knockdown cells compared with that in CPT-treated CON and CPT-treated N.CON cells at 6 hours and 12 hours after PEDF gene knockdown. However, the protein expression of Bcl-xL was lower in CPT-treated knockdown cells. Therefore, we assumed that high expression of Bcl-xL mRNA is a cellular adaptive response against decreased protein expression of Bcl-xL. The ubiquitin-proteasome pathway is a major mechanism for protein degradation. However, treatment with MG132, a proteasomal proteolysis inhibitor, showed no marked changes in expression of Bcl-xL in our study. In concordance with our results, Bcl-xL is not degraded by the ubiquitin-proteasome pathway in ischemic retinal injury.48 Autophagy is another major mechanism for protein degradation through lysosomal proteolysis.49 We found that treatment with bafilomycin A1, a lysosomal proteolysis inhibitor, prevented the CPT-induced decrease in expression of Bcl-xL. Treatment with bafilomycin A1 also prevented CPT-induced apoptosis in PEDF gene knockdown HepG2 cells. These findings led us to hypothesize that knockdown of the PEDF gene causes lysosomal proteolysis-mediated down-regulation of Bcl-xL and subsequent enhancement of CPT-induced apoptosis in HepG2 cells. In other words, PEDF may up-regulate Bcl-xL through inhibition of lysosomal proteolysis in HCC. Supporting our hypothesis, expression of Bcl-xL is known to be increased in human HCC.50 There are significant correlations between expression of Bcl-xL and disease-free survival, and Bcl-xL is an independent prognostic factor for disease-free survival in patients with HCC.51
In the present study, we also examined the expression of PEDF in normal hepatocytes and found that high expression of PEDF was found in cytosol of normal hepatocyte cell line, OUMS-29, but not in non-HCC tissue. Although the reason for this discrepancy is unclear, the following is a possible explanation. PEDF is a secretory protein,41,42 and cell-matrix interaction is one of the regulatory mechanisms for secretion of PEDF.52 There is no cell-matrix interaction in culture dish and PEDF was not secreted in the culture medium of OUMS-29. Thus, accumulation of unsecreted PEDF may be a reason for high expression of PEDF in OUMS-29 cells.
A limitation of this study is that a hepatoma xenograft study using nude mice was not conducted to prove the anti-apoptitic property of PEDF. However, continuous and specific down-regulation of PEDF gene of hepatoma cell in nude mice is impossible at this point because of following reasons: (1) because effects of knockdown only last 3 days in HepG2 cells (data not shown) and none of PEDF knockout hepatoma cells survived for more than 7 days; and (2) PEDF occurs in various organs53 and therefore, continuous knockdown by injection of siRNA for PEDF affects not only hepatoma cell line, but also various organ functions.
In conclusion, we found increased expression of PEDF in both human hepatoma cell lines and human HCC tissues. Knockdown of the PEDF gene enhanced CPT-induced apoptosis with down-regulation of Bcl-xL expression. Treatment with bafilomycin A1 prevented CPT-induced decreases in expression of Bcl-xL and increases in apoptosis in PEDF gene knockdown HepG2 cells. Thus, increased expression of PEDF may exert anti-apoptotic effects in HCC through inhibition of lysosomal degradation of Bcl-xL.
Acknowledgements
We thank Eiichi Hirano and Yumi Ogo for technical assistance.
Footnotes
Supported in part by the Vehicle Racing Commemorative Foundation, a Grant-in-Aid for Young Scientists (number 19790643 to T.K.) and a Grant-in-Aid for Scientific Research (number 21590865 to M.S.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, by Health and Labour Sciences Research Grants for Research on Hepatitis from the Ministry of Health, Labour, and Welfare of Japan, and by a Grant for Cancer Research from Fukuoka Cancer Society.
Supplemental material for this article can be found on http://ajp.amjpathol.org.
