Abstract
OBJECTIVE
Mutations in the HNF1A gene are the most common cause of maturity-onset diabetes of the young (MODY). There is a substantial variation in the age at diabetes diagnosis, even within families where diabetes is caused by the same mutation. We investigated the hypothesis that common polygenic variants that predispose to type 2 diabetes might account for the difference in age at diagnosis.
RESEARCH DESIGN AND METHODS
Fifteen robustly associated type 2 diabetes variants were successfully genotyped in 410 individuals from 203 HNF1A-MODY families, from two study centers in the U.K. and Norway. We assessed their effect on the age at diagnosis both individually and in a combined genetic score by summing the number of type 2 diabetes risk alleles carried by each patient.
RESULTS
We confirmed the effects of environmental and genetic factors known to modify the age at HNF1A-MODY diagnosis, namely intrauterine hyperglycemia (−5.1 years if present, P = 1.6 × 10−10) and HNF1A mutation position (−5.2 years if at least two isoforms affected, P = 1.8 × 10−2). Additionally, our data showed strong effects of sex (females diagnosed 3.0 years earlier, P = 6.0 × 10−4) and age at study (0.3 years later diagnosis per year increase in age, P = 4.7 × 10−38). There were no strong individual single nucleotide polymorphism effects; however, in the combined genetic score model, each additional risk allele was associated with 0.35 years earlier diabetes diagnosis (P = 5.1 × 10−3).
CONCLUSIONS
We show that type 2 diabetes risk variants of modest effect sizes reduce the age at diagnosis in HNF1A-MODY. This is one of the first studies to demonstrate that clinical characteristics of a monogenic disease can be modified by common polygenic variants.
Maturity-onset diabetes of the young (MODY) is a young-onset, dominantly inherited non–insulin-dependent diabetes resulting from β-cell dysfunction (1). There are at least eight genetic subgroups of MODY (1,2), with most patients having mutations in transcription factor genes. Hepatocyte nuclear factor 1α (HNF1A) mutations are the commonest cause of MODY in many series (3,4). HNF1A diabetes is characterized by progressive failure of β-cell function, resulting in increasing hyperglycemia throughout life (1). Initially, basal insulin secretion is maintained, but it cannot be increased in the presence of hyperglycemia (5).
The severity and clinical presentation of MODY varies according to MODY genetic subtype (6). In addition, there can be considerable variation both between and within families where diabetes is caused by mutations in the same gene. In HNF1A diabetes, the age of diagnosis is widely variable (4–74 years [7]), and, although the mutations are highly penetrant, only 63% of mutation carriers develop diabetes by the age of 25 years (8). The variation in diagnosis is influenced by social and environmental factors. Within families, early age at diagnosis tends to fall in the younger generations, in part owing to increased awareness of the familial nature of the condition (9,10). If the mother had diabetes during pregnancy, intrauterine exposure to hyperglycemia of maternal diabetes is associated with diabetes being diagnosed, on average, 12 years earlier compared with subjects not exposed to maternal hyperglycemia (9,10).
It is likely that there are genetic modifiers of the age of onset of HNF1A diabetes, namely the position of the HNF1A mutation (11,12). However, much of the variation in age at diagnosis within families, where diabetes is caused by the same mutation, cannot be explained by social or environmental factors, and this supports the notion that there are likely to be genetic modifiers independent of the HNF1A mutation. A genome-wide search for genetic modifiers of diagnosis age found no single large linkage peak (13), suggesting that the age of onset is a complex genetic trait. A previous study of one large pedigree has shown that severity of the HNF1A diabetes phenotype was increased (earlier age of diagnosis and more severe hyperglycemia) when type 2 diabetes was present in the noncarrier parent (14). We therefore hypothesized that common genetic variants that predispose to type 2 diabetes might modify the severity of the disease and explain some of the variation in the age at HNF1A diabetes diagnosis.
RESEARCH DESIGN AND METHODS
HNF1A mutation patients.
The subjects were 410 HNF1A mutation carriers with diabetes from two sources: the Department of Molecular Genetics, Royal Devon, and Exeter National Health Service Foundation Trust, Exeter, U.K. (n = 298 from 140 families) and the Center for Diabetes Genetics, Department of Pediatrics, Haukeland University Hospital, Bergen, Norway (n = 112 from 63 families). They were all established MODY patients previously recruited for HNF1A sequencing on the basis of clinical criteria, such as family history or first-degree relative with diabetes, onset of diabetes typically before age 25 years, or low-dose insulin requirement (full details are available at http://www.diabetesgenes.org and http://www.mody.no).
All patients gave consent for genetic testing and had HNF1A mutations identified by direct sequencing. To avoid population heterogeneity, individuals who were not Caucasian Europeans were excluded. Clinical characteristics of the patients are shown in Table 1. Two-thirds were female, and the majority of the probands had an HNF1A mutation in exons 1–6. Such mutations are regarded as clinically most severe as they affect all three isoforms of the gene product. The most common HNF1A mutation, P291fsinsC in exon 4, accounted for 32% of all cases in this study.
TABLE 1.
