SUMMARY
Enzymes from natural product biosynthetic pathways are attractive candidates for creating tailored biocatalysts to produce semisynthetic pharmaceutical compounds. LovD is an acyltransferase that converts the inactive monacolin J acid (MJA) into the cholesterol-lowering lovastatin. LovD can also synthesize the blockbuster drug simvastatin using MJA and a synthetic α-dimethylbutyryl thioester, albeit with suboptimal properties as a biocatalyst. Here we used directed evolution to improve the properties of LovD towards semisynthesis of simvastatin. Mutants with improved catalytic efficiency, solubility and thermal stability were obtained, with the best mutant displaying an ~11-fold increase in an Escherichia coli based biocatalytic platform. To understand the structural basis of LovD enzymology, seven X-ray crystal structures were determined, including the parent LovD, an improved mutant G5, and G5 co-crystallized with ligands. Comparisons between the structures reveal that beneficial mutations stabilize the structure of G5 in a more compact conformation that is favorable for catalysis.
INTRODUCTION
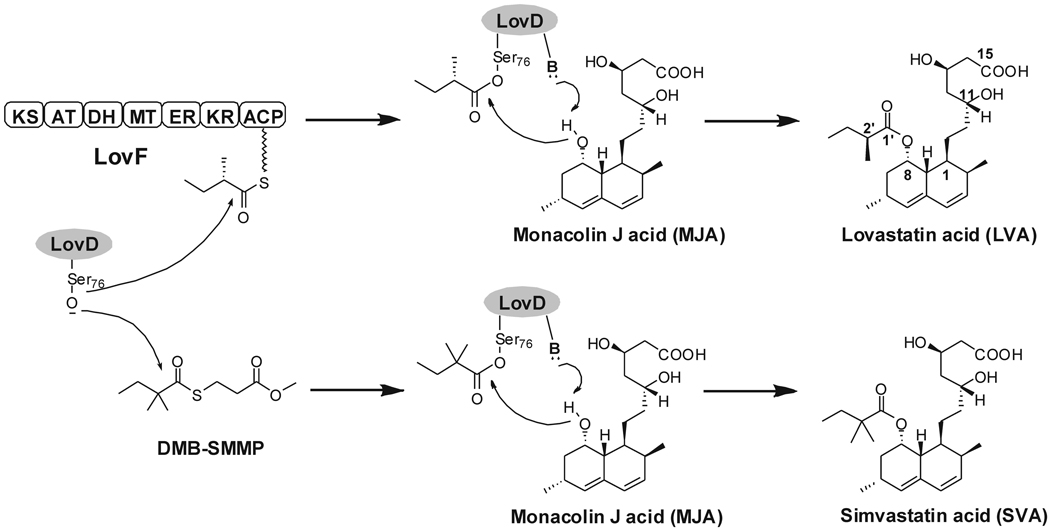
Tailoring enzymes found in natural product biosynthetic pathways catalyze a wide array of reactions, including acyltransfer (Loncaric, et al., 2006), glycosylation (Zhang, et al., 2006), hydroxylation (Rix, et al., 2002) and halogenation (Neumann, et al., 2008). A number of these enzymes decorate biologically inactive precursors into pharmaceutically active molecules via regioselective and stereoselective transformations. As a result, tailoring enzymes are attractive candidates as biocatalysts towards synthesis of semisynthetic derivatives and drug libraries (Zhou, et al., 2008). LovD is an acyltransferase found in Aspergillus terreus and is responsible for converting the inactive precursor monacolin J acid (MJA) into the cholesterol-lowering drug lovastatin (LV, acid form lovastatin acid: LVA) via acylation of the α-S-methylbutyrate side chain (Kennedy, et al., 1999; Xie, et al., 2006) (Figure 1). The importance of the hydrophobic α-S-methylbutyryl side chain for binding of LVA to HMG-CoA reductase has been structurally confirmed (Istvan and Deisenhofer, 2001). Chemical modification of the LV side chain to α-dimethylbutyrate yielded the semisynthetic derivative simvastatin (SV, acid form simvastatin acid: SVA), which is the active pharmaceutical ingredient in the blockbuster drug Zocor® (Hoffman, et al., 1986). Semisynthesis of SV from LV is a multiple-step chemical process and is therefore an intensely pursued target for devising an efficient biocatalytic approach (Berg, et al., 2009; Morgan, et al., 2006). As a result, LovD is a prime candidate to serve as such a biocatalyst.
Figure 1.
Reactions catalyzed by LovD. LovD is responsible for converting MJA into LVA via acylation of the α-S-methylbutyrate side chain and can also synthesize SVA using DMB-SMMP as an acyl donor.
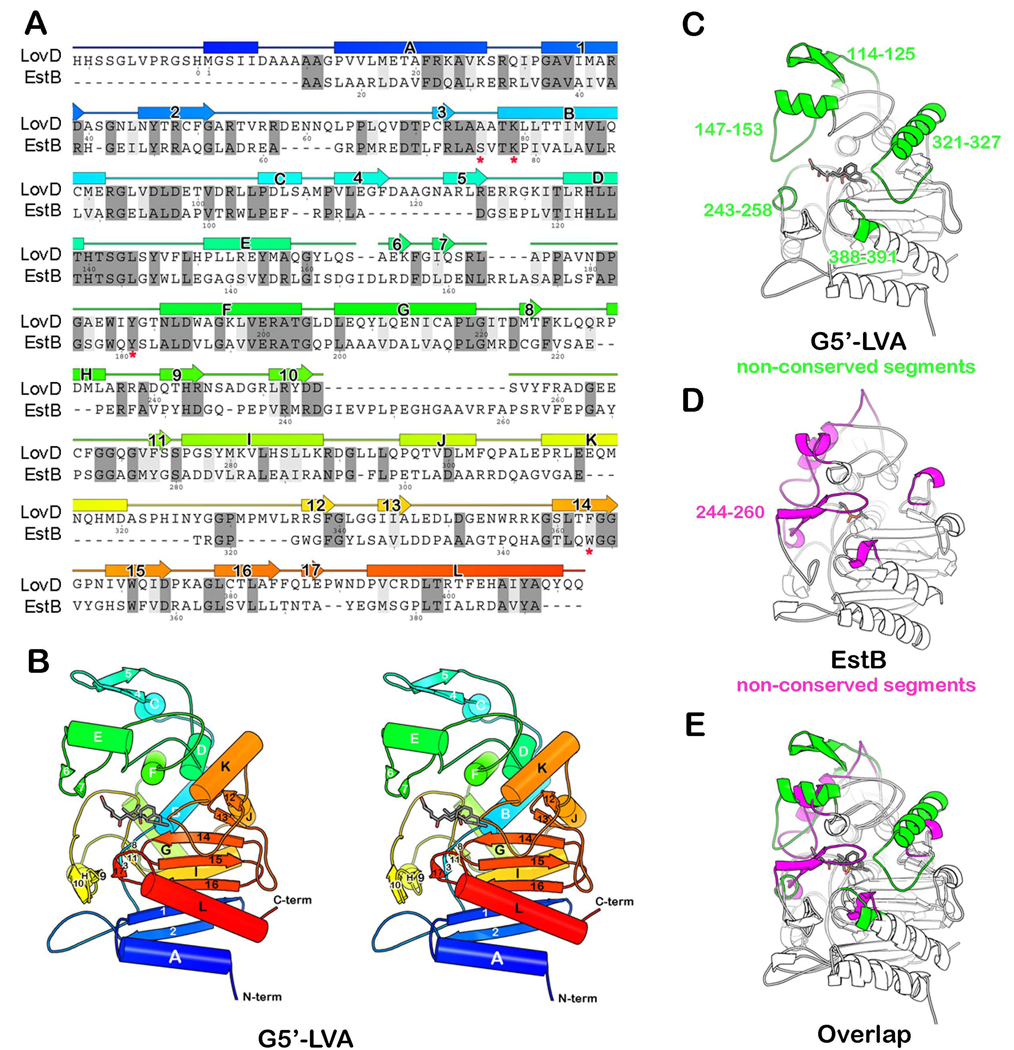
LovD is a 413-amino acid protein predicted to have an α/β hydrolase fold based on primary sequence analysis (Kennedy, et al., 1999). Among enzymes of known structure that are homologous to LovD is cephalosporin esterase, EstB (PDB ID 1CI9, 26% sequence identity) from Burkholderia gladioli (Wagner, et al., 2002). The likely general base Tyr188, as well as a conserved SXXK patch that contains the active site nucleophile Ser76, were indicated through alignment of LovD with EstB (Figure 2A) (Petersen, et al., 2001). During LVA biosynthesis, the α-S-methylbutyrate side chain is synthesized by the lovastatin diketide synthase (LDKS) LovF, and is then transferred by LovD regioselectively to the C8 hydroxyl of MJA via an unprecedented polyketide offloading mechanism (Xie, 2009). The protein-protein interaction between LovD and the acyl carrier protein (ACP) domain of LovF facilitates this highly efficient tailoring reaction in A. terreus. We have previously explored the substrate promiscuity of LovD and have shown that it can also synthesize SVA by using the small molecule substrate α-dimethylbutyryl-S-methyl-mercaptoproprionate (DMB-SMMP) as an acyl donor (Xie and Tang, 2007) (Figure 1). Using Escherichia coli as an expression host, a whole-cell biocatalytic platform for converting MJA to SVA was established that can produce SVA with low throughput (Xie and Tang, 2007). However, as with many enzymes that have been removed from their natural context, LovD is catalytically suboptimal as a biocatalyst and suffers from poor thermal stability (Arnold, 2001). The catalytic activity of SVA synthesis using DMB-SMMP is attenuated ~1,300 fold when compared to the natural substrate attached to LovF (Xie, 2009), indicating there is ample opportunity for optimization by protein engineering efforts. Furthermore, the structural basis of LovD function and substrate selection had not been elucidated, limiting our ability to rationally optimize the binding of the unnatural dimethylbutyryl substrate and improve LovD efficiency as a SV synthase.
