Abstract
Helicobacter species are among the most successful colonizers of the mammalian gastrointestinal and hepatobiliary tract. Colonization is usually lifelong, indicating that Helicobacter species have evolved intricate mechanisms of dealing with stresses encountered during colonization of host tissues, like restriction of essential metal ions. The recent availability of genome sequences of the human gastric pathogen Helicobacter pylori, the murine enterohepatic pathogen Helicobacter hepaticus and the unannotated genome sequence of the ferret gastric pathogen Helicobacter mustelae has allowed for comparitive genome analyses. In this review we present such analyses for metal transporters, metal-storage and metal-responsive regulators in these three Helicobacter species, and discuss possible contributions of the differences in metal metabolism in adaptation to the gastric or enterohepatic niches occupied by Helicobacter species.
Keywords: Helicobacter, Microbial pathogenesis, Gastrointestinal and hepatobiliary tract, Metal-responsive regulation, Metal transport
Introduction
Metals play an important role in the metabolism of all organisms, and this is reflected by the wide variety of chemical reactions in which they are involved. Metals can be cofactors of enzymes, catalyzing basic functions like electron transport, redox reactions and energy metabolism, and are essential for maintaining the osmotic pressure of the cell (van Vliet et al. 2001). Since both metal limitation and metal overload delay growth and can cause cell death, metal homeostasis is of critical importance to all living organisms. In bacteria, metal homeostasis is achieved by balancing import, efflux, metabolism and storage (Fig. 1).
Fig. 1.
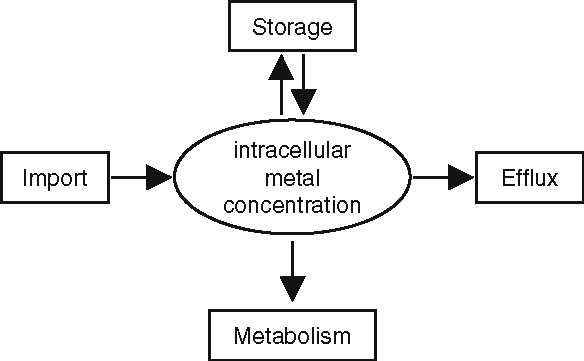
Schematic representation of the mechanisms involved in maintaining homeostasic control of the intracellular metal concentration via concerted action of import, efflux, storage and metabolism
The genus Helicobacter belongs to the epsilon subdivision of the proteobacteria, order Campylobacterales, family of the Helicobacteraceae. The best known related species is Campylobacter jejuni (Eppinger et al. 2004). Members of the genus Helicobacter are all microaerophilic organisms, and in most cases catalase- and oxidase-positive, and many are also urease-positive. Helicobacter species colonize the mucosal surfaces of the gastrointestinal and hepatobiliary tract of mammals, including humans. These mucosal surfaces provide a challenging environment with continuous changes in environmental conditions, including the availability of metal ions (van Vliet et al. 2001).
Colonization is usually lifelong, and often leads to the development of a wide range of inflammatory diseases (Blaser and Atherton 2004). Infection with Helicobacter species induces a strong proinflammatory response which includes the production of reactive oxygen species by the host immune system, and this necessitates the expression of systems detoxifying reactive oxygen species. Furthermore, the formation of reactive oxygen species is connected with iron metabolism, as oxygen radicals can be produced via the Fenton and Haber-Weiss reactions (Fe2+ + O2 → Fe3+ + O−2 and Fe2+ + H2O2 → Fe3+ + OH− + OH·). Helicobacter species are microaerophilic bacteria tolerating oxygen concentrations ranging from 2 to 10%. To combat oxidative stress, Helicobacter species expresses several key components of bacterial oxidative stress resistance, which are often metal-cofactored and controlled by metal-responsive regulatory systems (Baker et al. 2001; Seyler et al. 2001; Harris et al. 2002; Olczak et al. 2002; Wang et al. 2004; Ernst et al. 2005a; Hong et al. 2006). Many of the components of oxidative stress defense are essential for gastric colonization by H. pylori in animal models (Harris et al. 2003; Olczak et al. 2003; Barnard et al. 2004), and are likely to be of similar importance to colonization of other Helicobacter species.
Helicobacter species can be subdivided into two major lineages (Solnick and Schauer 2001; Fox 2002). The first lineage consists of the gastric Helicobacter species, which colonize the mucus layer overlaying the gastric epithelium. The best known example of a gastric Helicobacter species is Helicobacter pylori, the causative agent of gastritis, peptic ulcer disease and gastric adenocarcinoma (Blaser and Atherton 2004). However, many mammals are infected with gastric Helicobacter species, and they have been isolated from a wide variety of hosts, including dolphins, big cats and ferrets (Solnick and Schauer 2001; Fox 2002). The second lineage consists of the enteric or enterohepatic Helicobacter species, which colonize the mammalian intestinal and hepatobiliary tract, and have so far mostly been identified in rodents. The best characterized enteric Helicobacter species is Helicobacter hepaticus, which infects the intestines, bile ducts and liver of mice, and can cause hepatitis, hepatocellular carcinoma and possibly induce the formation of gallstones (Solnick and Schauer 2001; Fox 2002; Maurer et al. 2005).
The chronicity of infection suggests that Helicobacter species are well adapted to the conditions in their specific niches, and are more than capable of handling changes in these conditions (Blaser and Atherton 2004). Helicobacter species have a relatively small genome of 1.5–2.0 Mbp. The complete genome sequences of H. pylori (Tomb et al. 1997; Alm et al. 1999) and H. hepaticus (Suerbaum et al. 2003) have been published previously, whereas the almost finished, but unannotated genome sequence of the ferret gastric pathogen Helicobacter mustelae is available online (at http://www.sanger.ac.uk/Projects/H_mustelae). Together with the availability of the complete genome sequence of C. jejuni (Parkhill et al. 2000) this has allowed for comparative genomics, and here we will review the current knowledge on metal metabolism and metal-responsive gene regulation of H. pylori and H. hepaticus, focussing on the role of the metals iron, nickel and copper. We will also present hypotheses on metal-transport and metal-responsive regulation in H. mustelae, based on comparisons with H. pylori and H. hepaticus.
Metal-transport and storage of Helicobacter species
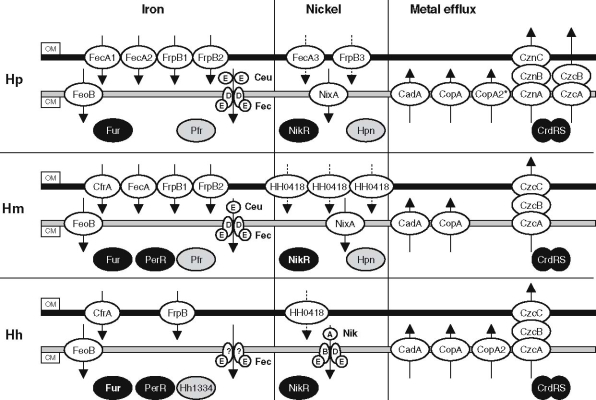
The gastric and enteric mucosa are niches with continuous changes in environmental conditions. For instance, the pH in the gastric mucosa is thought to range between 4 and 6.5, but acid shocks occasionally occur. Bioavailability of metals like iron and nickel is linked to the environmental pH, and thus Helicobacter species have to cope with both conditions of metal-restriction and metal-overload. Furthermore, the intestinal tract contains many bacterial species and thus enterohepatic Helicobacter species have to compete for metals and other nutrients. Metal metabolism has been studied intensively in H. pylori, but relatively little is known about metal metabolism of H. mustelae and H. hepaticus, and much of the information in this section is inferred from comparitive genomics using the H. pylori and C. jejuni genome sequences. In this section we will review the metal-transporter and metal-storage systems of the three selected Helicobacter species, of which an overview is given in Fig. 2.