Characteristics of the 410 HNF1A-MODY patients included in the analyses
| U.K. | Norway | |
|---|---|---|
| Families/singletons | 140/87 | 63/41 |
| Examined individuals | 298 | 112 |
| Male subjects | 100 (33.6) | 41 (36.6) |
| Average number of individuals in nonsingleton families (range) | 3.0 (2–9) | 3.1 (2–8) |
| Individuals with HNF1A mutations affecting: | ||
| Isoform A only (exons 8–10) | 30 | 3 |
| Isoforms A and B only (exon 7) | 22 | 4 |
| Isoforms A, B, and C (exons 1–6) | 246 | 105 |
| Age at study (years) | 37.1 ± 17.0 (8–87) | 33.4 + 17.6 (6–73) |
| Age at diabetes diagnosis (years) | 21.9 ± 11.2 (4–70) | 20.3 + 10.0 (6–60) |
| BMI (kg/m2)* | 24.1 ± 4.1 (15.9–50.7) | 23.9 + 3.7 (15.8–33.6) |
Data are n, n (%), and means ± SD (range).
*BMI was only available for 224 and 81 individuals in U.K. and Norway studies, respectively.
Classification and assessment of nonpolygenic modifiers.
We assessed the association between age at diagnosis and the following nonpolygenic factors: sex, age at study, BMI, exposure to in utero hyperglycemia, and the position of HNF1A mutation. Mother's diabetes status at pregnancy was calculated from the age of diagnosis and the date of birth of the mother and child. Where this information was incomplete (usually owing to mother's information not being available as deceased), we assumed that the patient was not exposed to in utero hyperglycemia. Previous studies have shown that mutation position impacts the severity of the disease by determining the number of the affected HNF1A isoforms: patients with mutations in exons 1–7, affecting two or all three isoforms, were diagnosed earlier than patients with mutations in exons 8–10, affecting only one isoform (11,12). To account for this effect, we classified mutations of patients in this study according to those two groups. Intronic mutations were assigned to a group according to the HNF1A isoform impacted.
Single nucleotide polymorphism selection and genotyping.
We decided to include in this study only those variants, or their proxies, robustly shown to predispose to type 2 diabetes in Caucasians. Seventeen common susceptibility variants had been identified and robustly replicated at the time of our study (15–20) (recently rev. in 21,22). These include single nucleotide polymophisms (SNPs) in or near PPARG, KCNJ11, TCF7L2, IGF2BP2, CDKN2A/2B, CDKAL1, SLC30A8, HHEX/IDE, FTO, WFS1, HNF1B (TCF2), MC4R, NOTCH2, ADAMTS9, THADA, TSPAN8/LGR5, CDC123/CAMK1D, and JAZF1 genes. Table 3 lists all 17 SNPs assessed in our study, of which we were able to combine 15 for the joint analysis, because JAZF1 and NOTCH2 loci failed genotyping in the Norwegian samples. At 4 of 15 loci, different SNPs were genotyped by the two study centers: rs757210 (U.K.) and rs4430796 (Norway) at the HNF1B locus (HapMap CEU r2 = 0.61, D′ = 0.96), rs10946398 and rs7754840 at CDKAL1 (r2 and D′ = 1), rs8050136 and rs9939609 at FTO (r2 and D′ = 1), and rs1111875 and rs5015480 at HHEX/IDE (r2 and D′ = 1). We combined genotypes for each of the four proxy pairs, coded with respect to the type 2 diabetes risk allele.
TABLE 3.
Effects of individual type 2 diabetes risk variants and the combined genetic scores on the age at diabetes diagnosis in 410 HNF1A-MODY patients
| Individual SNP effects | Unadjusted results |
Adjusted results* |
|||
|---|---|---|---|---|---|
| Effect size ± SE | P | Effect size ± SE | P | ||
| Gene region | SNP | ||||
| HNF1B (TCF2)† | rs757210/rs4430796 | −1.85 ± 0.58 | 0.0014 | −1.07 ± 0.43 | 0.014 |
| SLC30A8 | rs13266634 | −1.07 ± 0.64 | 0.095 | −0.90 ± 0.50 | 0.070 |
| CDKAL1† | rs10946398/rs7754840 | −1.22 ± 0.59 | 0.038 | −0.87 ± 0.46 | 0.059 |
| TCF7L2 | rs7903146 | −0.55 ± 0.63 | 0.39 | −0.65 ± 0.46 | 0.16 |
| ADAMTS9 | rs4607103 | 0.27 ± 0.69 | 0.70 | −0.59 ± 0.51 | 0.25 |
| TSPAN8 | rs7961581 | −0.97 ± 0.63 | 0.13 | −0.53 ± 0.44 | 0.22 |
| JAZF1‡ | rs864745 | −0.45 ± 0.72 | 0.53 | −0.46 ± 0.53 | 0.38 |
| FTO† | rs8050136/rs9939609 | −0.30 ± 0.63 | 0.63 | −0.42 ± 0.47 | 0.37 |
| KCNJ11 | rs5219 | −0.15 ± 0.63 | 0.82 | −0.34 ± 0.50 | 0.50 |
| CDKN2A/2B | rs10811661 | −0.89 ± 0.87 | 0.31 | −0.25 ± 0.65 | 0.70 |
| WFS1 | rs10010131 | −0.08 ± 0.61 | 0.89 | −0.21 ± 0.46 | 0.65 |
| CDC123 | rs12779790 | 0.83 ± 0.75 | 0.27 | 0.07 ± 0.55 | 0.91 |
| HHEX/IDE† | rs1111875/rs5015480 | −0.27 ± 0.61 | 0.66 | 0.19 ± 0.44 | 0.66 |
| PPARG | rs1801282 | −1.46 ± 0.99 | 0.14 | 0.36 ± 0.76 | 0.64 |
| IGF2BP2 | rs4402960 | 0.45 ± 0.64 | 0.48 | 0.43 ± 0.47 | 0.36 |
| THADA | rs7578597 | 0.50 ± 1.00 | 0.62 | 0.55 ± 0.78 | 0.48 |
| NOTCH2‡ | rs2934381 | 1.31 ± 1.32 | 0.32 | 0.82 ± 1.00 | 0.41 |
| Combined SNP effect | |||||
| Allele count score | −0.49 ± 0.17 | 0.0043 | −0.35 ± 0.13 | 0.0051 | |