Figure 2.
The crystal structure of LovD and its relationship to EstB. (A) A structure-based sequence alignment between LovD and EstB. Secondary structure elements assigned from the structure of LovD are shown above the sequence. The colors are ramped from blue at the N-terminus to red at the C-terminus. The active site residues in EstB are indicated by an asterisk “*” below the amino acid. (B) A ribbon diagram showing the G5’-LVA complex. (C) Structure of LovD. Highlighted in green are segments that are not conserved in EstB. These five loops project around the circumference of the active site like the fingers in a catcher’s mitt. (D) Structure of EstB. Highlighted in magenta are segments that are not conserved in LovD. (E) The overlay of LovD and EstB structures. Notably absent from LovD is a loop that covers the active site in EstB (residues 244–260).
In this article, we employed directed protein evolution (Arnold and Volkov, 1999) to improve the SV synthase activity of LovD. After seven rounds of screening, LovD mutants with significantly improved catalytic activities and higher thermal stability were isolated. In parallel, seven X-ray crystal structures including the parent LovD G0, an improved mutant G5, and the co-crystal structures of G5 with MJA, LVA and SVA were obtained. The crystal structures provide atomic resolution details regarding the mechanism of catalysis, substrate and product binding, protein-protein interactions with LovF, and a likely explanation for the effects of beneficial mutations on catalysis.
RESULTS
Development of an Agar-based Diffusion Screening Method
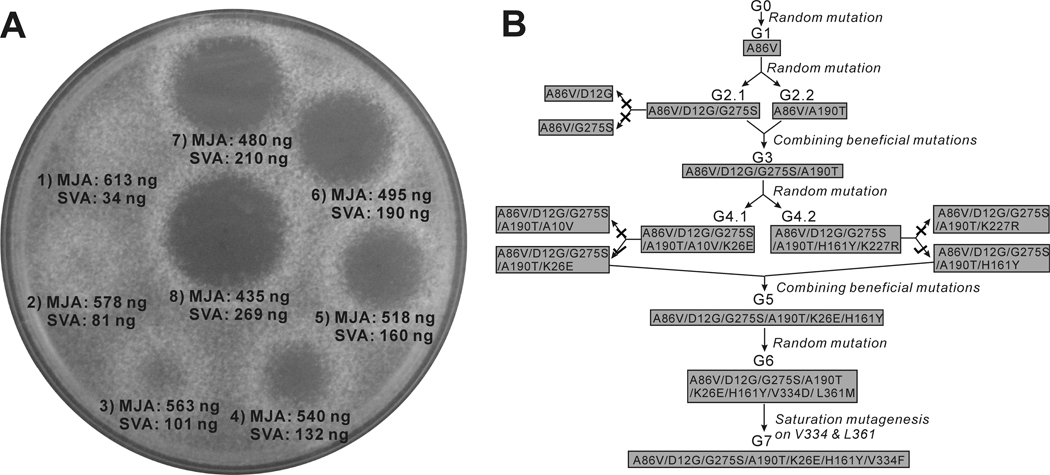
We developed an efficient screening method to assay for E. coli expressing LovD mutants with improved properties in the synthesis of SVA from MJA and DMB-SMMP. The assay relied on the growth inhibition of Neurospora crassa by statins, a property that was previously exploited in the screening of high LV producing A. terreus strains (Kumar, et al., 2000). We found that SVA can inhibit the growth of N. crassa at sub-microgram quantities, while inhibition by MJA requires hundred milligram quantities. To demonstrate the sensitivity and feasibility of the assay, an E. coli culture expressing wild type LovD was supplied with 10 mM MJA and 15 mM DMB-SMMP. At different time points, 2 µL aliquots were directly spotted on a Sabouraud’s dextrose agar (SDA) plate embedded with N. crassa at a density of 0.3~0.5 × 108 spores/L. After 16 hours of incubation at 30°C, different inhibition zones were observed for samples containing different degrees of conversion from MJA to SVA (as verified by HPLC) (Figure 3A). Based on this screening strategy, any significant contribution to whole cell LovD activity (defined as the rate of converting MJA to SVA by E. coli cultures expressing LovD variants; see Experimental Procedures), such as improvements in solubility, catalytic efficiency and stability can lead to a detectable phenotypical change.
Figure 3.
Directed evolution of LovD as a simvastatin synthase. (A) An agar diffusion-based assay was used to quantify the amount of SVA in the whole-cell activity experiments. N. crassa was embedded in the agar prior to spotting the reaction mixture. The numbers (1–8) designate different incubation times of 2.5, 4, 5.5, 7, 8, 9, 10, 12 hours following addition of MJA and DMB-SMMP to E. coli expressing wild type LovD. (B) Directed evolution of LovD mutants towards higher whole cell activities. There are a total of seven generations of LovD mutants. Four generations were derived from random mutagenesis including G1, G2.1, G2.2, G4.1, G4.2, and G6. Two generations were derived from combination of beneficial mutations from previous generation mutants including G3 and G5. G7 was derived though saturated mutagenesis of G6 at positions V334 and L361. All mutants with two amino acid changes were subjected to site directed mutagenesis to determine beneficial or deleterious mutations (G2.1, G4.1 and G4.2). (×) indicates that the mutant had lower whole cell activity comparing to the previous generation. (√) indicates that the mutant had higher whole cell activity comparing to the previous generation.
Screening of LovD Variants with Enhanced Whole Cell Activities
The starting LovD (generation zero or G0) used in directed evolution is the previously generated double mutant C40A/C60N from wild type LovD (Xie, et al., 2009). G0 was rationally engineered to be less prone to disulfide-mediated aggregation and was used for crystallization studies (Xie, et al., 2009). The mutant libraries were created by either saturation mutagenesis or error-prone Polymerase Chain Reaction (ep-PCR) that generated an average of 2.5 amino acid changes per round (Fromant, et al., 1995). During each round of screening, the mutant library was ligated into pET28(a) and electroplated into YT2 competent cells (Xie, et al., 2007). The individual mutants were cultured in 96-well plates, followed by induction of LovD expression, addition of MJA and DMB-SMMP, and spotting onto N. crassa embedded plates. Table 1 shows the gradual improvement in whole cell activity obtained following four rounds of ep-PCR (G1, G2, G4, G6), one round of saturated mutagenesis (G7), and two iterations of combining individual beneficial mutations (G3, G5), with the best mutant G7 displaying ~11 fold increase in whole cell activity as a SV synthase compared to G0 (Figure 3B). The mutant G2.1 contains amino acid changes at D12G and G275S. Construction of the corresponding single mutants using G1 as template showed that D12G alone had a large negative effect, while G275S alone had a weak positive effect compared to G1. This result suggests the two mutations in G2.1 act synergistically to enhance LovD activities. Combination of mutations from G2.1 and the A190T mutation in G2.2, which was recovered from the same round of ep-PCR, yielded the next generation mutant G3. Ep-PCR using G3 as a template yielded G4.1 and G4.2, each containing a different double mutation combination of A10V/K26E and H161Y/K227R, respectively. Site directed mutagenesis confirmed that both A10V and K227R had negative effects on the activities of LovD. Removal of these mutations and combination of K26E and H161Y yielded an improved mutant G5, which was ~6-fold improved in whole cell activity compared to G0. At this point, structural studies were performed on the G5 mutant to provide insights into the accumulated beneficial mutations. In parallel, an additional round of ep-PCR afforded G6 that contained the beneficial mutations V334D and L361M. Saturation mutagenesis was employed to optimize the combined effects of mutations at positions 334 and 361. The best mutant recovered was G7, of which the whole cell activity was increased an additional 20%. Surprisingly, we found that while position 334 was altered to phenylalanine, the previously deemed beneficial L361M mutation reverted back to leucine. Site-directed mutation of L361M in G7 confirmed that leucine was indeed the more favorable residue in the context of the V334F mutation.