Fig. 2.
Comparison of iron-transporters, nickel-transporters, metal-efflux systems, metal-storage and metal-responsive regulatory systems of Helicobacter pylori (Hp, top), Helicobacter mustelae (Hm, middle) and Helicobacter hepaticus (Hh, bottom). The ion transported and the direction of transport are indicated. OM, outer membrane; CM, cytoplasmic membrane. Ion transporters are grouped depending on the metal (putatively) transported. Arrows denotes the predicted direction of transport, dashed arrows indicate that this transport function is speculative. Note that the subdivision of outer membrane transporters for nickel is also tentative and speculative. Regulatory proteins are given in black, storage proteins in gray
Iron
Iron availability in the host tissues is mostly too low to support bacterial growth, as most iron is complexed into hemoglobin, or chelated by lactoferrin at mucosal surfaces (Andrews et al. 2003). Due to the insolubility of ferric iron, ferric iron transport requires an outer membrane receptor to transport the iron over the outer membrane, as well as a ABC transporter which transports the iron from the periplasm to the cytoplasm. In contrast, ferrous iron is soluble and only requires a inner membrane receptor.
Helicobacter pylori
Iron sources available in the gastric mucosa are lactoferrin, heme compounds, and iron derived from pepsin-degraded food. Iron is predicted to display increased solubility in the acidic conditions in the gastric mucosa, and eukaryotic iron-complexing proteins display lowered binding affinity at these conditions. H. pylori has been reported to be able to utilize heme compounds and ferric citrate as sole iron source (Dhaenens et al. 1999; Velayudhan et al. 2000), and it may be able to also use human lactoferrin, although this is still under debate (Bereswill et al. 1997). The H. pylori genome encodes eleven proteins predicted to be involved in iron transport and two proteins thought to function as iron storage proteins (Fig. 2).
H. pylori has three copies of the putative ferric citrate outer membrane receptor FecA, and three copies of the FrpB outer membrane receptor, which resembles a low-affinity enterochelin transporter in Neisseria species (Carson et al. 1999), for which in H. pylori the substrate is still unknown. There are two copies of the periplasmic binding protein CeuE, and finally a single inner membrane permease (FecD) and ATP-binding protein (FecE) (Fig. 2). Currently, mutants have only been described in the fecDE system and in one of the fecA genes (Velayudhan et al. 2000). Rather surprisingly, this did not affect iron transport. Thus the contribution of ferric iron uptake in H. pylori remains to be clarified. In the acidic, microaerobic gastric environment, ferrous iron (Fe2+) is thought to constitute the main form of free iron, and this is transported by H. pylori via the FeoB protein (Velayudhan et al. 2000). FeoB-mediated iron acquisition is of major importance to H. pylori, as isogenic feoB mutants were unable to colonize the gastric mucosa of mice (Velayudhan et al. 2000).
Two iron-storage proteins have been characterized in H. pylori, the Pfr ferritin and HP-NAP (Dps) bacterioferritin (Fig. 2) (Bereswill et al. 1998b; Tonello et al. 1999). The Pfr ferritin serves as an intracellular iron deposit and protects H. pylori against intracellular iron toxicity and free iron-mediated oxidative stress (Waidner et al. 2002a). Iron stored in Pfr can be released and reused to support growth under iron-limited conditions (Waidner et al. 2002a). The HP-NAP protein is both homologous to bacterioferritins as well as to the DNA-binding proteins of the Dps family (Tonello et al. 1999). A role of HP-NAP in H. pylori iron storage, although suggested, is yet to be demonstrated.
Helicobacter mustelae
H. mustelae has been reported to be able to utilize heme compounds and iron chloride as sole iron source (Dhaenens et al. 1999). In addition, it is also capable of using ferric citrate (Stoof et al. 2005). This is partially reflected in the putative outer membrane iron-uptake proteins identified in H. mustelae (Fig. 2). There is a single ortholog of the outer membrane ferric citrate transporter FecA, and mutation of the fecA gene significantly reduces growth when ferric citrate is used as sole iron source (Stoof and van Vliet unpublished results). Furthermore H. mustelae contains two genes in tandem encoding orthologs of the FrpB outer membrane receptor, and an ortholog of the C. jejuni CfrA outer membrane enterochelin receptor. Finally, there are three consecutive, > 80% identical genes encoding orthologs of the H. hepaticus HH0418 putative outer membrane protein (Fig. 2). The functions of the CfrA, FrpB and HH0418 orthologs are currently unclear. There is one ortholog of the periplasmic binding protein CeuE, and a single inner membrane FecDE ABC transporter system. Furthermore, there is an ortholog of the ferrous iron transporter FeoB present (Fig. 2). Finally, H. mustelae contains orthologs of the Pfr ferritin and HP-NAP (Dps) bacterioferritin, and it is likely that these have similar functions as the H. pylori counterparts.
Helicobacter hepaticus
H. hepaticus has been reported to be able to utilize lactoferrin, transferrin, heme compounds and iron chloride (Dhaenens et al. 1999). H. hepaticus colonizes the intestinal and hepatobiliary tract, and this is reflected in differences in its repertoire of putative iron-acquisition systems when compared to H. pylori and H. mustelae (Fig. 2). The H. hepaticus genome contains a single ortholog of the FrpB outer membrane receptor, for which it is tempting to speculate that this functions as heme receptor. Interestingly, despite the relatively close phylogenetic relationship and similar ecological niche as C. jejuni, there are surprising differences in the iron acquisition systems present. H. hepaticus and C. jejuni share a CfrA ortholog, which in C. jejuni functions as enterochelin uptake system (Palyada et al. 2004). However, it is not known whether H. hepaticus is able to use enterochelin as iron source. An ortholog of the C. jejuni hemin transporter ChuA is absent in the H. hepaticus genome sequence (van Vliet et al. 1998). Next to predicted outer membrane iron-transporters, H. hepaticus also contains a gene annotated as fecE, the ATPase component of the Fec ABC transporter. However, an ortholog of the permease protein FecD is not present in the H. hepaticus genome (Fig. 2), although this function may be fulfilled by a protein lacking sufficient homology to FecD. There is also an ortholog of the ferrous iron transporter FeoB present. Finally, there is also a ferritin present in H. hepaticus, encoded by the HH1334 gene. The bacterioferritin (Dps) ortholog of H. hepaticus also binds iron, but is reported to function in DNA-protection against oxidative stress (Hong et al. 2006).