| Weighted score (log odds) | −0.49 ± 0.15 | 0.0013 | −0.33 ± 0.12 | 0.0046 | |
All effect sizes are in years change of age at diagnosis per risk allele. The 410 patients were successfully genotyped for all 15 SNPs that were included in the combined genetic scores. All analyses took into account full family relationships and included a random family effect in the regression model. Individual SNP effects are based on risk allele count method. P values are unadjusted for multiple testing. Results are presented in order of the adjusted effect sizes.
*Adjusted results include study, sex, age at study, presence of intrauterine hyperglycemia, and mutation position (two groups, according to exon affected, 1–7 or 8–10) as covariates in the regression model.
†At 4 loci different SNPs representing the same signal were genotyped by the two study centers, in which case they are shown as U.K./Norway SNPs.
‡Results for JAZF1 and NOTCH2 SNPs were available only for U.K. samples (n = 296 and 297, respectively).
In the U.K. samples, genotyping of TCF7L2, KCNJ11, and PPARG SNPs was performed in house, using a TaqMan-based assay. The probes were supplied by Applied Biosystems (Foster City, CA). Genotyping of the remaining 14 variants was performed by K Biosciences (Herts, U.K.), who designed and used assays based on either their proprietary competitive allele-specific PCR (KASPar) method or a modified TaqMan assay (details of which are available on their website www.kbioscience.co.uk/chemistry/chemistry_Kasp_intro.htm). Genotyping success rate was >96% for each SNP, overall duplicate concordance rate was 99.9% (one discrepancy from 1,153 comparisons), and in the Hardy-Weinberg equilibrium test, used as an additional genotyping quality check, all P values were >0.01 for the full dataset and >0.05 for 140 unrelated probands.
In the Norwegian samples, genotyping was carried out by the multiplex MassARRAY iPLEX System (Sequenom, San Diego, CA) at the Norwegian national technology platform CIGENE. NOTCH2 SNP (rs2934381) failed the assay design, while JAZF1 SNP (rs864745) had a poor genotype call rate. For the remaining 15 SNPs, which we were able to combine with the U.K. data, genotype concordance rate was 100% for internal controls (n = 108 genotypes). Final genotyping call rate was 99.2% after exclusion of samples with bad quality or lacking DNA. All tests for Hardy-Weinberg equilibrium had P values >0.05.
Statistical methods.
We performed family-based association analyses using the ASSOC program from S.A.G.E. (Statistical Analysis for Genetic Epidemiology) software package, version 5.4.2, for Linux (23). Assuming randomly sampled independent pedigrees, ASSOC simultaneously tests for associations between a quantitative trait and one or more covariates of interest and estimates familial variance components from the given familial correlations. In our study, the trait of interest was age at diabetes diagnosis, while the main covariate of interest was the number of type 2 diabetes risk alleles. As one of the parameters for the ASSOC program, we set the family effect option to “true,” thus including the random nuclear family effect as an additional term in the regression model. Relationships between family members were fully established for most pedigrees. In some of the large pedigrees, we included parents and relatives that had no data for the analyses but were used by the program to accurately connect all related individuals. Singletons and five U.K. family members for whom we could not establish how they were related to other members of their pedigrees were automatically treated as one-person pedigrees and required no special handling in the model.
We assessed the effect of each risk variant on the age at diagnosis individually, jointly and by using an allele counting method to assign a genetic risk score to each patient (the sum of the number of risk alleles a person carries). The allele-counting method assumed equal and additive effects of the individual variants. We repeated this analysis using a weighted allele approach, where the genetic score was based on the previously reported odds ratios (ORs) for type 2 diabetes (obtained from a recent review [22]). For each patient, we first calculated the sum across SNPs of the number of risk alleles at each SNP multiplied by the log of the OR for that SNP (i.e., genotypes were coded as 0, log(OR), or 2xlog(OR), rather than 0, 1, 2 in the allele count model). To obtain a rescaled “weighted allele count” score, we multiplied each log(OR) score by 30 (maximum number of risk alleles) and divided the product by 1.81, the sum of the 15 risk homozygote log(OR) weights.