Table 1.
Amino Acid Substitutions and Characterization of LovD Variants
| Mutations | Whole cell Activitya |
kcat (min−1) | KM of MJA (mM) b | KM of DMB- SMMP (mM−1)c |
Soluble protein (mg/l)d |
Tm(°C)e | |
|---|---|---|---|---|---|---|---|
| G0 | 1 | 0.66 ± 0.03 | 0.77 ± 0.17 | 0.67 ± 0.12 | 138 ± 11 | 39.5 ± 0.4 | |
| G1 | A86V | 1.2 | 0.79 ± 0.03 | 0.74 ± 0.16 | 0.66 ± 0.19 | 140 ± 5.4 | 41 ± 0.7 |
| G2.1 | A86V D12G G275S | 1.9 | 1.14 ± 0.03 | 0.91 ± 0.17 | 0.62 ± 0.10 | 184 ± 8.7 | 40.5 ± 0.4 |
| G2.2 | A86V A190T | 1.8 | 1.20 ± 0.09 | 0.74 ± 0.21 | 0.69 ± 0.17 | 168 ± 17 | 41 ± 0.4 |
| G3 | A86V D12G A190T G275S | 3.6 | 1.86 ± 0.09 | 0.77 ± 0.11 | 0.70 ± 0.19 | 205 ± 23 | 41 ± 0.4 |
| G4.1 | A86V D12G A190T G275S A10V K26E |
4.8 | 2.13 ± 0.03 | 0.70 ± 0.24 | 0.66 ± 0.16 | 183 ± 18 | 43.5 ± 0.7 |
| G4.2 | A86V D12G A190T G275S H161Y K227R |
5.2 | 2.16 ± 0.12 | 0.80 ± 0.24 | 0.64 ± 0.16 | 221 ± 9.3 | 42.5 ± 1.9 |
| G5 | A86V D12G A190T G275S K26E H161Y |
6.4 | 2.61 ± 0.03 | 0.74 ± 0.03 | 0.69 ± 0.14 | 206 ± 5.7 | 46.5 ± 0.4 |
| G6 | A86V D12G A190T G275S K26E H161Y V334D L361M |
9.3 | 3.30 ± 0.06 | 0.70 ± 0.07 | 0.63 ± 0.15 | 212 ± 3.9 | 47 ± 0.1 |
| G7 | A86V D12G A190T G275S K26E H161Y V334F |
11.2 | 4.80 ± 0.06 | 0.70 ± 0.04 | 0.69 ± 0.17 | 214 ± 6.3 | 48.5 ± 0.7 |
The whole-cell activity of LovD mutants are compared to G0, which has a conversion rate of 1.7 mM/hr and is normalized to 1.
KM of MJA is derived at 25°C when DMB-SMMP is fixed at 2 mM.
KM of DMB-SMMP is derived when MJA is fixed at 2 mM.
The amounts of soluble proteins are measured from purified protein levels.
Tm is measure by circular dichroism. All results represent mean values of triplicate determinations and standard deviations.
In Vitro Characterization of LovD Variants
To dissect the contributions that led to the increases in whole cell activity, kinetic parameters (kcat, KM), soluble protein levels and thermal stability of all the improved mutants were characterized and listed in Table 1. The binding affinities (KM) of LovD mutants toward MJA and DMB-SMMP were each within a narrow range (0.7 mM to 0.9 mM for MJA and 0.6 mM to 0.7 mM for DMB-SMMP). The lack of improvement in KM towards either substrate is not surprising considering the high concentrations of substrates used in the screening assay (~10 KM of MJA and DMB-SMMP). The observed improvements in whole cell activity are mainly due to increases in the kcat of the mutants and levels of soluble proteins. The kcat and soluble protein levels were simultaneously increased ~3 fold and ~1.5 fold from G1 to G3, respectively. Impressively, the protein expression levels of G3 reached 205 mg/L. In contrast, improvements in kcat were the sole contribution to the increases in whole cell activities from round 4 to round 7. Most notably, a single V334F mutation from G5 to G7 nearly doubled the catalytic turnover rate. We also observed a gradual increase in the melting temperature (Tm) of the LovD mutants from 39.5°C to 48.5°C by using circular dichroism. The increases in thermal stability were also reflected in the whole cell activities of the mutants when expressed at elevated temperatures (Figure S1). Whereas the G0–G3 mutants have no detectable activity when expressed at 32°C, the later generation mutants retained significant SV synthase activities, with the G7 mutant exhibiting comparable activity to that of G0 at 25°C. Furthermore, the G7 mutant remained active even when expressed at 37°C. The increased thermal stabilities of the mutants have important practical implications in using LovD as a biocatalyst for SV semisynthesis.
To examine the activities of the mutants towards synthesis of the natural biological product LVA, we performed kinetic assays using α-methylbutyryl-SMMP (MB-SMMP) and MJA. A similar trend in the improvements of kcat toward LVA synthesis was observed (Figure S2), indicating the LovD relative substrate specificity towards the acyl group (either MB or DMB) has not changed. Interestingly, when LovF was used in the kinetic assay for LVA synthesis (Figure 1), we observed a progressive loss of activities of the LovD mutants (Figure S2). The G7 mutant exhibited a 27-fold decrease in activities towards LovF compared to the G0 parent, most likely attributed to the deterioration of the required protein-protein interactions for catalysis (Xie, 2009). Therefore, the mutations accumulated during directed evolution may have gradually altered the conformation of LovD to impair its communication with LovF, while not affecting binding of the SMMP-bound acyl group.
Overall Structure of LovD
Seven LovD crystal structures were determined to help illuminate the mechanism of the LovD-catalyzed reaction and possible basis for improved catalysis. These include (1) G0, (2) selenomethionyl G0 (G0-Semet), (3) the improved mutant G5, (4) G5 in complex with substrate MJA, the G5 with S76A active site mutated (called G5’) in complex with (5) LVA, (6) SVA, and (7) MJA. The resolution limits of the structures range from 2.5 to 2.0 Å except for G0. The native G0 structure was resolved at 3.4 Å, but was improved to 2.5 Å in the G0-Semet variant. Refinement statistics are provided in Table 2.
Table 2.