Web Extra Material
References
- 1.Burroughs A, Hochhauser D, Meyer T. Systemic treatment and liver transplantation for hepatocellular carcinoma: two ends of the therapeutic spectrum. Lancet Oncol. 2004;5:409–418. doi: 10.1016/S1470-2045(04)01508-6. [DOI] [PubMed] [Google Scholar]
- 2.Kulik LM, Mulcahy MF, Omary RA, Salem R. Emerging approaches in hepatocellular carcinoma. J Clin Gastroenterol. 2007;41:839–854. doi: 10.1097/MCG.0b013e318060ac52. [DOI] [PubMed] [Google Scholar]
- 3.Tombran-Tink J, Chader GG, Johnson LV. PEDF: a pigment epithelium-derived factor with potent neuronal differentiative activity. Exp Eye Res. 1991;53:411–414. doi: 10.1016/0014-4835(91)90248-d. [DOI] [PubMed] [Google Scholar]
- 4.Jimenez B, Volpert OV. Mechanistic insights on the inhibition of tumor angiogenesis. J Mol Med. 2001;78:663–672. doi: 10.1007/s001090000178. [DOI] [PubMed] [Google Scholar]
- 5.Abe R, Shimizu T, Yamagishi S, Shibaki A, Amano S, Inagaki Y, Watanabe H, Sugawara H, Nakamura H, Takeuchi M, Imaizumi T, Shimizu H. Overexpression of pigment epithelium-derived factor decreases angiogenesis and inhibits the growth of human malignant melanoma cells in vivo. Am J Pathol. 2004;164:1225–1232. doi: 10.1016/s0002-9440(10)63210-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garcia M, Fernandez-Garcia NI, Rivas V, Carretero M, Escamez MJ, Gonzalez-Martin A, Medrano EE, Volpert O, Jorcano JL, Jimenez B, Larcher F, Del Rio M. Inhibition of xenografted human melanoma growth and prevention of metastasis development by dual antiangiogenic/antitumor activities of pigment epithelium-derived factor. Cancer Res. 2004;64:5632–5642. doi: 10.1158/0008-5472.CAN-04-0230. [DOI] [PubMed] [Google Scholar]
- 7.Araki T, Taniwaki T, Becerra SP, Chader GJ, Schwartz JP. Pigment epithelium-derived factor (PEDF) differentially protects immature but not mature cerebellar granule cells against apoptotic cell death. J Neurosci Res. 1998;53:7–15. doi: 10.1002/(SICI)1097-4547(19980701)53:1<7::AID-JNR2>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 8.Amano S, Yamagishi S, Inagaki Y, Nakamura K, Takeuchi M, Inoue H, Imaizumi T. Pigment epithelium-derived factor inhibits oxidative stress-induced apoptosis and dysfunction of cultured retinal pericytes. Microvasc Res. 2005;69:45–55. doi: 10.1016/j.mvr.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 9.Yamagishi S, Inagaki Y, Amano S, Okamoto T, Takeuchi M, Makita Z. Pigment epithelium-derived factor protects cultured retinal pericytes from advanced glycation end product-induced injury through its antioxidative properties. Biochem Biophys Res Commun. 2002;296:877–882. doi: 10.1016/s0006-291x(02)00940-3. [DOI] [PubMed] [Google Scholar]
- 10.Imai D, Yoneya S, Gehlbach PL, Wei LL, Mori K. Intraocular gene transfer of pigment epithelium-derived factor rescues photoreceptors from light-induced cell death. J Cell Physiol. 2005;202:570–578. doi: 10.1002/jcp.20155. [DOI] [PubMed] [Google Scholar]
- 11.Miyazaki M, Ikeda Y, Yonemitsu Y, Goto Y, Sakamoto T, Tabata T, Ueda Y, Hasegawa M, Tobimatsu S, Ishibashi T, Sueishi K. Simian lentiviral vector-mediated retinal gene transfer of pigment epithelium-derived factor protects retinal degeneration and electrical defect in Royal College of Surgeons rats. Gene Ther. 2003;10:1503–1511. doi: 10.1038/sj.gt.3302028. [DOI] [PubMed] [Google Scholar]
- 12.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi N, Miyazaki M, Fukaya K, Inoue Y, Sakaguchi M, Noguchi H, Matsumura T, Watanabe T, Totsugawa T, Tanaka N, Namba M. Treatment of surgically induced acute liver failure with transplantation of highly differentiated immortalized human hepatocytes. Cell Transplant. 2000;9:733–735. doi: 10.1177/096368970000900524. [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi N, Miyazaki M, Fukaya K, Inoue Y, Sakaguchi M, Uemura T, Noguchi H, Kondo A, Tanaka N, Namba M. Transplantation of highly differentiated immortalized human hepatocytes to treat acute liver failure. Transplantation. 2000;69:202–207. doi: 10.1097/00007890-200001270-00002. [DOI] [PubMed] [Google Scholar]