Although family relationships were fully accounted for in the analyses, it is possible that the results could have been affected by the skewed allele distributions. Therefore, we analyzed the effects of both individual SNPs and the combined genetic score on age at diagnosis in 203 unrelated probands, using the youngest individual from each pedigree (see supplementary Table 1 in the online appendix [available at http://diabetes.diabetesjournals.org/cgi/content/full/db09-0555/DC1]).
We used StataSE version 10.0 for Windows (StataCorp LP, Brownsville, TX) to generate adjusted age at diagnosis, using the “predict” function after running linear regression that fitted family identification, study, sex, age at study, presence of intrauterine hyperglycemia, and mutation position in the same regression model. This enabled us to use the full dataset, with adjusted ages at diagnosis, for linear regression and cumulative diabetes incidence analyses (Figs. 1 and 2), rather than a much smaller sample of unrelated singletons and phenotypically homogeneous individuals. All figures were generated using SigmaPlot (Systat Software, San Jose, CA). Power calculations were performed using QUANTO power calculator, version 1.2.4 (24).
FIG. 1.
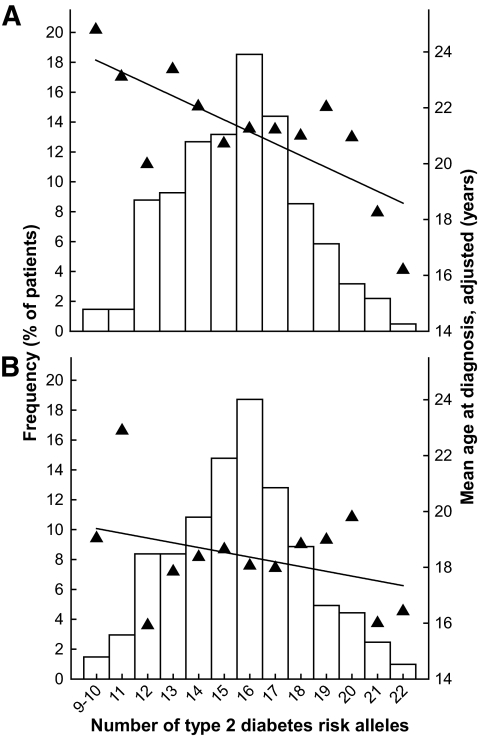
Mean age at diabetes diagnosis (▲) and frequency (□) of HNF1A-MODY patients at each number of the type 2 diabetes risk alleles carried. Only individuals genotyped for all 15 variants are included. A: Full dataset of 410 patients. B: 203 unrelated probands (youngest family members). Ages at diagnosis were adjusted for family (A only), study, sex, age at study, exposure to mother's hyperglycemia in utero, and position of HNF1A mutation. Black lines are the fitted age-at-diagnosis linear regression lines. Both y-axes are on the same scale in A and B.
FIG. 2.
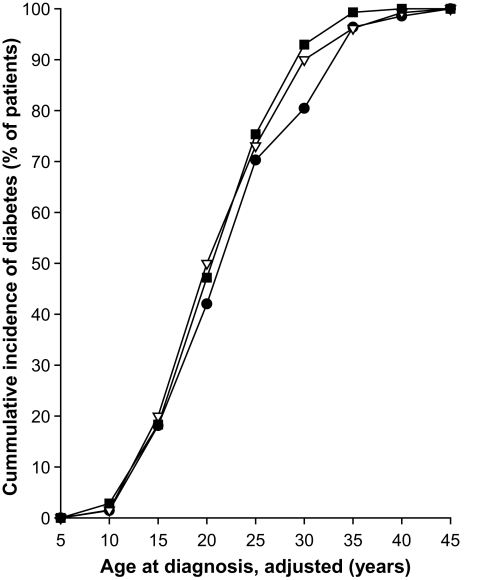
Cumulative incidence of diabetes in 410 HNF1A-MODY patients by type 2 diabetes risk allele count category. ●, 9–14 risk alleles, n = 138; △, 15–16 risk alleles, n = 130; ■, 17–22 risk alleles, n = 142. Only individuals genotyped for all 15 variants are included. The ages at diabetes diagnosis ware adjusted for family, study, sex, age at study, exposure to mother's hyperglycemia in utero, and position of HNF1A mutation.
RESULTS
The analyses included 410 diabetic HNF1A mutation carriers from 203 families who were successfully genotyped for all 15 type 2 diabetes risk variants.
We confirmed the strong effects of age at study, mutation position, and intrauterine hyperglycemia on the severity of HNF1A diabetes clinical presentation (Table 2). These associations were independent of the polygenic risk factors (Table 2). On average, patients were diagnosed 5.1 years earlier if the mother was diabetic during pregnancy (P = 1.6 × 10−10), 5.2 years earlier if the mutation affected at least two HNF1A isoforms (P = 1.8 × 10−2), and 0.3 years later for every additional year of their age at study (P = 4.7 × 10−38). In addition, we observed a strong effect of sex in our data, where female subjects were diagnosed 3.0 years earlier than male subjects (P = 6.0 × 10−4), but there was no association with BMI (available for 305 subjects; P = 0.99).