Nickel
Nickel plays an important role in metabolism of Helicobacter species and in pathogenesis of infection. This is mostly due to the role of nickel as cofactor for the urease and hydrogenase enzymes, which are both involved in acid resistance, mucosal colonization and metabolism. Nickel availability in human serum is very low (2–11 nM) (Christensen et al. 1999), and the nickel concentration in ingested food varies significantly depending on the diet and on food sources (Sunderman 1993). Therefore Helicobacter species require efficient high-affinity transport of nickel to cater for the urease and hydrogenase enzymes.
Helicobacter pylori
The adaptation of H. pylori to life in the gastric mucosa seems to have led to a more pronounced role of nickel in its metabolism. The major nickel-transporter of H. pylori is the NixA protein, a 37 kDa protein of the HoxN-type of nickel/cobalt transporters (Mobley et al. 1995). Mutation of the nixA gene results in clear reduction of nickel transport and lowered urease activity (Bauerfeind et al. 1996), and mutation of nixA complemented nickel-sensitivity of a H. pylori nikR mutant (Ernst et al. 2005c). Absence of nixA also affected colonization efficiency in a mouse model of H. pylori infection, presumably due to the reduced urease activity (Nolan et al. 2002). A second putative nickel transport system is the abcCD locus, since abcC nixA double mutants showed only residual urease activity (Hendricks and Mobley 1997). However, nickel transport by the AbcCD system is yet to be demonstrated. A third system thought to be involved in nickel transport is the Dpp dipeptide permease, but mutation of this gene did not affect overall urease activity (Davis and Mobley 2005). Finally, two TonB-dependent outer membrane proteins (FecA3 [HP1400] and FrpB3 [HP1512, also called FrpB4]) have recently been shown to be regulated by nickel and the NikR-regulator (Ernst et al. 2006), and may thus represent a novel type of nickel-transporters.
Several urease- and hydrogenase-associated systems have adapted to the central role of nickel. The accessory proteins involved in hydrogenase biogenesis also affect urease activity, and the HspA (GroES) chaperone contains a nickel-binding motif (Kansau et al. 1996). Next to the nickel-transporters and urease/hydrogenase accessory proteins, H. pylori also expresses one or two small, very histidine-rich proteins (Hpn) which show strong binding to nickel (Gilbert et al. 1995; Ge et al. 2006). Mutation of the hpn gene rendered H. pylori sensitive to nickel (Mobley et al. 1999), which is suggestive of a role of Hpn in nickel storage or nickel sequestration, but this remains to be proven experimentally.
Helicobacter mustelae
Nickel transport in H. mustelae looks very similar to H. pylori. Both organisms have highly active urease and hydrogenase enzymes, and thus require efficient transport of nickel to cater for the high demand of nickel. H. mustelae contains a NixA ortholog, which probably represents the major nickel transporter. Interestingly, there are no clear orthologs of the FrpB3 and FecA3 proteins implicated in nickel transport of H. pylori (Ernst et al. 2006), but H. mustelae contains three tandem copies of the putative outer membrane receptor HH0418, which may fulfill a similar role as FecA3/FrpB3 in H. pylori. However, this awaits experimental validation. Finally, H. mustelae also expresses the histidine-rich Hpn protein (Gilbert et al. 1995), which is thought to function in nickel storage.
Helicobacter hepaticus
H. hepaticus expresses two nickel-cofactored enzymes, urease and the Ni/Fe hydrogenase (Maier et al. 2003; Mehta et al. 2005). This has necessitated the efficient transport of nickel, and this is likely to be mediated by the NikABDE ABC transporter system, which displays homology with the E. coli NikABCDE nickel transport system (De Pina et al. 1999). NikA is an ortholog of the E. coli nickel-binding periplasmic protein, and NikB is the predicted cytoplasmic membrane permease. NikD and NikE are predicted to function as ATPases which energize the transport (Navarro et al. 1993). There is no NixA ortholog present in H. hepaticus. The H. hepaticus genome contains a gene encoding a putative siderophore outer membrane receptor (HH0418), which is divergently orientated to the nikABDE operon encoding the predicted nickel-uptake ABC transporter system (Beckwith et al. 2001). It is therefore tempting to speculate that the HH0418 protein may function in nickel uptake, although this hypothesis may be farfetched and awaits rigorous experimental validation.
Copper
Copper is a cofactor for several proteins involved in electron transport, oxidases, and hydroxylases, but may also contribute to the formation of reactive oxygen species.
Helicobacter pylori
H. pylori expresses several proteins which are either involved in either copper transport or may act as copper chaperones. It is currently unclear whether there is a specific import system for copper, or whether it is transported by other metal transporters like FeoB or NixA (Mobley et al. 1999). However, H. pylori expresses several proteins involved in copper export, including the CopA and CopA2 P-type ATPases (Table 1), and the Crd copper resistance determinant (Melchers et al. 1998; Waidner et al. 2002b). Furthermore, H. pylori also expresses a small protein, CopP, that may function as copper chaperone (Beier et al. 1997; Bayle et al. 1998).
Table 1.
Metal-transport and metaboli-associated genes of selected Helicobacter species
| Gene | H. pyloria | H. hepaticusb | H. mustelae | Proposed function |
|---|---|---|---|---|
| fecA1 | 0686 | – | Present | Outer membrane receptor |
| fecA2 | 0807 | – | – | Outer membrane receptor |
| fecA3 | 1400 | – | – | Outer membrane receptor |
| frpB1 | 0876 | 1847 | Present | Outer membrane receptor |
| frpB2 | 0916/0915 | – | Present | Outer membrane receptor |
| frpB3c | 1512 | – | – | Outer membrane receptor |
| cfrA | – | 0721 | Present | Outer membrane receptor |
| hh0418 | – | 0418 | 3 copies | Outer membrane receptor |
| fecD | 0889 | ? | Present | Inner membrane permease |
| fecE | 0888 | 1759 | Present | ATPase |
| ceuE1 | 1562 | ? | Present | Periplasmic binding protein |
| ceuE2 | 1561 | – | – | Periplasmic binding protein |
| feoB | 0687 | 0033 | Present | Ferrous iron transporter |
| pfr | 0653 | 1334 | Present | Ferritin |
| dpsd | 0243 | 0210 | Present | Bacterioferritin/Dps ortholog |
| nixA | 1077 | – | Present | Inner membrane nickel transporter |
| nikABDE | – | 0417–0414 | – | Inner membrane nickel ABC transporter |
| cadA | 0791 | 0586 | Present | P-type ATPase |
| copA | 1072 | 0682 | Present | P-type ATPase |
| copA2 | 1502 | 1022 | – | P-type ATPase |
| cznABC | 0969–0971 | 0625–0623 | Present | Cobalt-zinc-nickel resistance determinant |
| crdAB | 1326–1327 | – | – | Copper resistance determinant |
| fur | 1027 | 0893 | Present | Ferric uptake regulator |
| perR | – | 0942 | Present | Peroxide stress regulator |
| nikR | 1338 | 0352 | Present | Nickel-responsive regulator |
| crdRS | 1365/1364 | 1656–1657 | ? | Two-component copper-regulatory system |
a Gene numbers from the genome sequence of H. pylori strain 26695 (Tomb et al. 1997)
b Gene numbers from the genome sequence of H. hepaticus strain ATCC51449 (Suerbaum et al. 2003)
c Alternative designation frpB4
d The H. pylori Dps protein is also known as Neutrophil Activating Protein (HP-NAP)
Helicobacter mustelae and Helicobacter hepaticus
Very little is known about copper metabolism of both H. mustelae and H. hepaticus. Both genomes contain orthologs of the CrdRS two-component regulatory system mediating copper-responsive induction of a Czc-like metal efflux system (Waidner et al. 2002b, 2005), but a role in copper metabolism is not yet established.