Statistics of X-ray Data Collection and Atomic Refinement
| Protein | G0 | SeMet G0 | G5 | G5 | G5′ | G5′ | G5′ |
|---|---|---|---|---|---|---|---|
| Ligand | none | none | none | MJA | MJA | SVA | LVA |
| Data collection | |||||||
| Space group | P1 | C2 | P212121 | P212121 | P212121 | P212121 | P212121 |
| Cell dimensions | |||||||
| a, b, c (Å) | 56.7,79.7,104.1 | 209.5,85.2,104.0 | 58.2, 75.0, 131.6 | 58.5, 75.2, 133.5 | 58.0, 75.2, 132.7 | 58.5, 75.1, 131.8 | 59.0, 75.2, 131.7 |
| α, β, γ (°) | 94.1,91.6,106.8 | 90.0,117.5,90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| protomers/ asymmetric unit |
4 | 4 | 1 | 1 | 1 | 1 | 1 |
| Resolution (Å) | 100.00-3.40 | 80.00-2.50 | 90.00-2.00 | 60.00-2.00 | 60.00-2.05 | 60.00-2.05 | 60.00-2.00 |
| (3.66-3.40) | (2.59-2.50) | (2.07-2.00) | (2.07-2.00) | (2.12-2.05) | (2.07-2.00) | (2.07-2.00) | |
| Rmerge(%) | 20.8 (43.4) | 11.8 (45.3) | 4.9 (37.8) | 15.6 (39.6) | 6.8 (40.2) | 6.9 (31.4) | 11.8 (39.6) |
| I / σI | 5.1 (2.9) | 10.2 (1.7) | 47.2 (6.5) | 13.0 (4.8) | 38.3 (6.6) | 19.5 (3.9) | 13.33(4.64) |
| Completeness (%) | 99.3(98.9) | 89.9 (46.7) | 99.9 (100.0) | 99.8 (100.0) | 99.9 (99.8) | 97.7 (99.6) | 98.5 (99.5) |
| Redundancy | 3.7(3.7) | 5.3 (3.0) | 11.6 (11.7) | 6.2 (6.4) | 15.4 (12.9) | 4.9( 4.9) | 6.4( 6.5) |
| wavelength (Å) | 0.9793 | 0.9793 | 0.9794 | 0.9794 | 0.9792 &1.5418 | 0.9717 | 0.9795 |
| Refinement | |||||||
| Resolution (Å) | 103.7- 3.4 | 65.9- 2.50 | 65.80- 2.00 | 46.18- 2.00 | 49.86- 2.05 | 49.51- 2.00 | 46.42- 2.00 |
| (3.5- 3.4) | (2.56- 2.50) | (2.05- 2.00) | (2.05- 2.00) | (2.10- 2.05) | (2.05- 2.00) | (2.06- 2.00) | |
| No. reflections | 22457 (1669) | 47728 (1657) | 37539 (2837) | 38325 (2910) | 35223 (2686) | 36938 (2714) | 37005 (2671) |
| Rwork / Rfree (%) | 23.1/27.5 | 24.8/29.0 | 17.9/20.6 | 17.9/21.3 | 17.7/20.9 | 17.6/20.9 | 16.2/18.9 |
| (31.9/35.5) | (35.6/40.5) | (20.4/24.4) | (21.5/26.1) | (19.8/23.7) | (23.1/25.1) | (18.3/21.8) | |
| Number of atoms | |||||||
| Protein | 12328 | 12547 | 3289 | 3332 | 3355 | 3351 | 3343 |
| Ligand/ion 1 | 0 | 65 (sulfate) | 13 (PEG) | 48 (MJA) | 24 (MJA) | 31(SVA) | 30 |
| Ligand/ion 2 | 0 | 0 | 6 (glycerol) | 12 (formate) | 8 (dithiothreitol) | 0 | 0 |
| Water | 0 | 21 | 151 | 275 | 157 | 179 | 230 |
| B-factors (Å2) | |||||||
| Protein | 6.3 | 51.7 | 34.4 | 27.6 | 35.4 | 32.2 | 31.4 |
| Ligand/ion 1 | n/a | 68.0 (SO4) | 58.6 (PEG) | 58.8 (MJA) | 46.5 (MJA) | 45.5 (SVA) | 38.3 (LVA) |
| Ligand/ion 2 | n/a | n/a | 58.2 (glycerol) | 38.6 (formate) | 74.2 (dithiothreitol) |
n/a | n/a |
| Water | n/a | 39.4 | 36.9 | 32.8 | 37.6 | 36.1 | 37.2 |
| Rmsds | |||||||
| Bond lengths (Å) | 0.011 | 0.006 | 0.009 | 0.009 | 0.008 | 0.008 | 0.008 |
| Bond angles (°) | 1.3 | 0.995 | 1.199 | 1.230 | 1.202 | 1.193 | 1.193 |
| Ramachandran Plot (%) |
|||||||
| most favored | 88.0 | 90.0 | 91.2 | 89.4 | 89.7 | 90.3 | 90.6 |
| Additionally allowed |
11.4 | 9.7 | 8.5 | 10.3 | 10.0 | 9.4 | 9.1 |
| Generously allowed |
0.6 | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 |
| Disallowed | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| PDB ID code | 3HL9 | 3HLB | 3HLC | 3HLD | 3HLE | 3HLF | 3HLG |
Rmerge = ∑| I- <I>|2/∑I2, where I is the observed intensity. Both summations involve all input reflections for which more than one symmetry equivalent is averaged. Rwork = ∑||Fo| - |Fc||/∑|Fo|, where Fo and Fc refer to observed and calculated structure factors, respectively. Rfree is similar to Rwork, but is based on a subset of the reflections, which were withheld from refinement for cross validation. Numbers in parentheses refer to the outer shell of data.
The crystal structure of LovD G0-Semet revealed a variation of the α/β hydrolase fold (Heikinheimo, et al., 1999; Nardini and Dijkstra, 1999). It consists of two domains. The first domain (residues 1–92 and 204–413) is a central seven-stranded antiparallel β-sheet flanked by α-helices on either face (Figure 2B and Figure S3A). A cis-peptide bond is formed between Glu388 and Pro389, contributing to a kink in the sheet. The second domain is smaller (residues 93–203) and primarily α-helical. A deep and narrow cleft (11 × 6 Å) is formed at the interface between the two domains. At the bottom of the cleft is the catalytic Ser76 that acts as the nucleophile in the acyltransfer reaction.
Comparison between LovD and EstB
Encircling the active site cleft is a broad, ring-shaped ridge, which is absent from the homologous enzyme EstB. Their structures are superimposable with RMS deviation of only 1.5 Å over 270 pairs of α-carbons (about 2/3 of the structure) (Figure 2E). The similarity is striking for the core of the two enzymes, but they differ notably in the loops peripheral to the active site, both in size and architecture. In LovD, these loops give the impression of a ring-shaped ridge or baseball catcher’s mitt over the active site with fingers composed of five loops: residues 114–125, 147–173, 243–258, 321–327, and 388–391 (Figure 2C). The first and last of these loops are longer in LovD than EstB by 11 and 19 residues, respectively. The second loop is displaced 7 Å from the active site compared to EstB, extending the grasp of the “mitt”. Most notably absent from the LovD molecule is the 23-residue loop that if present would obstruct the grasp of the mitt and cover the active site entrance (corresponding to residues 244–260 in EstB) (Figure 2D, Figure 3B). The shape and diameter of the ridge surrounding the active site (a circle of 17 Å diameter) satisfies the requirement of accommodating LovD’s natural binding partner, the ACP of LovF (Xie, 2009). The distance between the rim of LovD and active site Ser76 is ~20 Å, which is roughly the same as the length of the phosphopantetheine (Ppant) arm of the ACP domain of LovF.
Crystal Structures of the Mutant G5
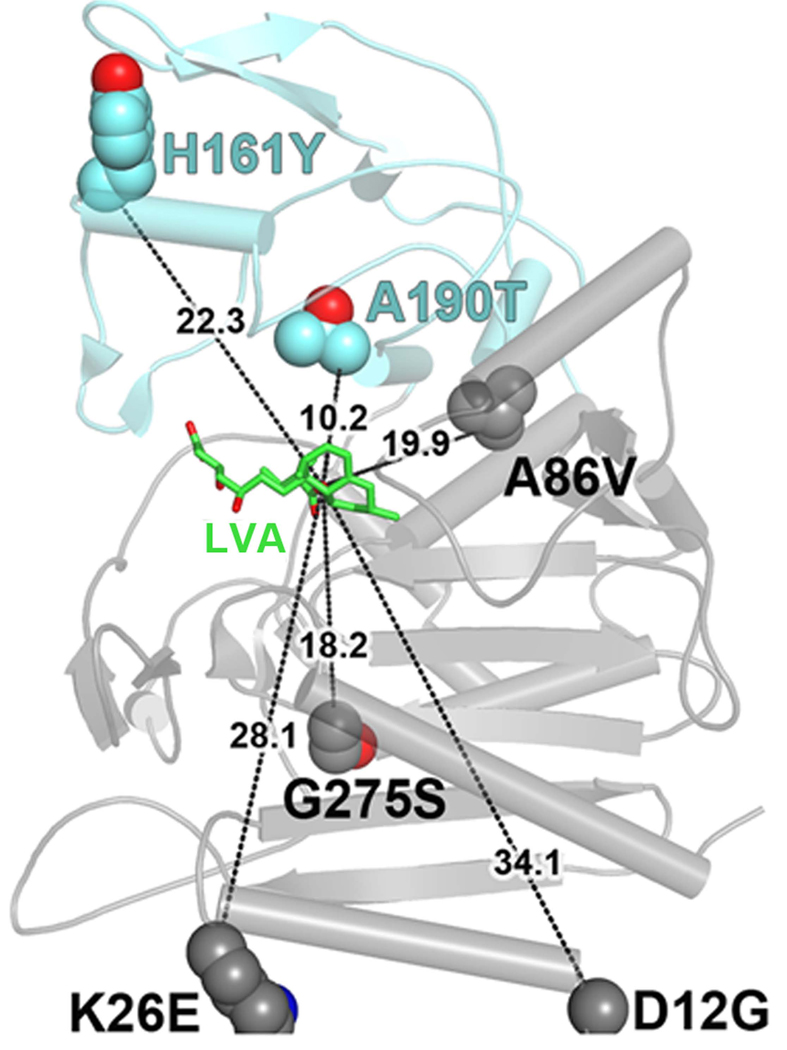
The LovD G0-Semet structure enables us to identify residues important for catalytic efficiency which, when mutated to LovD G5, improved the kcat ~4 fold. These residues are scattered widely over space, forming no mutual contacts. Further, because they are located in both buried and solvent exposed regions, they do not share a common physical environment. Moreover, distances of these residues to the active site Ser76 are relatively large, ranging from 10 to 32 Å for Thr190 and Gly12, respectively (Figure 4). The lack of connectivity of the residues to each other and to the active site is a common phenomenon in numerous directed evolution experiments (Hsu, et al., 2005; Oue, et al., 1999; Zhao and Arnold, 1999).