- 15.Kawaguchi T, Osatomi K, Yamashita H, Kabashima T, Uyeda K. Mechanism for fatty acid “sparing” effect on glucose-induced transcription: regulation of carbohydrate-responsive element-binding protein by AMP-activated protein kinase. J Biol Chem. 2002;277:3829–3835. doi: 10.1074/jbc.M107895200. [DOI] [PubMed] [Google Scholar]
- 16.Kawaguchi T, Takenoshita M, Kabashima T, Uyeda K. Glucose and cAMP regulate the L-type pyruvate kinase gene by phosphorylation/dephosphorylation of the carbohydrate response element binding protein. Proc Natl Acad Sci USA. 2001;98:13710–13715. doi: 10.1073/pnas.231370798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawaguchi T, Sakisaka S, Sata M, Mori M, Tanikawa K. Different lobular distributions of altered hepatocyte tight junctions in rat models of intrahepatic and extrahepatic cholestasis. Hepatology. 1999;29:205–216. doi: 10.1002/hep.510290115. [DOI] [PubMed] [Google Scholar]
- 18.Kawaguchi T, Sakisaka S, Mitsuyama K, Harada M, Koga H, Taniguchi E, Sasatomi K, Kimura R, Ueno T, Sawada N, Mori M, Sata M. Cholestasis with altered structure and function of hepatocyte tight junction and decreased expression of canalicular multispecific organic anion transporter in a rat model of colitis. Hepatology. 2000;31:1285–1295. doi: 10.1053/jhep.2000.7435. [DOI] [PubMed] [Google Scholar]
- 19.Yamagishi S, Adachi H, Abe A, Yashiro T, Enomoto M, Furuki K, Hino A, Jinnouchi Y, Takenaka K, Matsui T, Nakamura K, Imaizumi T. Elevated serum levels of pigment epithelium-derived factor in the metabolic syndrome. J Clin Endocrinol Metab. 2006;91:2447–2450. doi: 10.1210/jc.2005-2654. [DOI] [PubMed] [Google Scholar]
- 20.Doll JA, Stellmach VM, Bouck NP, Bergh AR, Lee C, Abramson LP, Cornwell ML, Pins MR, Borensztajn J, Crawford SE. Pigment epithelium-derived factor regulates the vasculature and mass of the prostate and pancreas. Nat Med. 2003;9:774–780. doi: 10.1038/nm870. [DOI] [PubMed] [Google Scholar]
- 21.Uehara H, Miyamoto M, Kato K, Ebihara Y, Kaneko H, Hashimoto H, Murakami Y, Hase R, Takahashi R, Mega S, Shichinohe T, Kawarada Y, Itoh T, Okushiba S, Kondo S, Katoh H. Expression of pigment epithelium-derived factor decreases liver metastasis and correlates with favorable prognosis for patients with ductal pancreatic adenocarcinoma. Cancer Res. 2004;64:3533–3537. doi: 10.1158/0008-5472.CAN-03-3725. [DOI] [PubMed] [Google Scholar]
- 22.Cai J, Parr C, Watkins G, Jiang WG, Boulton M. Decreased pigment epithelium-derived factor expression in human breast cancer progression. Clin Cancer Res. 2006;12:3510–3517. doi: 10.1158/1078-0432.CCR-06-0094. [DOI] [PubMed] [Google Scholar]
- 23.Cheung LW, Au SC, Cheung AN, Ngan HY, Tombran-Tink J, Auersperg N, Wong AS. Pigment epithelium-derived factor is estrogen sensitive and inhibits the growth of human ovarian cancer and ovarian surface epithelial cells. Endocrinology. 2006;147:4179–4191. doi: 10.1210/en.2006-0168. [DOI] [PubMed] [Google Scholar]
- 24.Dawson DW, Volpert OV, Gillis P, Crawford SE, Xu H, Benedict W, Bouck NP. Pigment epithelium-derived factor: a potent inhibitor of angiogenesis. Science. 1999;285:245–248. doi: 10.1126/science.285.5425.245. [DOI] [PubMed] [Google Scholar]
- 25.Ek ET, Dass CR, Choong PF. PEDF: a potential molecular therapeutic target with multiple anti-cancer activities. Trends Mol Med. 2006;12:497–502. doi: 10.1016/j.molmed.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 26.Crawford SE, Stellmach V, Ranalli M, Huang X, Huang L, Volpert O, De Vries GH, Abramson LP, Bouck N. Pigment epithelium-derived factor (PEDF) in neuroblastoma: a multifunctional mediator of Schwann cell antitumor activity. J Cell Sci. 2001;114:4421–4428. doi: 10.1242/jcs.114.24.4421. [DOI] [PubMed] [Google Scholar]