Metal efflux systems
Helicobacter pylori
Metal efflux is of great importance to H. pylori. The urease enzyme of H. pylori is sensitive to transition metals (Perez-Perez et al. 1994; Herrmann et al. 1999), and thus effective homeostasis of transition metals is necessary to prevent acid-sensitivity. H. pylori expresses three P-type ATPases, which can function in efflux of toxic metals. Two of these P-type ATPases are suggested to be CopA homologs which are involved in copper metabolism (see previous section). The third P-type ATPase is the H. pylori CadA protein, which is homologous to bacterial cadmium and copper P-type ATPases, and contains eight transmembrane domains (Melchers et al. 1996). Mutational studies demonstrated that CadA is involved in resistance to cadmium, zinc and cobalt, but not to copper or nickel. This indicates that the CadA exporter is not specific for a single metal. Mutation of cadA also reduced urease activity and nickel accumulation in some but not all cadA mutants (Herrmann et al. 1999). The other type of metal-efflux systems in H. pylori consists of the Czc system. These are cation-proton antiporters, and usually contain an cytoplasmic membrane transporter (CzcA), a periplasm-spanning protein (CzcB) and an OM protein thought to be involved in transport over the OM (CzcC). The H. pylori genome contains two Czc-like systems, the Crd copper resistance determinant (see previous section), and a cobalt-zinc-nickel (Czn) resistance determinant (Table 1) (Stähler et al. 2006). The latter system is also implicated in acid-resistance (possibly via nickel efflux) (Stähler et al. 2006) and efflux of metronidazole (van Amsterdam et al. 2005).
Helicobacter mustelae and Helicobacter hepaticus
The H. mustelae and H. hepaticus genomes contains, respectively, two and three genes encoding P-type ATPase orthologs, which are tentatively named CadA, CopA and CopA2, like in H. pylori. Similarly, there is a single CzcABC metal efflux system present. The function of these systems has not been investigated yet.
Metal-responsive regulatory systems of Helicobacter species
Rapid responses to stressful changes in environmental conditions are often mediated via changes in transcription of sets of genes, that encode some factor involved in the dealing with these stresses. Examples of this are the expression of oxidative stress defense genes in response to oxidative stress. In many bacteria, such stress-responsive systems are often encoded by genes organized in an operon, and the transcription is regulated by one or two regulatory proteins.
The ion-responsive regulatory systems of bacteria usually consist of a single regulatory protein, that combines sensor and effector functions in one molecule (Escolar et al. 1999). It senses the cytoplasmic ion concentration and, when activated, can induce or repress transcription of the genes encoding the corresponding uptake, efflux and/or storage systems (Andrews et al. 2003). Four types of metal-responsive proteins have been identified in Helicobacter species: the iron-responsive regulator Fur (Bereswill et al. 1998a), the peroxide-regulatory protein PerR (Suerbaum et al. 2003; Belzer et al. 2005b), the nickel-responsive regulator NikR (van Vliet et al. 2002a) and the copper-responsive two-component regulatory system CrdRS (Waidner et al. 2005). Comparison of the three Helicobacter genome sequences with that of C. jejuni has indicated an interesting distribution of metal-responsive regulators (Fig. 3A). The Fur regulator is present in all Helicobacter species and in C. jejuni, although its functions and regulons differ (see below). The PerR regulator is present in H. mustelae, H. hepaticus and C. jejuni, but absent in H. pylori (Fig. 3A). This is rather surprising since the main proteins regulated by PerR are catalase and AhpC (Bsat et al. 1998; van Vliet et al. 1999), and these are present in all four species. The NikR regulator is present in all three Helicobacter species, but is absent in C. jejuni. This is likely to be linked to the presence of the nickel-cofactored urease system in the investigated Helicobacter species, and the absence of this enzyme in C. jejuni. Finally, the distribution of the CrdRS is difficult to predict, since two-component regulatory systems share significant sequence homology both for the response regulator and the sensory histidine kinase, and this makes predictions based on homology searches risky.
Fig. 3.
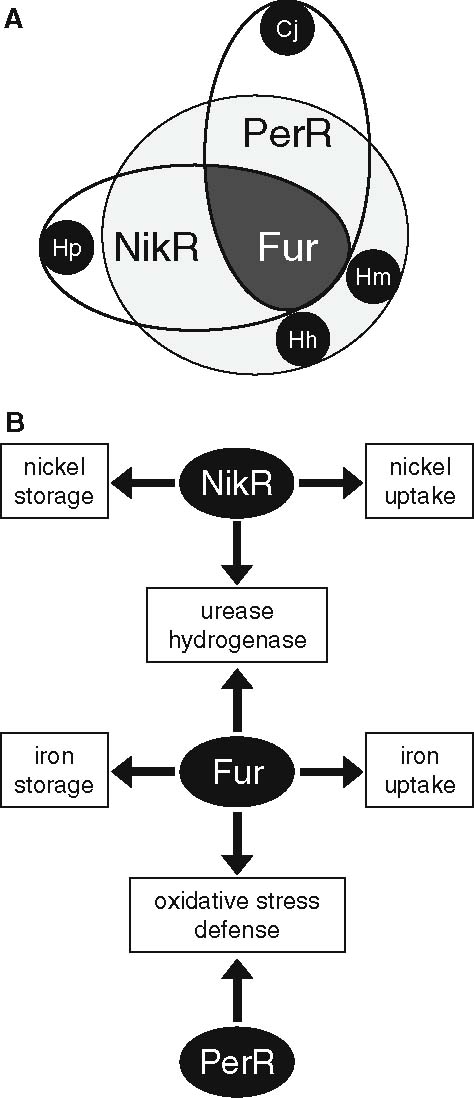
Distribution and overlapping functions of the Fur, NikR and PerR regulatory systems in Helicobacter and Campylobacter species. (A) Distribution of the regulators in Helicobacter pylori (Hp), Helicobacter mustelae (Hm), Helicobacter hepaticus (Hh) and Campylobacter jejuni (Cj). The Fur protein is ubiquitous in all four species (indicated by the dark gray area), whereas the PerR and NikR proteins are only present in 3/4 species (indicated in light gray). (B) Overlapping functions of the Fur, NikR and PerR regulatory systems in Helicobacter
Fur
The Fur protein is primarily known for its role as repressor of iron acquisition systems (Hantke 2001). It fullfils this classical function in all three Helicobacter species as well, as the H. pylori Fur mediates regulation of iron acquisition outer membrane proteins (Delany et al. 2001a; van Vliet et al. 2002b). However, the Fur protein also regulates part of the nitrogen metabolism of H. pylori, via iron-responsive repression of the AmiE aliphatic amidase (van Vliet et al. 2003). Interestingly, the H. pylori Fur protein is bifunctional, since it also functions in its iron-free (apo)form. Iron-free Fur represses the expression of the Pfr ferritin and the iron-cofactored superoxide dismutase SodB (Delany et al. 2001b; Ernst et al. 2005a). This form of regulation may compensate for the absence of iron-responsive regulation via small RNAs like RyhB (Masse and Gottesman 2002), which is of great importance in several pathogenic bacteria, but has so far not been reported for H. pylori.