Figure 4.
Structure of the G5 mutant provides insight into improved catalysis. Positions of the amino acid changes present in the improved mutant G5, highlighting their generally large distances from the active site. Distances are drawn from the amino acid α-carbons to the nucleophilic hydroxyl (C8) of LVA (shown in green).
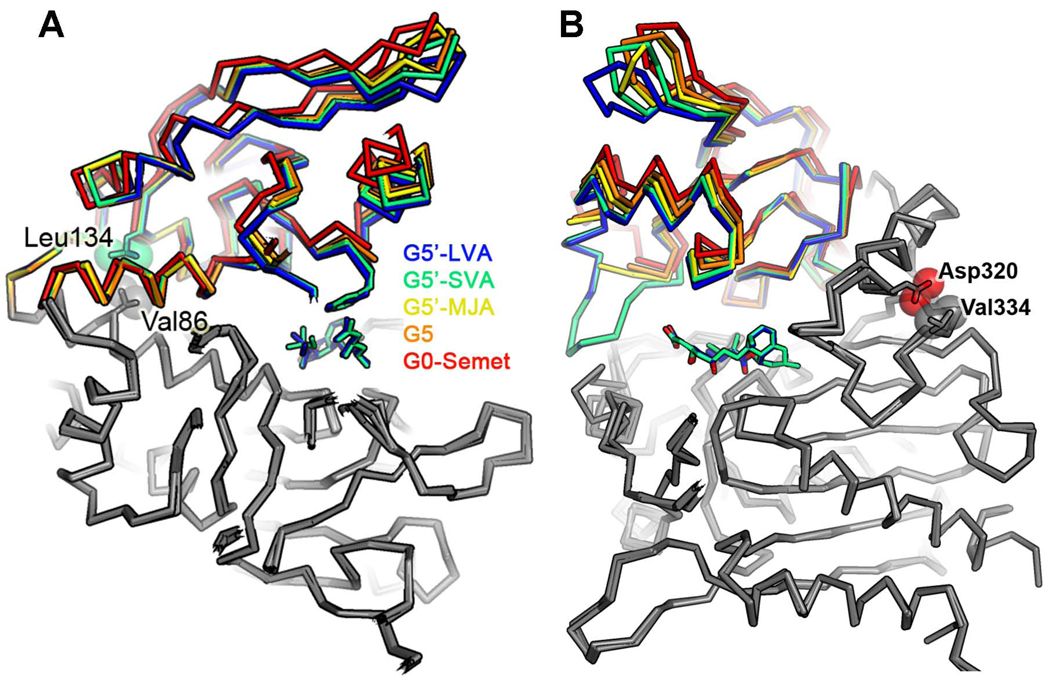
The crystal structure of G5, however, offered evidence to suggest that the increased activity afforded by its six mutations can be attributed to their ability to stabilize a more closed form of the active site cleft. Comparison of the G0-Semet structure with G5 revealed a rotation about the domain-domain hinge of 5°, narrowing the cleft by about 0.5 Å and producing motions up to 3 Å for atoms furthest from the hinge (Figure 5A). Subsequent structures of G5’ bound to the LVA showed larger movements along the same trajectory arising from a 14° hinge rotation. That observation suggests that the beneficial mutations in the G5 variant help promote a conformational change required for catalysis.
Figure 5.
Side view of the overlap of five LovD structures. This representation indicates a hinge rotation between two domains due to G5 mutations and the presence of bound ligands. (A) Two important residues (Val86 and Leu134), which may stabilize closure of the hinge, are shown in spheres. (B) Another two residues (Val334 and Asp320) are directly in contact with each other. Mutation V334D in G6 would result in electrostatic repulsion between Asp334 and Asp320 while mutation V334F in G7 could result in steric clash between the phenyl side chain and Asp320. Both could stabilize closure of the hinge.
Stabilization of the closed conformation of LovD might enhance activity by positioning residues critical for catalysis. For example, when the large domains of G0-Semet, G5, and G5-MJA are superimposed (residues 14–92 and 204–405), domain rotation from G0-Semet to G5 closes the gap between the guanido group of Arg173 and the C15 carboxylate of MJA by 0.5 Å. This rotation could be attributed to the G5 mutations (and perhaps to a difference in crystal packing) but not to ligand binding, since the comparison is between two unliganded structures. Ligand binding produces a further rotation from G5 to G5-MJA which closes the gap between Arg173 and MJA by an additional 2.2 Å, so that a hydrogen bond is formed between the two groups. Similarly, G5 mutations bring Phe148 and Tyr188 side chains from the G0-Semet position closer to substrate (G5-MJA), although their motion is smaller since they lie closer to the hinge axis (Figure S4A).
The A86V mutation in particular appears responsible for stabilizing closure of the hinge. Its two additional methyl groups buried in the boundary between domains act as a wedge pushing against Leu134 on the distal side of the hinge axis, thereby closing the active site cleft on the proximal side of the hinge axis (Figure 5A). The beneficial effects of the K26E and G275S mutations are less obvious. The K26E mutation might improve stability of the enzyme by breaking up a patch of positively charged residues (R22, K23, K26, and R28) on the surface of helix A (Figure S5A) (Schweiker, et al., 2007). The G275S mutation appears to improve stability of the enzyme by adding a hydrogen bond with the N-terminal end of helix I and decreasing torsional flexibility of the backbone (Figure S5B).
Co-crystallization of LovD G5’ with MJA, LVA and SVA
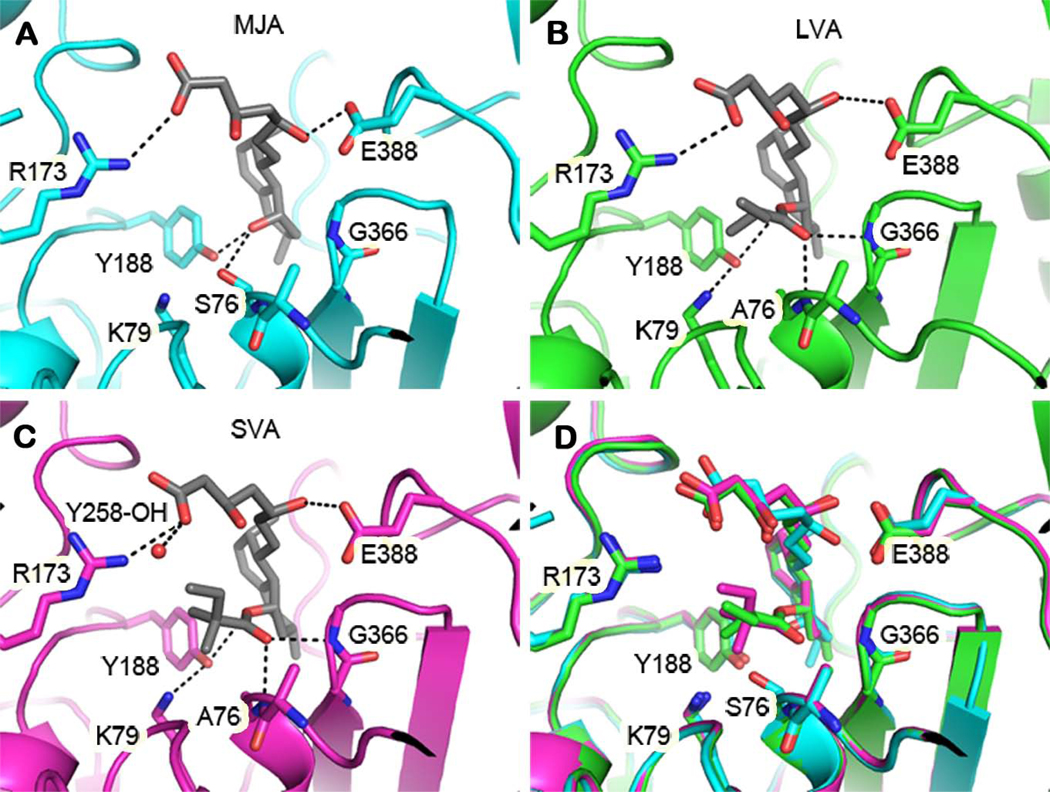
Structures of LovD G5’ in complex with substrate and products illustrate the mode of binding of these ligands and suggest a catalytic mechanism for acyl transfer. The substrate MJA binds with its C8-hydroxyl group deep inside the cleft between domains, forming hydrogen bonds with Ser76, Tyr188, and a fortuitously bound formate molecule (used as a cryoprotectant) (Figure 6A). The proximity of the C8-hydroxyl to the Ser76 hydroxyl is consistent with the expectation that both hydroxyl groups initiate a nucleophilic attack on the same acyl group during different steps in the reaction sequence (Figure S6B). The two faces of the decalin ring system of MJA are sandwiched between the aromatic rings of Trp390 and Tyr188 (Figure S3C). Additional hydrophobic and van der Waals interactions with the edges of the ring system are observed with Phe363, Ile325, Tyr327, Phe148, Ler149, and the peptide planes of Gly364, Gly365, and Gly366 (Figure S3C). The hydrophilic tail of MJA (i.e. the C1 substituent on the decalin ring) extends away from the active site into bulk solvent. The C11 hydroxyl group hydrogen-bonds with the Glu388 side chain and the backbone amide of Trp390, the latter being mediated by a water molecule. The C15 carboxylic acid forms a salt bridge with Arg173.