TABLE 2.
Results of regression analyses of nonpolygenic factors on the age at diabetes diagnosis in 410 HNF1A-MODY patients with and without the inclusion of the polygenic risk score in the regression model
| Without genetic score |
With genetic score |
|||
|---|---|---|---|---|
| Effect size ± SE | P | Effect size ± SE | P | |
| Study (Norway = 0, U.K. = 1) | 1.31 ± 0.99 | 0.18 | 1.44 ± 0.97 | 0.14 |
| Sex (effect with respect to female subjects) | −2.97 ± 0.86 | 6.0 × 10−4 | −2.93 ± 0.85 | 5.7 × 10−4 |
| BMI (kg/m2) (per unit increase)* | 0.0009 ± 0.13 | 0.99 | 0.033 ± 0.13 | 0.80 |
| Presence of intrauterine hyperglycemia | −5.06 ± 0.79 | 1.6 × 10−10 | −4.86 ± 0.79 | 6.5 × 10−10 |
| Position of HNF1A mutation† | −5.22 ± 2.21 | 1.8 × 10−2 | −5.67 ± 2.18 | 9.4 × 10−3 |
| Age at study (per year increase) | 0.29 ± 0.02 | 4.7 × 10−38 | 0.29 ± 0.02 | 1.5 × 10−37 |
All effect sizes are in years. Genetic score is the number of risk alleles, carried by each patient, from the 15 type 2 diabetes susceptibility variants.
*BMI was only available for 305 individuals.
†The position of the mutation has been dichotomized into those affecting exons 8–10 (isomer A only; n = 33) versus those affecting exons 1–7 (n = 377). The age at diagnosis is lower for patients with mutations affecting exons 1–7.
We included those variables that had individual effect on age at diagnosis (i.e., all of the above apart from BMI) as covariates in the individual and joint SNP models to reduce the remaining variance in the age at diagnosis and therefore increase our power to detect the effect of polygenic modifiers. We repeated these analyses excluding age at study, to make sure that its strong association with age at diagnosis did not drive the SNP association (supplementary Table 2). As expected, the results were not statistically significantly different to the fully adjusted model (all t test P > 0.32). Although for some of the SNPs the effects on age at diagnosis were slightly stronger when age at study was excluded, the SEs were larger, resulting in similar overall P values.
Individual type 2 diabetes risk variants were not strongly associated with the age at diagnosis, as shown in Table 3. However, of 15 variants, 11 risk alleles for type 2 diabetes in the unadjusted analyses and 10 in the adjusted analyses were associated with reduced age at diagnosis. When we included all 15 variants in the regression model, there was borderline evidence of an overall joint effect on the age at diagnosis (P = 0.062). The 15 variants explain 6.4% of the total proportion of age at diagnosis variation, while the nonpolygenic factors (sex, age at study, mutation position, and presence of intrauterine hyperglycemia) explain 37.9%; combining these together, they explain 42.1% of the total variance in the HNF1A-MODY age at diagnosis in these families.
We then generated a single genetic risk score representing the combined genetic susceptibility for type 2 diabetes (Table 3). In the allele count model, each additional risk allele was associated with a 0.35-year reduction in age at diagnosis (P = 0.005). The association strength was weaker when we used unrelated probands (0.28 years earlier age at diagnosis per one additional risk allele; P = 0.094; supplementary Table 1), which most probably reflects reduced power. The correlation between the decreasing age at diagnosis and the increasing number of risk alleles appears to be linear for the full dataset of 410 patients (Fig. 1A). Figure 1B presents the results for 203 unrelated probands only. Looking at the impact of risk alleles on the cumulative incidence of diabetes, the effect was most noticeable around age 30 years, where diabetes developed in 80% of HNF1A mutation carriers with 9–14 polygenic risk alleles, compared with 93% with 17–22 risk alleles (Fig. 2).
The weighted allele score yielded similar results to the allele count model (P = 0.005). Stratified analysis showed that the impact of the allele count score was of similar magnitude in the two cohorts individually, with all t test P values >0.1 (supplementary Table 3).
DISCUSSION
We have shown that type 2 diabetes risk variants of modest effect sizes when combined are associated with a reduced age at diagnosis in monogenic HNF1A diabetes. This association is independent of other genetic and environmental modifiers, namely the HNF1A mutation position, age at study, sex, and mother's diabetes status during pregnancy. Thus, this is one of the first studies to demonstrate that clinical characteristics of a monogenic disease can be influenced by common variants that predispose to the polygenic form of that disease. To our knowledge, only two other studies, of breast cancer (25) and Alzheimer's disease (26) have identified polygenic variants that act as modifiers of disease onset age.