In H. hepaticus, Fur co-regulates expression of the peroxide stress defense genes AhpC and KatA with PerR (see below) (Belzer et al. 2005b), and also mediates iron-responsive regulation of the urease system (Belzer et al. 2007). The rationale behind Fur-regulation of urease expression is not directly apparent, but may be linked to Fur functioning as a sensory system for iron-restricted conditions encountered in the natural niche of H. hepaticus.
PerR
The peroxide stress regulator PerR is a Fur homolog and is responsible for iron- and manganese-dependent regulation of ahpC and katA in Bacillus subtilis and C. jejuni (Bsat et al. 1998; van Vliet et al. 1999). The H. hepaticus and H. mustelae genomes contain a gene encoding a PerR homolog, and this may serve as a replacement system for the OxyR regulator, which is absent in Helicobacter species and C. jejuni. However, PerR is not present in H. pylori, and this raises the interesting question whether the PerR system was either lost by H. pylori or gained by the other species.
In C. jejuni, perR mutants were shown to express very high levels of both catalase and alkyl hydroperoxide reductase and to be highly resistant to peroxide stress (van Vliet et al. 1999). Like Fur repressors, PerR repressors also recognize a specific sequence upstream of their target genes although the consensus sequence has only been confirmed in B. subtilis (Herbig and Helmann 2001). Identification of PerR-binding sites is complicated by their similarity to Fur-binding sequences. Although PerR responds to iron, it has been suggested that PerR may use manganese as its metal cofactor (Herbig and Helmann 2001). Preliminary analysis of Fur and PerR expression in H. hepaticus suggests that Fur and PerR co-regulate expression of catalase and AhpC, in a similar level to what was described for C. jejuni (Belzer et al. 2005b).
NikR
The NikR regulator belongs to the class of ribbon-helix-helix (RHH) proteins. It was first described in E. coli as repressor of the nickel uptake operon nikABCDE (De Pina et al. 1999). Binding of nickel-cofactored NikR to a 5′-GTATGA-N16-TCATAC-3′ sequence in the nikA promoter region results in repression of transcription (Chivers and Sauer 2000), and this allows NikR to maintain intracellular nickel homeostasis in a similar manner as Fur and iron. NikR orthologs are present in all the Helicobacter genomes sequenced thusfar, but has only been studied in H. pylori (van Vliet et al. 2004a). NikR was shown to function as a repressor as well as an inducer of transcription, since nickel-dependent binding of NikR to 5′-TATWATT-N11-AATWATA-3′ sequences in the ureA and nixA promoters resulted in induction and repression of transcription, respectively (Delany et al. 2005; Ernst et al. 2005c). Furthermore, NikR and Fur display overlapping regulons and each regulates the expression of the other protein and the corresponding regulon (Bury-Mone et al. 2004; van Vliet et al. 2004b), allowing intricate regulation of gene expression in different environmental conditions.
In H. pylori NikR controls utilization of nickel, by mediating nickel-responsive regulation of urease expression (van Vliet et al. 2002a; Ernst et al. 2005c). A similar regulation of urease expression has been observed in H. mustelae (Stoof and van Vliet, unpublished results). Interestingly, the urease system of H. hepaticus is not nickel-responsive at the transcriptional level, but only at the post-translational level (Belzer et al. 2005a). This indicates that NikR may not be involved in controlling urease expression in H. hepaticus, and thus this suggests that NikR-controlled nickel-responsive regulation of urease expression represents a specific adaptation to the gastric lifestyle.
CrdRS
The CrdRS system was identified in H. pylori as the two-component regulatory system required for copper-responsive induction of the copper-resistance determinant CrdAB-CzcBA (Waidner et al. 2002b, 2005). It is not yet known how the CrdRS system senses copper, but absence of either CrdR or CrdS disturbs regulation of Crd (Waidner et al. 2005). The CrdRS system plays an important role in gastric colonization in a mouse infection model for H. pylori (Panthel et al. 2003) and is also implicated in acid resistance of H. pylori (Loh and Cover 2006). Whether H. mustelae and H. hepaticus contain orthologs of CrdRS is not known, although both genomes contain two-component regulatory systems with sequence homology to CrdRS.
Conclusions
Although they are considered to be fastidious, Helicobacter species are well adapted to their ecological niche. From genomic comparisons it is apparent that while clustered in a single species, there are considerable differences in genomic content (Eppinger et al. 2004) which are also reflected in the components of metal-transport systems as reviewed here. There seems to be no specific evolution of metal-transporters equipped for either the gastric or enteric environment, with possibly the exception of the NixA vs. the Nik-ABDE nickel transporters, the Hpn nickel-binding protein and possibly the presence of a ferric citrate outer membrane transporter, which are only present in the gastric Helicobacter species. Therefore differences in niche adaptation may well be more at the level of gene regulation and biochemical properties of the regulated proteins.
The considerable differences between H. pylori and H. mustelae also need to be taken into account when using the H. mustelae ferret infection model, which has been proposed as suitable animal model for H. pylori infection. Although pathogenesis of H. mustelae infection does mimic that of H. pylori infection of humans, there are still many differences between H. pylori and H. mustelae as well as between man and ferret that need to be taken into account, and will make direct application of this model difficult. Similarly, the differences between host, niche and genetics of H. hepaticus and H. pylori make direct comparisons very difficult.
In conclusion, the availability of genome sequences of different Helicobacter species has allowed for comparative genomic analyses of metal transport systems, and this shows the significant diversity in these systems within a single genus. It also displays the versatility of these bacteria to adapt to the diversity of conditions encountered in their natural gastric or enteric niches in mammals.
Acknowledgement
We thank Dr Paul O’Toole and Dr Julian Parkhill for access to the unpublished genome sequence of H. mustelae. The sequencing of the H. mustelae genome is funded by the Wellcome Trust.