Figure 6.
Comparison of the LovD active site bound with different ligands. (A) G5 in complex with substrate MJA. (B) G5’ in complex with product LVA. (C) G5’ in complex with product SVA. (D) Overlay of the three structures showing conformational changes, particularly at the nucleophilic serine, associated with binding to different ligands. Dashed lines represent hydrogen bonds. The active site entrance is at the top of each figure. Some residues involved in ligand binding are not shown.
The position of the α-S-methylbutyryl group is revealed in the crystal structure of the LovD G5’ mutant in complex with LVA. As in the MJA complex, the decalin ring and hydrophilic tail of LVA bind with similar geometry. Interestingly, the additional methylbutyryl group extends parallel to the MJA hydrophilic tail (Figure 6B). The proximity of the two tails gives LVA a hairpin shape, with the decalin ring forming the hairpin turn between the two tails. The hydrophobic side chain binding position is likely the site at which the acyl donor binds. The proximity of the tails also suggests how MJA competitively inhibits the acyl transfer reaction when it binds prior to the methylbutyryl substrate (Xie, et al., 2006), because the hydrophilic tail of MJA partly obstructs access of the methylbutyryl group to the active site, ordered binding of the substrates is required.
Structural comparisons between complexes of LovD G5’ with LVA and SVA suggest some strain is involved in accommodating the non-natural product, SVA (Figure 6C). SVA contains an additional methyl group compared to LVA, located on the α-S-methylbutyryl moiety. Superimposition using only α-carbons in the large domains of the two structures shows nearly identical arrangements of atoms in the large domains and the decalin rings (Figure 6D). But contact between the additional methyl group (attached at C2’) and the side chain of Phe148 appears to push open the cleft between domains. As a result, Phe148 moves approximately 0.8 Å away from its position in the LVA complex (Figure S4B). There is also a 30° rotation about the C1’–C2’ bond of the α-dimethylbutyryl moiety. The consequence of these rotations for the relative catalytic rates of the two substrates appears minor; the movements near the atoms directly involved in acyl transfer are small. This is consistent with the ability of LovD to catalyze acyl transfer using an α-dimethylbutyryl group as substrate instead of the natural α-methylbutyryl group. However, further amino acid mutations, such as the aforementioned Phe148, could improve the fit to the α-dimethylbutyryl substrate, or other variations. The structures presented here provide a framework for such design efforts.
DISCUSSION
In this work, we show that seven amino acid changes led to the ~ 11 fold increase in the SV synthase activity of LovD. This level of enhancement is significant considering G0 was already an adequate SV synthase following our previous efforts in substrate and strain optimization. Although the kinetic activity of G7 is far below that of the natural reaction catalyzed by LovD using acyl-LovF, G7 is a robust mutant for high volume synthesis of SVA using the whole cell platform. When G7 was applied in a high density fermentation environment, more than 30g/L of MJA was quantitatively converted to SVA within one day. The relatively few rounds of direct evolution to achieve the activities of G7 also demonstrate that LovD is highly evolvable as a biocatalyst.
The X-ray crystal structures solved in this work provides insight into different facets of LovD enzymology. Among these, the spatial arrangement of the LovD catalytic triad Ser76-Lys79-Tyr188 was captured and is shown to be consistent with that of the esterase EstB (Figure S6). Tyr188 (as the phenolate) appears to be the general base in initiating the two nucleophilic attacks required for completion of the acyl transfer reaction (Oefner, et al., 1990). The first nucleophilic attack is by Ser76 on the α-S-methybutyryl group and the second attack is by the C8 hydroxyl of MJA on the acylated enzyme intermediate. In both reactions, the attacking hydroxyl group must be activated by deprotonation (Figure S6C). Tyr188 is well-positioned to deprotonate both hydroxyl groups, forming hydrogen bonds with Ser76 in the apo-enzyme and with MJA in the G5-MJA complex. Lys79 is also well-positioned to aid in activating the two hydroxyl groups by forming a hydrogen bond relay with Tyr188 in the G5-MJA complex. Site-directed mutation of either Tyr188 or Lys79 to alanine resulted in complete loss of activity.
Details of the α-S-methylbutyryl binding pocket suggest how the transition states for the acyl transfer reactions are stabilized. As in the MJA complex, the C8 oxygen of LVA maintains a hydrogen bond to Tyr188, but the neighboring water molecule has been displaced by the carbonyl oxygen of the α-S-methylbutyryl group. This carbonyl oxygen forms a pair of hydrogen bonds with the backbone amides of Ala76 (i.e. Ser76 in the G0-Semet) and Gly366. The geometry of the hydrogen bonds appears ideal, with the amide hydrogen atoms pointed directly at the two lone electron pairs on the carbonyl oxygen. These hydrogen bonds would appear well suited to stabilize a tetrahedral transition state. The closest protein contacts with the aliphatic portion of the methylbutyryl group are aliphatic or aromatic carbons: Ala75 (Cβ), Phe148 (Cζ), Tyr146 (Cζ), and Asn270 (Cβ). Notably, the α-S-methylbutyryl aliphatic carbons are also surrounded by three positively charged side chains, Arg73, Lys79, and Arg173, all within 4.1 Å. These positive charges might help stabilize the negative charge of the oxyanion hole that forms during acyl transfer. Indeed, the pocket’s affinity for negative charges is demonstrated by the fortuitous presence of a bound formate anion in this position in the G5-MJA complex.
The crystal structures provide plausible explanations for the basis of enhanced catalytic efficiency. Upon ligand binding, LovD undergoes a conformational change analogous to the closing of the catcher’s mitt. Movement of the domains shown in Figure 5A positions the catalytic residues in closer proximity to each other and to the ligands, and serves to enhance the rate of catalysis. The G5 structure suggested that beneficial mutations afforded an alternative way to pre-position the active site residues to increase the catalytic efficiency. Although the V334D and V334F mutations of G6 and G7, respectively, were discovered subsequent to the structural work of G5, the molecular explanation above can also be applied to rationalize the beneficial nature of the additional single mutations. Val334 is located in the middle of a loop between helix K and sheet 12. The two side chain methyl groups of Val334 are directly in contact with the side chain of Asp320 on helix K, which serves as one of the domain-domain hinges and is in contact with the loop where Tyr188 is located (Figure 5B). Mutation of V334 could therefore result in movement of the domains around the helix K hinge. For example, the mutation V334D in G6 would result in electrostatic repulsion between Asp334 and Asp320. Similarly, the mutation V334F in G7 could result in steric clash between the phenyl side chain and the side chain of Asp320, further closing the active site cleft and bringing key residues (e.g. Tyr188) into more optimal positions for catalysis. On the other hand, the more compact conformations of LovD mutants are less compatible with binding to LovF, which apparently favors the open conformation of LovD.