- 27.Tsuru M, Arima N, Toyozumi Y, Kato S. Pigment epithelium-derived factor as a new diagnostic marker for melanocytic tumors. Kurume Med J. 2005;52:81–87. doi: 10.2739/kurumemedj.52.81. [DOI] [PubMed] [Google Scholar]
- 28.Dor I, Namba M, Sato J. Establishment and some biological characteristics of human hepatoma cell lines. Gann. 1975;66:385–392. [PubMed] [Google Scholar]
- 29.Matsumoto K, Ishikawa H, Nishimura D, Hamasaki K, Nakao K, Eguchi K. Antiangiogenic property of pigment epithelium-derived factor in hepatocellular carcinoma. Hepatology. 2004;40:252–259. doi: 10.1002/hep.20259. [DOI] [PubMed] [Google Scholar]
- 30.Chung C, Doll JA, Gattu AK, Shugrue C, Cornwell M, Fitchev P, Crawford SE. Anti-angiogenic pigment epithelium-derived factor regulates hepatocyte triglyceride content through adipose triglyceride lipase (ATGL) J Hepatol. 2008;48:471–478. doi: 10.1016/j.jhep.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 31.Chung C, Shugrue C, Nagar A, Doll JA, Cornwell M, Gattu A, Kolodecik T, Pandol SJ, Gorelick F. Ethanol exposure depletes hepatic pigment epithelium-derived factor, a novel lipid regulator. Gastroenterology. 2009;136:331–340. doi: 10.1053/j.gastro.2008.09.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kratchmarova I, Kalume DE, Blagoev B, Scherer PE, Podtelejnikov AV, Molina H, Bickel PE, Andersen JS, Fernandez MM, Bunkenborg J, Roepstorff P, Kristiansen K, Lodish HF, Mann M, Pandey A. A proteomic approach for identification of secreted proteins during the differentiation of 3T3-L1 preadipocytes to adipocytes. Mol Cell Proteomics. 2002;1:213–222. doi: 10.1074/mcp.m200006-mcp200. [DOI] [PubMed] [Google Scholar]
- 33.Nakamura K, Yamagishi S, Adachi H, Kurita-Nakamura Y, Matsui T, Inoue H. Serum levels of pigment epithelium-derived factor (PEDF) are positively associated with visceral adiposity in Japanese patients with type 2 diabetes. Diabetes Metab Res Rev. 2009;25:52–56. doi: 10.1002/dmrr.820. [DOI] [PubMed] [Google Scholar]
- 34.Nakamura K, Yamagishi SI, Adachi H, Matsui T, Kurita Y, Imaizumi T. Serum levels of pigment epithelium-derived factor (PEDF) are an independent determinant of insulin resistance in patients with essential hypertension. Int J Cardiol. 2008 doi: 10.1016/j.ijcard.2008.11.169. [DOI] [PubMed] [Google Scholar]
- 35.Cao W, Tombran-Tink J, Chen W, Mrazek D, Elias R, McGinnis JF. Pigment epithelium-derived factor protects cultured retinal neurons against hydrogen peroxide-induced cell death. J Neurosci Res. 1999;57:789–800. [PubMed] [Google Scholar]
- 36.Taniwaki T, Becerra SP, Chader GJ, Schwartz JP. Pigment epithelium-derived factor is a survival factor for cerebellar granule cells in culture. J Neurochem. 1995;64:2509–2517. doi: 10.1046/j.1471-4159.1995.64062509.x. [DOI] [PubMed] [Google Scholar]
- 37.Taniwaki T, Hirashima N, Becerra SP, Chader GJ, Etcheberrigaray R, Schwartz JP. Pigment epithelium-derived factor protects cultured cerebellar granule cells against glutamate-induced neurotoxicity. J Neurochem. 1997;68:26–32. doi: 10.1046/j.1471-4159.1997.68010026.x. [DOI] [PubMed] [Google Scholar]
- 38.Yabe T, Kanemitsu K, Sanagi T, Schwartz JP, Yamada H. Pigment epithelium-derived factor induces pro-survival genes through cyclic AMP-responsive element binding protein and nuclear factor kappa B activation in rat cultured cerebellar granule cells: implication for its neuroprotective effect. Neuroscience. 2005;133:691–700. doi: 10.1016/j.neuroscience.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 39.Yabe T, Wilson D, Schwartz JP. NFkappaB activation is required for the neuroprotective effects of pigment epithelium-derived factor (PEDF) on cerebellar granule neurons. J Biol Chem. 2001;276:43313–43319. doi: 10.1074/jbc.M107831200. [DOI] [PubMed] [Google Scholar]