References
- Alm RA, Ling LS, Moir DT, King BL, Brown ED, Doig PC, Smith DR, Noonan B, Guild BC, deJonge BL, Carmel G, Tummino PJ, Caruso A, Uria-Nickelsen M, Mills DM, Ives C, Gibson R, Merberg D, Mills SD, Jiang Q, Taylor DE, Vovis GF, Trust TJ (1999) Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176–180 [DOI] [PubMed]
- Andrews SC, Robinson AK, Rodriguez-Quinones F (2003) Bacterial iron homeostasis. FEMS Microbiol Rev 27:215–237 [DOI] [PubMed]
- Baker LM, Raudonikiene A, Hoffman PS, Poole LB (2001) Essential thioredoxin-dependent peroxiredoxin system from Helicobacter pylori: genetic and kinetic characterization. J Bacteriol 183:1961–1973 [DOI] [PMC free article] [PubMed]
- Barnard FM, Loughlin MF, Fainberg HP, Messenger MP, Ussery DW, Williams P, Jenks PJ (2004) Global regulation of virulence and the stress response by CsrA in the highly adapted human gastric pathogen Helicobacter pylori. Mol Microbiol 51:15–32 [DOI] [PubMed]
- Bauerfeind P, Garner RM, Mobley HLT (1996) Allelic exchange mutagenesis of nixA in Helicobacter pylori results in reduced nickel transport and urease activity. Infect Immun 64:2877–2880 [DOI] [PMC free article] [PubMed]
- Bayle D, Wangler S, Weitzenegger T, Steinhilber W, Volz J, Przybylski M, Schafer KP, Sachs G, Melchers K (1998) Properties of the P-type ATPases encoded by the copAP operons of Helicobacter pylori and Helicobacter felis. J Bacteriol 180:317–329 [DOI] [PMC free article] [PubMed]
- Beckwith CS, McGee DJ, Mobley HL, Riley LK (2001) Cloning, expression, and catalytic activity of Helicobacter hepaticus urease. Infect Immun 69:5914–5920 [DOI] [PMC free article] [PubMed]
- Beier D, Spohn G, Rappuoli R, Scarlato V (1997) Identification and characterization of an operon of Helicobacter pylori that is involved in motility and stress adaptation. J Bacteriol 179:4676–4683 [DOI] [PMC free article] [PubMed]
- Belzer C, Stoof J, Beckwith CS, Kuipers EJ, Kusters JG, van Vliet AHM (2005a) Differential regulation of urease activity in Helicobacter hepaticus and Helicobacter pylori. Microbiology 151:3989–3995 [DOI] [PubMed]
- Belzer C, van Schendel BAM, Kuipers EJ, Hoogenboezem T, Hermans PWM, Kusters JG, van Vliet AHM (2005b) PerR is a regulator of oxidative stress defense in Helicobacter hepaticus. Helicobacter 10:548 [DOI] [PMC free article] [PubMed]
- Belzer C, van Schendel BAM, Kuipers EJ, Kusters JG, van Vliet AHM (2007) Iron-responsive repression of urease expression in Helicobacter hepaticus is mediated by the transcriptional regulator Fur. Infect Immun 75:745–752 [DOI] [PMC free article] [PubMed]
- Bereswill S, Strobel S, Lichte F, Fassbinder F, Hantke K, Kist M (1997) Evaluation of microbiological parameters for the study of iron metabolism in Helicobacter pylori. Gut 41(S1):A10
- Bereswill S, Lichte F, Vey T, Fassbinder F, Kist M (1998a) Cloning and characterization of the fur gene from Helicobacter pylori. FEMS Microbiol Lett 159:193–200 [DOI] [PubMed]
- Bereswill S, Waidner U, Odenbreit S, Lichte F, Fassbinder F, Bode G, Kist M (1998b) Structural, functional and mutational analysis of the pfr gene encoding a ferritin from Helicobacter pylori. Microbiology 144:2505–2516 [DOI] [PubMed]
- Blaser MJ, Atherton JC (2004) Helicobacter pylori persistence: biology and disease. J Clin Invest 113:321–333 [DOI] [PMC free article] [PubMed]
- Bsat N, Herbig A, Casillas-Martinez L, Setlow P, Helmann JD (1998) Bacillus subtilis contains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol Microbiol 29:189–198 [DOI] [PubMed]
- Bury-Mone S, Thiberge JM, Contreras M, Maitournam A, Labigne A, De Reuse H (2004) Responsiveness to acidity via metal ion regulators mediates virulence in the gastric pathogen Helicobacter pylori. Mol Microbiol 53:623–638 [DOI] [PubMed]
- Carson SD, Klebba PE, Newton SM, Sparling PF (1999) Ferric enterobactin binding and utilization by Neisseria gonorrhoeae. J Bacteriol 181:2895–2901 [DOI] [PMC free article] [PubMed]
- Chivers PT, Sauer RT (2000) Regulation of high-affinity nickel uptake in bacteria: Ni2+- dependent interaction of NikR with wild-type and mutant operator sites. J␣Biol Chem 275:19735–19741 [DOI] [PubMed]
- Christensen JM, Kristiansen J, Nielsen NH, Menne T, Byrialsen K (1999) Nickel concentrations in serum and urine of patients with nickel eczema. Toxicol Lett 108:185–189 [DOI] [PubMed]
- Davis GS, Mobley HL (2005) Contribution of dppA to urease activity in Helicobacter pylori 26695. Helicobacter 10:416–423 [DOI] [PubMed]
- De Pina K, Desjardin V, Mandrand-Berthelot MA, Giordano G, Wu LF (1999) Isolation and characterization of the nikR gene encoding a nickel-responsive regulator in Escherichia coli. J Bacteriol 181:670–674 [DOI] [PMC free article] [PubMed]
- Delany I, Pacheco ABF, Spohn G, Rappuoli R, Scarlato V (2001a) Iron-dependent transcription of the frpB gene of Helicobacter pylori is controlled by the Fur protein. J Bacteriol 183:4932–4937 [DOI] [PMC free article] [PubMed]
- Delany I, Spohn G, Rappuoli R, Scarlato V (2001b) The Fur repressor controls transcription of iron-activated and -repressed genes in Helicobacter pylori. Mol Microbiol 42:1297–1309 [DOI] [PubMed]
- Delany I, Ieva R, Soragni A, Hilleringmann M, Rappuoli R, Scarlato V (2005) In vitro analysis of protein-operator interactions of the NikR and Fur metal-responsive regulators of coregulated genes in Helicobacter pylori. J Bacteriol 187:7703–7715 [DOI] [PMC free article] [PubMed]
- Dhaenens L, Szczebara F, Van Nieuwenhuyse S, Husson MO (1999) Comparison of iron uptake in different Helicobacter species. Res Microbiol 150:475–481 [DOI] [PubMed]
- Eppinger M, Baar C, Raddatz G, Huson DH, Schuster SC (2004) Comparative analysis of four Campylobacterales. Nat Rev Microbiol 2:872–885 [DOI] [PubMed]
- Ernst FD, Homuth G, Stoof J, Mader U, Waidner B, Kuipers EJ, Kist M, Kusters JG, Bereswill S, van Vliet AHM (2005a) Iron-responsive regulation of the Helicobacter pylori iron-cofactored superoxide dismutase SodB is mediated by Fur. J Bacteriol 187:3687–3692 [DOI] [PMC free article] [PubMed]