SIGNIFICANCE
SV is the active pharmaceutical ingredient of the blockbuster cholesterol-lowering drug Zocor®. Semisynthesis of SV from LV is therefore an intensely pursued target for devising an efficient biocatalytic approach. Our previously developed platform for the biosynthesis of SV was powerful, but still suboptimal due to the lack of a robust biocatalyst. In this work, we employed directed evolution to engineer LovD. Several better mutants were obtained through a well designed screening method and the best mutant “G7” displaying an ~11-fold increase in whole cell biosynthesis of SVA compared to the parent G0. Catalytic efficiency, solubility and thermostability were improved simultaneously showing the power of our selection system. More strikingly, we have determined seven X-ray crystal structures including the parent LovD G0, an improved mutant G5, and the co-crystal structures of G5’ with MJA, LVA and SVA. The structure information not only aided our understanding of the catalytic mechanism of LovD, but also afforded a great insight into how mutations affected the overall properties of LovD. Comparing the structures between LovD G0 and G5 suggests the beneficial mutations help promote a more compact conformation required for catalysis. The co-crystallization of LovD G5’ with substrate MJA, product LVA and SVA reveals how acyl transfer reaction proceeds via a ping-pong mechanism, how MJA becomes a competitive inhibitor and how the catalytic cavity is adapted to accommodate its non-natural product. Our work, therefore, can have significant impact on biocatalyst development and provides deep insights into fundamental understanding of enzymology.
EXPERIMENTAL PROCEDURES
Ep-PCR and Construction of Mutant Library
Ep-PCR procedure was modified from established protocols (Fromant, et al., 1995). The reaction consisted of 0.35 mM dATP, 0.4 mM dCTP, 0.2 mM dGTP, 1.35 mM dTTP, 4 mM MgCl2, 0.25 mM MnCl2, and 2.5 U Taq polymerase. The reaction mixture was submitted to 25 cycles PCR: 94°C for 1 min, 55°C for 1 min and 72°C for 3 mins. The resulting PCR products were digested with DpnI, further digested with EcoRI and NdeI, and ligated to pET28(a). The ligation mixture was transformed to YT2 and plated on LB agar containing 35 mg/L kanamycin.
Selection of High Activity Mutants
N. crassa was grown on SDA slants for 10 days and spores were harvested with 1% Tween-80. 100 ml of molten SDA was seeded with 0.3–0.5×108 spores and poured into a 230×230 mm plate. Colonies from mutation library were cultured in 96 well plates containing 250 µl LB medium (with 35 mg/L kanamycin). The cells were grown at 37°C to saturation and transferred to duplicated plates. Protein expression was induced with 0.1 mM IPTG at OD600 of 0.5 and the expression was performed at 25°C for 16 h. 5 mM MJA and 10 mM DMB-SMMP were added to initiate the reaction. After a certain reaction time (45 mins to 4 hours depending on the activity of the parent), cells were removed with centrifugation (2,000 g, 4°C, 5 min). The amount of supernatant spotted on the SDA plate was typically 1~3 µl. The plates were incubated at 30°C for 16~18 hours. The improved mutants were selected following visual comparison of the inhibition zones. Normally, one or two improved mutants can be obtained from screening ~ 2,000 mutants in two weeks.
Site-Directed Mutagenesis
Site-directed mutations were performed using the standard Quickchange® strategy using relevant templates. The primers were ordered from IDT (Integrated DNA Technologies). All mutations were verified by DNA sequencing (Laragen, Los Angeles, CA).
Saturation Mutagenesis
The LovD G6 gene was randomly mutated at positions of V334 and L361. Since the two residues are close to each other, the two random mutations were introduced in a single pair of primers. Two segments were amplified by PCR and linked together using slice-by-overlap extension (SOE) PCR to give intact LovD gene, which was subsequently introduced to pET28(a).
Determining Whole-cell Biocatalysis Activity
Parent LovD G0 and all mutants were cultured in parallel for comparison. A single colony of the freshly transformed YT2 competent cells was used to inoculate a 5 mL LB culture supplemented with 35 mg/L kanamycin. Following overnight growth at 37°C, 100 µl of the culture was inoculated into 50 mL LB medium supplemented with 35 mg/L kanamycin. When OD600 reached 0.4~0.6, 0.1 mM IPTG was added to the cultures and expression of all LovD variants was performed at 25°C for 16 hr. To mimic the high density fermentation conditions, the cells were then concentrated 10-fold before addition of substrates. A 10 ml aliquot of each culture was collected by centrifugation (4°C, 2,000 g, 10 min). The cell pellet was gently resuspended in 1 ml of the medium supernatant, followed by addition of 70 µl of MJA (300 mM stock) to a final concentration of 15 mM. The concentrated culture was then divided into seven 200 µl aliquots and 1 µl of pure DMB-SMMP was added to each sample to a final concentration of 20 mM. The small cultures were then shaken at 300 rpm at 25 °C. At each time point, a complete extraction of one culture aliquot was performed by adding 10 µl of 20% SDS for cells lysis, followed by extraction with 500 µl ethyl acetate containing 1% trifluoroacetic acid TFA. The organic phase was removed, evaporated, and redissolved in 500 µl acetonitrile for HPLC analysis. The whole-cell activity was determined by fitting the linear regions of the conversion time course plot.
Kinetic Assay of LovD Variants towards MJA and DMB-SMMP
To obtain KM values for MJA and kcat, the DMB-SMMP concentration was fixed at 2 mM, while the concentration of MJA was varied from 0.25 to 5 mM. To obtain KM values for DMB-SMMP and kcat, the MJA concentration was fixed at 2 mM, while the concentration of DMB-SMMP was varied from 0.5 to 5 mM. Dimethyl sulfoxide (DMSO) was added to a final concentration of 10% to facilitate the solubilization of DMB-SMMP. At different time points of the kinetic assay, an aliquot of the reaction mixture was removed, quenched with 1% TFA and extracted with EA containing 1% acetic acid. The organic phase was separated, dried, resolubilized by acetonitrile (ACN) and analyzed by a Beckman Gold HPLC using a reverse phase C18 column (Alltech Apollo 5µ, 150 mm × 4.6 mm) and a linear gradient: 60% ACN in water (0.1% trifluoroacetic acid [TFA]) to 95% ACN in water (0.1% TFA) for 10 min, 1 mL/min. Conversion of MJA to SVA was measured by integration of the peaks at 238 nm.
Kinetic Assay of LovD Variants towards MB-SMMP
To compare the kcat of LovD mutants towards lovastatin synthesis using MB-SMMP as the substrate, both MJA and MB-SMMP were fixed at 2 mM. DMSO was added to a final concentration of 10% to facilitate the solubilization of MB-SMMP. At different time points of the kinetic assay, an aliquot of the reaction mixture was removed, quenched with 1% TFA and extracted with EA containing 1% acetic acid. The organic phase was separated, dried, resolubilized by ACN and analyzed HPLC using the same program described above.
In Vitro Assay of LovD Variants towards LovF (Xie, 2009)
50 µM LovF was incubated with 1 µM LovD variants, 2 mM MJA, 2 mM malonyl-CoA, 2 mM S-(5’-adenosyl)-L-methionine chloride (SAM), 2 mM NADPH in 100 mM PBS, pH 7.4. At 1 and 2 hours time points, an aliquot of the reaction mixture was removed, quenched with 1% TFA and extracted with EA containing 1% acetic acid. The organic phase was separated, dried, resolubilized by ACN and analyzed HPLC using the same program described above.
Comparing Expression Levels of Soluble LovD
Each expression plasmid encoding LovD mutant was transformed into E. coli BL21(DE3). The transformant was cultured in 50 mL LB medium containing 35 mg/L Kan at 37°C to optical density (OD600) value of 0.4~0.6. Protein expression was induced with 0.1 mM IPTG and the subsequent expression was performed at 25°C for 16 h. Cells were collected by centrifugation (2,000g, 4°C, 15 min), resuspended in 7 mL Buffer A (50 mM Tris–HCl, pH 8.0, 2 mM DTT, 2 mM EDTA), and lysed by sonication. Cell debris and insoluble proteins were removed by centrifugation (20,000g, 4°C, 1 h). To the cleared cell lysate, excess amount (0.5 mL) of Ni-NTA resin (Qiagen, Valencia, CA) was added to each sample. The mutants were then purified using a step gradient of Buffer A with increasing concentration of imidazole (10, 20, and 250 mM). LovD variants were eluted with 5 mL Buffer A containing 250 mM imidazole. No LovD protein was found in other fractions. The protein concentrations were qualitatively assessed by SDS–PAGE and quantitatively determined by the Bradford protein assay using bovine serum albumin (BSA) as the standard.
Tm Measurement by Circular Dichroism
Samples were prepared by adding 50 µg of proteins to 250µl 10 mM Tris-HCl buffer (pH 7.0). The sample was placed in a quartz cuvette with a 1 cm path length and heated in a Peltier-controlled cell at a rate of 1°C per min. Ellipticity was monitored at 222 nm in a Jasco spectropolarimeter (Jasco Inc., Easton, MD). The midpoint of the denaturation curve was determined with Microcal Origin 5.0 software (OriginLab Corporation, Northampton, MA).