- 40.Pang IH, Zeng H, Fleenor DL, Clark AF. Pigment epithelium-derived factor protects retinal ganglion cells. BMC Neurosci. 2007;8:11. doi: 10.1186/1471-2202-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tombran-Tink J, Barnstable CJ. PEDF: a multifaceted neurotrophic factor. Nat Rev Neurosci. 2003;4:628–636. doi: 10.1038/nrn1176. [DOI] [PubMed] [Google Scholar]
- 42.Tombran-Tink J, Shivaram SM, Chader GJ, Johnson LV, Bok D. Expression, secretion, and age-related downregulation of pigment epithelium-derived factor, a serpin with neurotrophic activity. J Neurosci. 1995;15:4992–5003. doi: 10.1523/JNEUROSCI.15-07-04992.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ho TC, Chen SL, Yang YC, Liao CL, Cheng HC, Tsao YP. PEDF induces p53-mediated apoptosis through PPAR gamma signaling in human umbilical vein endothelial cells. Cardiovasc Res. 2007;76:213–223. doi: 10.1016/j.cardiores.2007.06.032. [DOI] [PubMed] [Google Scholar]
- 44.Nakamura K, Yamagishi S, Matsui T, Yoshida T, Takenaka K, Jinnouchi Y, Yoshida Y, Ueda S, Adachi H, Imaizumi T. Pigment epithelium-derived factor inhibits neointimal hyperplasia after vascular injury by blocking NADPH oxidase-mediated reactive oxygen species generation. Am J Pathol. 2007;170:2159–2170. doi: 10.2353/ajpath.2007.060838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sasaki Y, Naishiro Y, Oshima Y, Imai K, Nakamura Y, Tokino T. Identification of pigment epithelium-derived factor as a direct target of the p53 family member genes. Oncogene. 2005;24:5131–5136. doi: 10.1038/sj.onc.1208695. [DOI] [PubMed] [Google Scholar]
- 46.Tombran-Tink J. The neuroprotective and angiogenesis inhibitory serpin. PEDF: new insights into phylogeny, function, and signaling. Front Biosci. 2005;10:2131–2149. doi: 10.2741/1686. [DOI] [PubMed] [Google Scholar]
- 47.Zhang T, Guan M, Xu C, Chen Y, Lu Y. Pigment epithelium-derived factor inhibits glioma cell growth in vitro and in vivo. Life Sci. 2007;81:1256–1263. doi: 10.1016/j.lfs.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 48.Harada T, Harada C, Wang YL, Osaka H, Amanai K, Tanaka K, Takizawa S, Setsuie R, Sakurai M, Sato Y, Noda M, Wada K. Role of ubiquitin carboxy terminal hydrolase-L1 in neural cell apoptosis induced by ischemic retinal injury in vivo. Am J Pathol. 2004;164:59–64. doi: 10.1016/S0002-9440(10)63096-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kiffin R, Christian C, Knecht E, Cuervo AM. Activation of chaperone-mediated autophagy during oxidative stress. Mol Biol Cell. 2004;15:4829–4840. doi: 10.1091/mbc.E04-06-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takehara T, Liu X, Fujimoto J, Friedman SL, Takahashi H. Expression and role of Bcl-xL in human hepatocellular carcinomas. Hepatology. 2001;34:55–61. doi: 10.1053/jhep.2001.25387. [DOI] [PubMed] [Google Scholar]
- 51.Watanabe J, Kushihata F, Honda K, Sugita A, Tateishi N, Mominoki K, Matsuda S, Kobayashi N. Prognostic significance of Bcl-xL in human hepatocellular carcinoma. Surgery. 2004;135:604–612. doi: 10.1016/j.surg.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 52.Ma DH, Yao JY, Yeh LK, Liang ST, See LC, Chen HT, Lin KY, Liang CC, Lin KK, Chen JK. In vitro antiangiogenic activity in ex vivo expanded human limbocorneal epithelial cells cultivated on human amniotic membrane. Invest Ophthalmol Vis Sci. 2004;45:2586–2595. doi: 10.1167/iovs.03-1338. [DOI] [PubMed] [Google Scholar]
- 53.Singh VK, Chader GJ, Rodriguez IR. Structural and comparative analysis of the mouse gene for pigment epithelium-derived factor (PEDF) Mol Vis. 1998;4:7. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