- Ernst FD, Stoof J, Horrevoets JM, Kuipers EJ, Kusters JG, van Vliet AHM (2006) NikR mediates nickel-responsive transcriptional repression of the Helicobacter pylori outer merribrane proteins FecA3 (HP1400) and FrpB4 (HP1512). Infect Immun 74: 6821–6828 [DOI] [PMC free article] [PubMed]
- Ernst FD, Kuipers EJ, Heijens A, Sarwari R, Stoof J, Penn CW, Kusters JG, van Vliet AHM (2005c) The nickel-responsive regulator NikR controls activation and repression of gene transcription in Helicobacter pylori. Infect Immun 73:7252–7258 [DOI] [PMC free article] [PubMed]
- Escolar L, Perez-Martin J, de Lorenzo V (1999) Opening the iron-box: transcriptional metalloregulation by the Fur protein. J Bacteriol 181:6223–6229 [DOI] [PMC free article] [PubMed]
- Fox JG (2002) The non- H. pylori helicobacters: their expanding role in gastrointestinal and systemic diseases. Gut 50:273–283 [DOI] [PMC free article] [PubMed]
- Ge R, Watt RM, Sun X, Tanner JA, He QY, Huang JD, Sun H (2006) Expression and characterization of a histidine-rich protein, Hpn: potential for Ni2+ storage in Helicobacter pylori. Biochem J 393:285–293 [DOI] [PMC free article] [PubMed]
- Gilbert JV, Ramakrishna J, Sunderman FW Jr, Wright A, Plaut AG (1995) Protein Hpn: cloning and characterization of a histidine-rich metal–binding polypeptide in Helicobacter pylori and Helicobacter mustelae. Infect Immun 63:2682–2688 [DOI] [PMC free article] [PubMed]
- Hantke K (2001) Iron and metal regulation in bacteria. Curr Opin Microbiol 4:172–177 [DOI] [PubMed]
- Harris AG, Hinds FE, Beckhouse AG, Kolesnikow T, Hazell SL (2002) Resistance to hydrogen peroxide in Helicobacter pylori: role of catalase (KatA) and Fur, and functional analysis of a novel gene product designated ‘KatA-associated protein’ KapA (HP0874). Microbiology 148:3813–3825 [DOI] [PubMed]
- Harris AG, Wilson JE, Danon SJ, Dixon MF, Donegan K, Hazell SL (2003) Catalase (KatA) and KatA-associated protein (KapA) are essential to persistent colonization in the Helicobacter pylori SS1 mouse model. Microbiology 149:665–672 [DOI] [PubMed]
- Hendricks JK, Mobley HL (1997) Helicobacter pylori ABC transporter: effect of allelic exchange mutagenesis on urease activity. J Bacteriol 179:5892–5902 [DOI] [PMC free article] [PubMed]
- Herbig AF, Helmann JD (2001) Roles of metal ions and hydrogen peroxide in modulating the interaction of the Bacillus subtilis PerR peroxide regulon repressor with operator DNA. Mol Microbiol 41:849–859 [DOI] [PubMed]
- Herrmann L, Schwan D, Garner R, Mobley HL, Haas R, Schafer KP, Melchers K (1999) Helicobacter pylori cadA encodes an essential Cd(II)-Zn(II)-Co(II) resistance factor influencing urease activity. Mol Microbiol 33:524–536 [DOI] [PubMed]
- Hong Y, Wang G, Maier RJ (2006) Helicobacter hepaticus Dps protein plays an important role in protecting DNA from oxidative damage. Free Radic Res Suppl:1 [DOI] [PubMed]
- Kansau I, Guillain F, Thiberge JM, Labigne A (1996) Nickel binding and immunological properties of the C-terminal domain of the Helicobacter pylori GroES homologue (HspA). Mol Microbiol 22:1013–1023 [DOI] [PubMed]
- Loh JT, Cover TL (2006) Requirement of histidine kinases HP0165 and HP1364 for acid resistance in Helicobacter pylori. Infect Immun 74:3052–3059 [DOI] [PMC free article] [PubMed]
- Maier RJ, Olson J, Olczak A (2003) Hydrogen-oxidizing capabilities of Helicobacter hepaticus and in␣vivo availability of the substrate. J Bacteriol 185:2680–2682 [DOI] [PMC free article] [PubMed]
- Masse E, Gottesman S (2002) A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci USA 99:4620–4625 [DOI] [PMC free article] [PubMed]
- Maurer KJ, Ihrig MM, Rogers AB, Ng V, Bouchard G, Leonard MR, Carey MC, Fox JG (2005) Identification of cholelithogenic enterohepatic Helicobacter species and their role in murine cholesterol gallstone formation. Gastroenterology 128:1023–1033 [DOI] [PubMed]
- Mehta NS, Benoit S, Mysore JV, Sousa RS, Maier RJ (2005) Helicobacter hepaticus hydrogenase mutants are deficient in hydrogen-supported amino acid uptake and in causing liver lesions in A/J mice. Infect Immun 73:5311–5318 [DOI] [PMC free article] [PubMed]
- Melchers K, Weitzenegger T, Buhmann A, Steinhilber W, Sachs G, Schafer KP (1996) Cloning and membrane topology of a P type ATPase from Helicobacter pylori. J Biol Chem 271:446–457 [DOI] [PubMed]
- Melchers K, Herrmann L, Mauch F, Bayle D, Heuermann D, Weitzenegger T, Schuhmacher A, Sachs G, Haas R, Bode G, Bensch K, Schafer KP (1998) Properties and function of the P type ion pumps cloned from Helicobacter pylori. Acta Physiol Scand Suppl 643:123–135 [PubMed]
- Mobley HL, Garner RM, Bauerfeind P (1995) Helicobacter pylori nickel-transport gene nixA: synthesis of catalytically active urease in Escherichia coli independent of growth conditions. Mol Microbiol 16:97–109 [DOI] [PubMed]
- Mobley HL, Garner RM, Chippendale GR, Gilbert JV, Kane AV, Plaut AG (1999) Role of Hpn and NixA of Helicobacter pylori in susceptibility and resistance to bismuth and other metal ions. Helicobacter 4:162–169 [DOI] [PubMed]
- Navarro C, Wu LF, Mandrand-Berthelot MA (1993) The nik operon of Escherichia coli encodes a periplasmic binding-protein-dependent transport system for nickel. Mol Microbiol 9:1181–1191 [DOI] [PubMed]
- Nolan KJ, McGee DJ, Mitchell HM, Kolesnikow T, Harro JM, O’Rourke J, Wilson JE, Danon SJ, Moss ND, Mobley HL, Lee A (2002) In vivo behavior of a Helicobacter pylori SS1 nixA mutant with reduced urease activity. Infect Immun 70:685–691 [DOI] [PMC free article] [PubMed]
- Olczak AA, Olson JW, Maier RJ (2002) Oxidative-stress resistance mutants of Helicobacter pylori. J Bacteriol 184:3186–3193 [DOI] [PMC free article] [PubMed]
- Olczak AA, Seyler RW Jr, Olson JW, Maier RJ (2003) Association of Helicobacter pylori antioxidant activities with host colonization proficiency. Infect Immun 71:580–583 [DOI] [PMC free article] [PubMed]
- Palyada K, Threadgill D, Stintzi A (2004) Iron acquisition and regulation in Campylobacter jejuni. J Bacteriol 186:4714–4729 [DOI] [PMC free article] [PubMed]
- Panthel K, Dietz P, Haas R, Beier D (2003) Two-component systems of Helicobacter pylori contribute to virulence in a mouse infection model. Infect Immun 71:5381–5385 [DOI] [PMC free article] [PubMed]
- Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, Basham D, Chillingworth T, Davies RM, Feltwell T, Holroyd S, Jagels K, Karlyshev AV, Moule S, Pallen MJ, Penn CW, Quail MA, Rajandream MA, Rutherford KM, van Vliet AHM, Whitehead S, Barrell BG (2000) The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665–668 [DOI] [PubMed]
- Perez-Perez GI, Gower CB, Blaser MJ (1994) Effects of cations on Helicobacter pylori urease activity, release, and stability. Infect Immun 62:299–302 [DOI] [PMC free article] [PubMed]
- Seyler RW Jr, Olson JW, Maier RJ (2001) Superoxide dismutase-deficient mutants of Helicobacter pylori are hypersensitive to oxidative stress and defective in host colonization. Infect Immun 69:4034–4040 [DOI] [PMC free article] [PubMed]
- Solnick JV, Schauer DB (2001) Emergence of diverse Helicobacter species in the pathogenesis of gastric and enterohepatic diseases. Clin Microbiol Rev 14:59–97 [DOI] [PMC free article] [PubMed]
- Stähler FN, Odenbreit S, Haas R, Wilrich J, van Vliet AHM, Kusters JG, Kist M, Bereswill S (2006) The novel Helicobacter pylori CznABC metal efflux pump is required for cadmium, zinc and nickel resistance, for urease modulation and for gastric colonization. Infect Immun 74:3845–3852 [DOI] [PMC free article] [PubMed]
- Stoof J, Kuipers EJ, Kusters JG, van Vliet AHM (2005) Fur mediates regulation of iron-uptake genes in Helicobacter mustelae. Helicobacter 10:465
- Suerbaum S, Josenhans C, Sterzenbach T, Drescher B, Brandt P, Bell M, Droge M, Fartmann B, Fischer HP, Ge Z, Horster A, Holland R, Klein K, Konig J, Macko L, Mendz GL, Nyakatura G, Schauer DB, Shen Z, Weber J, Frosch M, Fox JG (2003) The complete genome sequence of the carcinogenic bacterium Helicobacter hepaticus. Proc Natl Acad Sci USA 100:7901–7906 [DOI] [PMC free article] [PubMed]
- Sunderman FW (1993) Biological monitoring of nickel in humans. Scand J Work Environm Health 19:34–38 [PubMed]
- Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, Nelson K, Quackenbush J, Zhou L, Kirkness EF, Peterson S, Loftus B, Richardson D, Dodson R, Khalak HG, Glodek A, McKenney K, Fitzegerald LM, Lee N, Adams MD, Hickey EK, Berg DE, Gocayne JD, Utterback TR, Peterson JD, Kelley JM, Cotton MD, Weidman JM, Fujii C, Bowman C, Watthey L, Wallin E, Hayes WS, Borodovsky M, Karpk PD, Smith HO, Fraser CM, Venter JC (1997) The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539–547 [DOI] [PubMed]
- Tonello F, Dundon WG, Satin B, Molinari M, Tognon G, Grandi G, del Guicide G, Rappuoli R, Montecucco C (1999) The Helicobacter pylori neutrophil-activating protein is an iron-binding protein with dodecameric structure. Mol Microbiol 34:238–246 [DOI] [PubMed]
- van Amsterdam K, Bart A, van der Ende A (2005) A Helicobacter pylori TolC efflux pump confers resistance to metronidazole. Antimicrob Agents Chemother 49:1477–1482 [DOI] [PMC free article] [PubMed]
- van Vliet AHM, Wooldridge KG, Ketley JM (1998) Iron-responsive gene regulation in a Campylobacter jejuni fur mutant. J Bacteriol 180:5291–5298 [DOI] [PMC free article] [PubMed]
- van Vliet AH, Baillon ML, Penn CW, Ketley JM (1999) Campylobacter jejuni contains two Fur homologs: characterization of iron-responsive regulation of peroxide stress defense genes by the PerR repressor. J␣Bacteriol 181:6371–6376 [DOI] [PMC free article] [PubMed]
- van Vliet AHM, Bereswill S, Kusters JG (2001) Ion metabolism and transport. In: Mobley HLT, Mendz GL, Hazell SL (eds) Helicobacter pylori: physiology and genetics. ASM Press, Washington, DC, pp 193–206
- van Vliet AHM, Poppelaars SW, Davies BJ, Stoof J, Bereswill S, Kist M, Kuipers EJ, Penn CW, Kusters JG (2002a) NikR mediates nickel-responsive transcriptional induction of urease expression in Helicobacter pylori. Infect Immun 70:2846–2852 [DOI] [PMC free article] [PubMed]
- van Vliet AHM, Stoof J, Vlasblom R, Wainwright SA, Hughes NJ, Kelly DJ, Bereswill S, Bijlsma JJE, Hoogenboezem T, Vandenbroucke-Grauls CMJE, Kist M, Kuipers EJ, Kusters JG (2002b) The role of the ferric uptake regulator (Fur) in regulation of Helicobacter pylori iron uptake. Helicobacter 7:237–244 [DOI] [PubMed]
- van Vliet AHM, Stoof J, Poppelaars SW, Bereswill S, Homuth G, Kist M, Kuipers EJ, Kusters JG (2003) Differential regulation of amidase- and formamidase-mediated ammonia production by the Helicobacter pylori Fur repressor. J Biol Chem 278:9052–9057 [DOI] [PubMed]
- van Vliet AHM, Ernst FD, Kusters JG (2004a) NikR-mediated regulation of Helicobacter pylori acid adaptation. Trends Microbiol 12:489–494 [DOI] [PubMed]
- van Vliet AHM, Kuipers EJ, Stoof J, Poppelaars SW, Kusters JG (2004b) Acid-responsive gene induction of ammonia-producing enzymes in Helicobacter pylori is mediated via a metal-responsive repressor cascade. Infect Immun 72:766–773 [DOI] [PMC free article] [PubMed]
- Velayudhan J, Hughes NJ, McColm AA, Bagshaw J, Clayton CL, Andrews SC, Kelly DJ (2000) Iron acquisition and virulence in Helicobacter pylori: a major role for FeoB, a high-affinity ferrous iron transporter. Mol Microbiol 37:274–286 [DOI] [PubMed]
- Waidner B, Greiner S, Odenbreit S, Kavermann H, Velayudhan J, Stähler F, Bisse E, van Vliet AHM, Andrews SC, Kusters JG, Kelly DJ, Haas R, Kist M, Bereswill S (2002a) Essential role of ferritin Pfr in Helicobacter pylori iron metabolism and gastric colonization. Infect Immun 70:3923–3929 [DOI] [PMC free article] [PubMed]
- Waidner B, Melchers K, Ivanov I, Loferer H, Bensch KW, Kist M, Bereswill S (2002b) Identification by RNA profiling and mutational analysis of the novel copper resistance determinants CrdA (HP1326), CrdB (HP1327), and CzcB (HP1328) in Helicobacter pylori. J Bacteriol 184:6700–6708 [DOI] [PMC free article] [PubMed]
- Waidner B, Melchers K, Stahler FN, Kist M, Bereswill S (2005) The Helicobacter pylori CrdRS two-component regulation system (HP1364/HP1365) is required for copper-mediated induction of the copper resistance determinant CrdA. J Bacteriol 187:4683–4688 [DOI] [PMC free article] [PubMed]
- Wang G, Conover RC, Benoit S, Olczak AA, Olson JW, Johnson MK, Maier RJ (2004) Role of a bacterial organic hydroperoxide detoxification system in preventing catalase inactivation. J Biol Chem 279:51908–51914 [DOI] [PubMed]