Supplementary Material
Supplemental Data include six figures and Supplemental Experimental Procedures.
ACKNOWLEDGEMENTS
This work was supported by American Heart Association (0535069N) to YT and the National Institutes of Health (1R21HL091197) to YT and TOY. The authors thank Dr. Duilio Cascio and the staff of the Advance Photon Source beamline 24-ID-C for assistance with data collection.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
The coordinates and structure factors of G0, SeMet G0, G5, G5-MJA, G5’-MJA, G5’-LVA and G5’-SVA have been deposited into the Protein Data Bank under code 3HL9, 3HLB, 3HLC, 3HLD, 3HLE, 3HLF and 3HLG, respectively.
REFERENCES
- Arnold FH. Combinatorial and computational challenges for biocatalyst design. Nature. 2001;409:253–257. doi: 10.1038/35051731. [DOI] [PubMed] [Google Scholar]
- Arnold FH, Volkov AA. Directed evolution of biocatalysts. Curr. Opin. Chem. Biol. 1999;3:54–59. doi: 10.1016/s1367-5931(99)80010-6. [DOI] [PubMed] [Google Scholar]
- Berg VA, Hans M, Steekstra H. Method for the production of simvastatin. 2009 WO 2007147801 (A1) [Google Scholar]
- Fromant M, Blanquet S, Plateau P. Direct random mutagenesis of gene-sized DNA fragments using polymerase chain-reaction. Anal. Biochem. 1995;224:347–353. doi: 10.1006/abio.1995.1050. [DOI] [PubMed] [Google Scholar]
- Heikinheimo P, Goldman A, Jeffries C, Ollis DL. Of barn owls and bankers: a lush variety of alpha/beta hydrolases. Structure. 1999;7:141–146. doi: 10.1016/s0969-2126(99)80079-3. [DOI] [PubMed] [Google Scholar]
- Hoffman WF, Alberts AW, Anderson PS, Chen JS, Smith RL, Willard AK. 3-Hydroxy-3-methylglutaryl-coenzyme A reductase inhibitors .4. side-chain ester derivatives of mevinolin. J. Med. Chem. 1986;29:849–852. doi: 10.1021/jm00155a040. [DOI] [PubMed] [Google Scholar]
- Hsu CC, Hong ZY, Wada M, Franke D, Wong CH. Directed evolution of D-sialic acid aldolase to L-3-deoxy-manno-2-octulosonic acid (L-KDO) aldolase. P. Natl. Acad. Sci. USA. 2005;102:9122–9126. doi: 10.1073/pnas.0504033102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Istvan ES, Deisenhofer J. Structural mechanism for statin inhibition of HMG-CoA reductase. Science. 2001;292:1160–1164. doi: 10.1126/science.1059344. [DOI] [PubMed] [Google Scholar]
- Kennedy J, Auclair K, Kendrew SG, Park C, Vederas JC, Hutchinson CR. Modulation of polyketide synthase activity by accessory proteins during lovastatin biosynthesis. Science. 1999;284:1368–1372. doi: 10.1126/science.284.5418.1368. [DOI] [PubMed] [Google Scholar]
- Kumar MS, Kumar PM, Sarnaik HM, Sadhukhan AK. A rapid technique for screening of lovastatin-producing strains of Aspergillus terreus by agar plug and Neurospora crassa bioassay. J. Microbiol. Methods. 2000;40:99–104. doi: 10.1016/s0167-7012(99)00135-9. [DOI] [PubMed] [Google Scholar]
- Loncaric C, Merriweather E, Walker KD. Profiling a taxol pathway 10 beta-acetyltransferase: Assessment of the specificity and the production of baccatin III by in vivo acetylation in E. coli. Chem. Biol. 2006;13:309–317. doi: 10.1016/j.chembiol.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Morgan B, Burk M, Levin M, Zhu Z, Chaplin J, Kustedjo K, Huang Z, Greenberg W. Methods for making simvastatin and intermediates. 2006 WO 2005040107 (A2) [Google Scholar]
- Nardini M, Dijkstra BW. Alpha/beta hydrolase fold enzymes: the family keeps growing. Curr. Opin. Struct. Biol. 1999;9:732–737. doi: 10.1016/s0959-440x(99)00037-8. [DOI] [PubMed] [Google Scholar]
- Neumann CS, Fujimori DG, Walsh CT. Halogenation strategies in natural product biosynthesis. Chem. Biol. 2008;15:99–109. doi: 10.1016/j.chembiol.2008.01.006. [DOI] [PubMed] [Google Scholar]
- Oefner C, Darcy A, Daly JJ, Gubernator K, Charnas RL, Heinze I, Hubschwerlen C, Winkler FK. Refined crystal-structure of beta-lactamase from citrobacter-freundii Indicates a mechanism for beta-lactam hydrolysis. Nature. 1990;343:284–288. doi: 10.1038/343284a0. [DOI] [PubMed] [Google Scholar]
- Oue S, Okamoto A, Yano T, Kagamiyama H. Redesigning the substrate specificity of an enzyme by cumulative effects of the mutations of non-active site residues. J. Biol. Chem. 1999;274:2344–2349. doi: 10.1074/jbc.274.4.2344. [DOI] [PubMed] [Google Scholar]
- Petersen EI, Valinger G, Solkner B, Stubenrauch G, Schwab H. A novel esterase from Burkholderia gladioli which shows high deacetylation activity on cephalosporins is related to beta-lactamases and DD-peptidases. J. Biotechnol. 2001;89:11–25. doi: 10.1016/s0168-1656(01)00284-x. [DOI] [PubMed] [Google Scholar]
- Rix U, Fischer C, Remsing LL, Rohr J. Modification of post-PKS tailoring steps through combinatorial biosynthesis. Nat. Prod. Rep. 2002;19:542–580. doi: 10.1039/b103920m. [DOI] [PubMed] [Google Scholar]
- Schweiker KL, Zarrine-Afsar A, Davidson AR, Makhatadze GI. Computational design of the Fyn SH3 domain with increased stability through optimization of surface charge charge interactions. Protein Sci. 2007;16:2694–2702. doi: 10.1110/ps.073091607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner UG, Petersen EI, Schwab H, Kratky C. EstB from Burkholderia gladioli: a novel esterase with a beta-lactamase fold reveals steric factors to discriminate between esterolytic and beta-lactam cleaving activity. Protein Sci. 2002;11:467–478. doi: 10.1110/ps.33002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Meehan MJ, Xu W, Dorrecstein PC, Tang Y. Acyltransferase Mediated Polyketide Release from a Fungal Megasynthase. J. Am. Chem. Soc. 2009;131:8388–8389. doi: 10.1021/ja903203g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Pashkov I, Gao X, Guerrero JL, Yeates TO, Tang Y. Rational improvement of simvastatin synthase solubility in Escherichia coli leads to higher whole-cell biocatalytic activity. Biotechnol. Bioeng. 2009;102:20–28. doi: 10.1002/bit.22028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Tang Y. Efficient synthesis of simvastatin by use of whole-cell biocatalysis. Appl. Environ. Microbiol. 2007;73:2054–2060. doi: 10.1128/AEM.02820-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Watanabe K, Wojcicki WA, Wang CC, Tang Y. Biosynthesis of lovastatin analogs with a broadly specific acyltransferase. Chem. Biol. 2006;13:1161–1169. doi: 10.1016/j.chembiol.2006.09.008. [DOI] [PubMed] [Google Scholar]
- Xie X, Wong WW, Tang Y. Improving simvastatin bioconversion in Escherichia coli by deletion of bioH. Metab. Eng. 2007;9:379–386. doi: 10.1016/j.ymben.2007.05.006. [DOI] [PubMed] [Google Scholar]
- Zhang CS, Griffith BR, Fu Q, Albermann C, Fu X, Lee IK, Li LJ, Thorson JS. Exploiting the reversibility of natural product glycosyltransferase-catalyzed reactions. Science. 2006;313:1291–1294. doi: 10.1126/science.1130028. [DOI] [PubMed] [Google Scholar]
- Zhao HM, Arnold FH. Directed evolution converts subtilisin E into a functional equivalent of thermitase. Protein Eng. 1999;12:47–53. doi: 10.1093/protein/12.1.47. [DOI] [PubMed] [Google Scholar]
- Zhou H, Xie X, Tang Y. Engineering natural products using combinatorial biosynthesis and biocatalysis. Curr. Opin. Biotech. 2008;19:590–596. doi: 10.1016/j.copbio.2008.10.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data include six figures and Supplemental Experimental Procedures.