In support of previous findings, an increase in the age of patients at the time of genetic testing is strongly associated with an older age at diabetes diagnosis. It is not known if this represents an earlier diagnosis as a result of the increasing awareness of diabetes in the family by their physicians or a genuine decrease in age of onset in succeeding generations. The former is likely to be a large contributor. In addition, there is strong evidence that the age at diagnosis is affected by genetic factors. We confirm previous findings by Harries et al. (11) and Bellanné-Chantelot et al. (12) that patients with mutations affecting at least two of the three known HNF1A isoforms were diagnosed earlier than patients with mutations affecting only one HNF1A isoform.
In our study, we provide evidence for additional genetic modifiers, the robustly replicated type 2 diabetes risk variants. Combining the effect of the variants by adding up the total number of risk alleles carried, each additional risk allele was associated with 0.49 and 0.35 years earlier age at diagnosis in the unadjusted and adjusted models, respectively. Most of the genetic variants predisposing to type 2 diabetes act through reducing β-cell function rather than increasing insulin resistance. This is true of the three risk variants with strongest effects observed in this study, the SNPs in the HNF1B, SLC30A8, and CDKAL1 genes. It is possible that they interact with the β-cell dysfunction resulting from the HNF1A mutation, leading to an increased rate of β-cell destruction and, therefore, earlier onset of diabetes. Furthermore, mutations of HNF1B, also known as TCF2, are another known cause of MODY, accounting for ∼2% of cases (27).
This study does have limitations. Although we included 410 subjects, one of the largest cohorts of HNF1A patients ever reported, some simple power calculations suggest that we were still underpowered to detect the impact of individual loci. For example, we had only 29% power to detect an individual SNP explaining 1% of the variation in age at diagnosis (and this is assuming independence of the individuals in the study) at P < 0.01; in singleton-only analysis the power was 13%. These patients were not studied prospectively, and, therefore, the age at diagnosis does not accurately reflect the age at onset of diabetes. We would anticipate that if age of onset was studied using prospective data, the impact of these type 2 diabetes loci would be greater.
In conclusion, we show that type 2 diabetes risk variants of modest effect sizes act as an additional modifier of age at diagnosis in HNF1A-MODY. This is one of the first studies to demonstrate that common variants associated with a polygenic disease can also influence clinical characteristics of a monogenic form of the disease.
Supplementary Material
Acknowledgments
M.N.W. is a Vandervell Foundation Research Fellow. B.S. and S.E. are employed as core staff within the National Institute for Health Research–funded Peninsula Clinical Research Facility. Parts of this study were supported by the University of Bergen, Haukeland University Hospital, Helse Vest, Innovest, and the Research Council of Norway (to P.R.N.).
No potential conflicts of interest relevant to this article were reported.
Parts of the preliminary results of this study were presented at the European Association for the Study of Diabetes–Study Group on Genetics of Diabetes 2nd Meeting, 22–25 April 2009, Bergen, Norway.
We thank all MODY patients and nurses who took part and contributed to this study.
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Murphy R, Ellard S, Hattersley AT: Clinical implications of a molecular genetic classification of monogenic beta-cell diabetes. Nat Clin Pract Endocrinol Metab 2008;4:200–213 [DOI] [PubMed] [Google Scholar]
- 2.Hattersley A, Bruining J, Shield J, Njolstad P, Donaghue K: ISPAD Clinical Practice Consensus Guidelines 2006–2007: the diagnosis and management of monogenic diabetes in children. Pediatr Diabetes 2006;7:352–360 [DOI] [PubMed] [Google Scholar]
- 3.Frayling T, Bulman MP, Ellard S, Appleton M, Dronsfield M, Mackie A, Baird J, Kaisaki P, Yamagata K, Bell G, Bain S, Hattersley A: Mutations in the Hepatocyte Nuclear Factor 1 Alpha gene are a common cause of maturity-onset diabetes of the young in the United Kingdom. Diabetes 1997;46:720–725 [DOI] [PubMed] [Google Scholar]
- 4.Bjorkhaug L, Sagen JV, Thorsby P, Sovik O, Molven A, Njolstad PR: Hepatocyte nuclear factor-1 alpha gene mutations and diabetes in Norway. J Clin Endocrinol Metab 2003;88:920–931 [DOI] [PubMed] [Google Scholar]
- 5.Byrne MM, Sturis J, Menzel S, Yamagata K, Fajans SS, Dronsfield MJ, Bain SC, Hattersley AT, Velho G, Froguel P, Bell GI, Polonsky KS: Altered insulin secretory responses to glucose in diabetic and nondiabetic subjects with mutations in the diabetes susceptibility gene MODY3 on Chromosome 12. Diabetes 1996;45:1503–1510 [DOI] [PubMed] [Google Scholar]
- 6.Hattersley AT: Maturity-onset diabetes of the young: clinical heterogeneity explained by genetic heterogeneity. Diabet Med 1998;15:15–24 [DOI] [PubMed] [Google Scholar]
- 7.Frayling TM, Evans JC, Bulman MP, Pearson E, Allen L, Owen K, Bingham C, Hannemann M, Shepherd M, Ellard S, Hattersley AT: Â-Cell genes and diabetes: molecular and clinical characterization of mutations in transcription factors. Diabetes 2001;50:S94–100 [DOI] [PubMed] [Google Scholar]
- 8.Shepherd M, Hattersley AT, Sparkes A: Genetic testing in maturity onset diabetes of the young (MODY): a new challenge for the diabetic clinic. Pract Diabetes 2001;18:16–21 [DOI] [PubMed] [Google Scholar]
- 9.Stride A, Shepherd M, Frayling TM, Bulman MP, Ellard S, Hattersley AT: Intrauterine hyperglycemia is associated with an earlier diagnosis of diabetes in HNF-1α gene mutation carriers. Diabetes Care 2002;25:2287–2291 [DOI] [PubMed] [Google Scholar]
- 10.Klupa T, Warram JH, Antonellis A, Pezzolesi M, Nam M, Malecki MT, Doria A, Rich SS, Krolewski AS: Determinants of the development of diabetes (maturity-onset diabetes of the young-3) in carriers of HNF-1α mutations: evidence for parent-of-origin effect. Diabetes Care 2002;25:2292–2301 [DOI] [PubMed] [Google Scholar]
- 11.Harries LW, Ellard S, Stride A, The European Mc. Morgan NG, Hattersley AT: Isomers of the TCF1 gene encoding hepatocyte nuclear factor-1 alpha show differential expression in the pancreas and define the relationship between mutation position and clinical phenotype in monogenic diabetes. Hum Mol Genet 2006;15:2216–2224 [DOI] [PubMed] [Google Scholar]
- 12.Bellanne-Chantelot C, Carette C, Riveline JP, Valero R, Gautier JF, Larger E, Reznik Y, Ducluzeau PH, Sola A, Hartemann-Heurtier A, Lecomte P, Chaillous L, Laloi-Michelin M, Wilhem JM, Cuny P, Duron F, Guerci B, Jeandidier N, Mosnier-Pudar H, Assayag M, Dubois-Laforgue D, Velho G, Timsit J: The type and the position of HNF1A mutation modulate age at diagnosis of diabetes in patients with maturity-onset diabetes of the young (MODY)-3. Diabetes 2008;57:503–508 [DOI] [PubMed] [Google Scholar]
- 13.Kim SH, Ma XW, Klupa T, Powers C, Pezzolesi M, Warram JH, Rich SS, Krolewski AS, Doria A: Genetic modifiers of the age at diagnosis of diabetes (MODY3) in carriers of hepatocyte nuclear factor-1α mutations map to chromosomes 5p15, 9q22, and 14q24. Diabetes 2003;52:2182–2186 [DOI] [PubMed] [Google Scholar]
- 14.Tack CJJ, Ellard S, Hattersley AT: A severe clinical phenotype results from the co-inheritance of type 2 susceptibility genes and a hepatocyte nuclear factor-1α mutation. Diabetes Care 2000;23:424–425 [DOI] [PubMed] [Google Scholar]
- 15.Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PIW, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ, Hughes TE, Groop L, Altshuler D, Almgren P, Florez JC, Meyer J, Ardlie K, Bostrom KB, Isomaa B, Lettre G, Lindblad U, Lyon HN, Melander O, Newton-Cheh C, Nilsson P, Orho-Melander M, Rastam L, Speliotes EK, Taskinen MR, Tuomi T, Guiducci C, Berglund A, Carlson J, Gianniny L, Hackett R, Hall L, Holmkvist J, Laurila E, Sjogren M, Sterner M, Surti A, Svensson M, Svensson M, Tewhey R, Blumenstiel B, Parkin M, DeFelice M, Barry R, Brodeur W, Camarata J, Chia N, Fava M, Gibbons J, Handsaker B, Healy C, Nguyen K, Gates C, Sougnez C, Gage D, Nizzari M, Gabriel SB, Chirn GW, Ma QC, Parikh H, Richardson D, Ricke D, Purcell S: Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 2007;316:1331–1336 [DOI] [PubMed] [Google Scholar]
- 16.Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU, Prokunina-Olsson L, Ding CJ, Swift AJ, Narisu N, Hu T, Pruim R, Xiao R, Li XY, Conneely KN, Riebow NL, Sprau AG, Tong M, White PP, Hetrick KN, Barnhart MW, Bark CW, Goldstein JL, Watkins L, Xiang F, Saramies J, Buchanan TA, Watanabe RM, Valle TT, Kinnunen L, Abecasis GR, Pugh EW, Doheny KF, Bergman RN, Tuomilehto J, Collins FS, Boehnke M: A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 2007;316:1341–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S, Balkau B, Heude B, Charpentier G, Hudson TJ, Montpetit A, Pshezhetsky AV, Prentki M, Posner BI, Balding DJ, Meyre D, Polychronakos C, Froguel P: A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 2007;445:881. [DOI] [PubMed] [Google Scholar]
- 18.Zeggini E, Scott LJ, Saxena R, Voight BF, Marchini JL, Hu T, de Bakker PIW, Abecasis GR, Almgren P, Andersen G, Ardlie K, Bostroem KB, Bergman RN, Bonnycastle LL, Borch-Johnsen K, Burtt NP, Chen H, Chines PS, Daly MJ, Deodhar P, Ding CJ, Doney ASF, Duren WL, Elliott KS, Erdos MR, Frayling TM, Freathy RM, Gianniny L, Grallert H, Grarup N, Groves CJ, Guiducci C, Hansen T, Herder C, Hitman GA, Hughes TE, Isomaa B, Jackson AU, Jorgensen T, Kong A, Kubalanza K, Kuruvilla FG, Kuusisto J, Langenberg C, Lango H, Lauritzen T, Li Y, Lindgren CM, Lyssenko V, Marvelle AF, Meisinger C, Midthjell K, Mohlke KL, Morken MA, Morris AD, Narisu N, Nilsson P, Owen KR, Palmer CNA, Payne F, Perry JRB, Pettersen E, Platou C, Prokopenko I, Qi L, Qin L, Rayner NW, Rees M, Roix JJ, Sandbaek A, Shields B, Sjogren M, Steinthorsdottir V, Stringham HM, Swift AJ, Thorleifsson G, Thorsteinsdottir U, Timpson NJ, Tuomi T, Tuomilehto J, Walker M, Watanabe RM, Weedon MN, Willer CJ, Illig T, Hveem K, Hu FB, Laakso M, Stefansson K, Pedersen O, Wareham NJ, Barroso I, Hattersley AT, Collins FS, Groop L, McCarthy MI, Boehnke M, Altshuler D: Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet 2008;40:638–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, Timpson NJ, Perry JRB, Rayner NW, Freathy RM, Barrett JC, Shields B, Morris AP, Ellard S, Groves CJ, Harries LW, Marchini JL, Owen KR, Knight B, Cardon LR, Walker M, Hitman GA, Morris AD, Doney ASF, McCarthy MI, Hattersley AT: Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 2007;316:1336–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hertel JK, Johansson S, Raeder H, Midthjell K, Lyssenko V, Groop L, Molven A, Njolstad PR: Genetic analysis of recently identified type 2 diabetes loci in 1,638 unselected patients with type 2 diabetes and 1,858 control participants from a Norwegian population-based cohort (the HUNT study). Diabetologia 2008;51:971–977 [DOI] [PubMed] [Google Scholar]
- 21.Frayling TM: Genome-wide association studies provide new insights into type 2 diabetes aetiology. Nat Rev Genet 2007;8:657–662 [DOI] [PubMed] [Google Scholar]
- 22.Prokopenko I, McCarthy MI, Lindgren CM: Type 2 diabetes: new genes, new understanding. Trends Genet 2008;24:613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.S.A.G.E. Statistical Analysis for Genetic Epidemiology, Release 5.4.2. [Accessed 11 December 2008]. Available at http://darwin.cwru.edu/
- 24.Gauderman W, Morrison J: QUANTO 1.1: a computer program for power and sample size calculations for genetic-epidemiology studies. Available at http://hydra.usc.edu/gxe Accessed 11 May 2009
- 25.Antoniou AC, Sinilnikova OM, Simard J, Leone M, Dumont M, Neuhausen SL, Struewing JP, Stoppa-Lyonnet D, Barjhoux L, Hughes DJ, Coupier I, Belotti M, Lasset C, Rebbeck TR, Wagner T, Lynch HT, Domchek SM, Nathanson KL, Garber JE, Weitzel J, Narod SA, Tomlinson G, Olopade OI, Godwin A, Isaacs C, Jakubowska A, Lubinski J, Gronwald J, Gorski B, Byrski T, Huzarski T, Peock S, Cook M, Baynes C, Murray A, Rogers M, Daly PA, Dorkins H, Schmutzler RK, Versmold B, Engel C, Meindl A, Arnold N, Niederacher D, Deissler H, Spurdle AB, Chen XQ, Waddell N, Cloonan N, Kirchhoff T, Offit K, Friedman E, Kaufmann B, Laitman Y, Galore G, Rennert G, Lejbkowicz F, Raskin L, Andrulis IL, Ilyushik E, Ozcelik H, Devilee P, Vreeswijk MPG, Greene MH, Prindiville SA, Osorio A, Benitez J, Zikan M, Szabo CI, Kilpivaara O, Nevanlinna H, Hamann U, Durocher F, Arason A, Couch FJ, Easton DF, Chenevix-Trench G: RADS1 135G -> C modifies breast cancer risk among BRCA2 mutation carriers: results from a combined analysis of 19 studies. Am J Hum Genet 2007;81:1186–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bertram L, Lange C, Mullin K, Parkinson M, Hsiao M, Hogan MF, Schjeide BMM, Hooli B, DiVito J, Ionita L, Jiang HY, Laird N, Moscarillo T, Ohlsen KL, Elliott K, Wang X, Hu-Lince D, Ryder M, Murphy A, Wagner SL, Blacker D, Becker KD, Tanzi RE: Genome-wide association analysis reveals putative Alzheimer's disease susceptibility loci in addition to APOE. Am J Hum Genet 2008;83:623–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCarthy MI, Hattersley AT: Learning from molecular genetics: novel insights arising from the definition of genes for monogenic and type 2 diabetes. Diabetes 2008;57:2889–2898 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.